Analytic Correlation Filtration: A New Tool to Reduce Analytical Complexity of Metabolomic Datasets
Abstract
1. Introduction
2. Materials and Methods
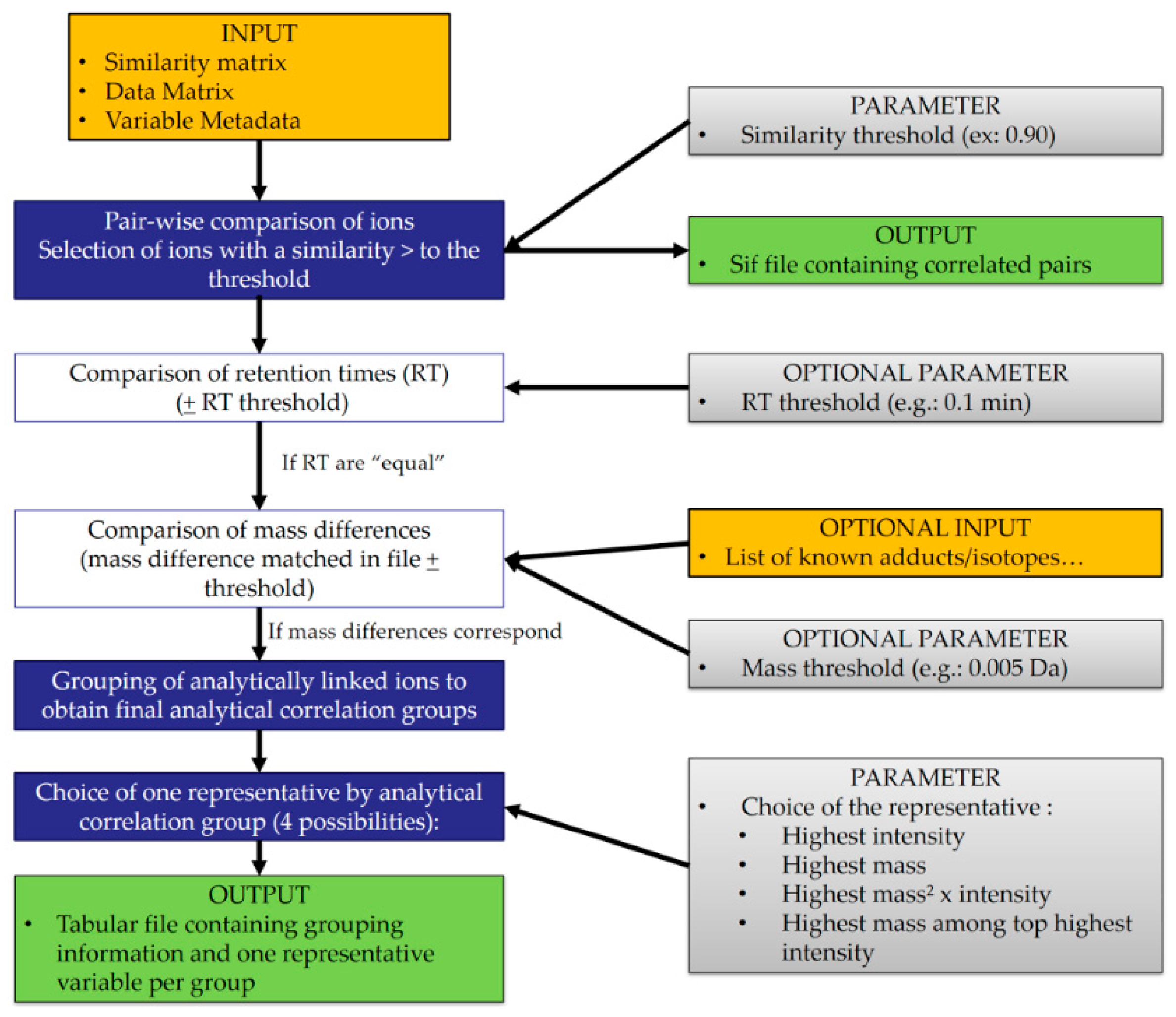
2.1. Algorithm Description
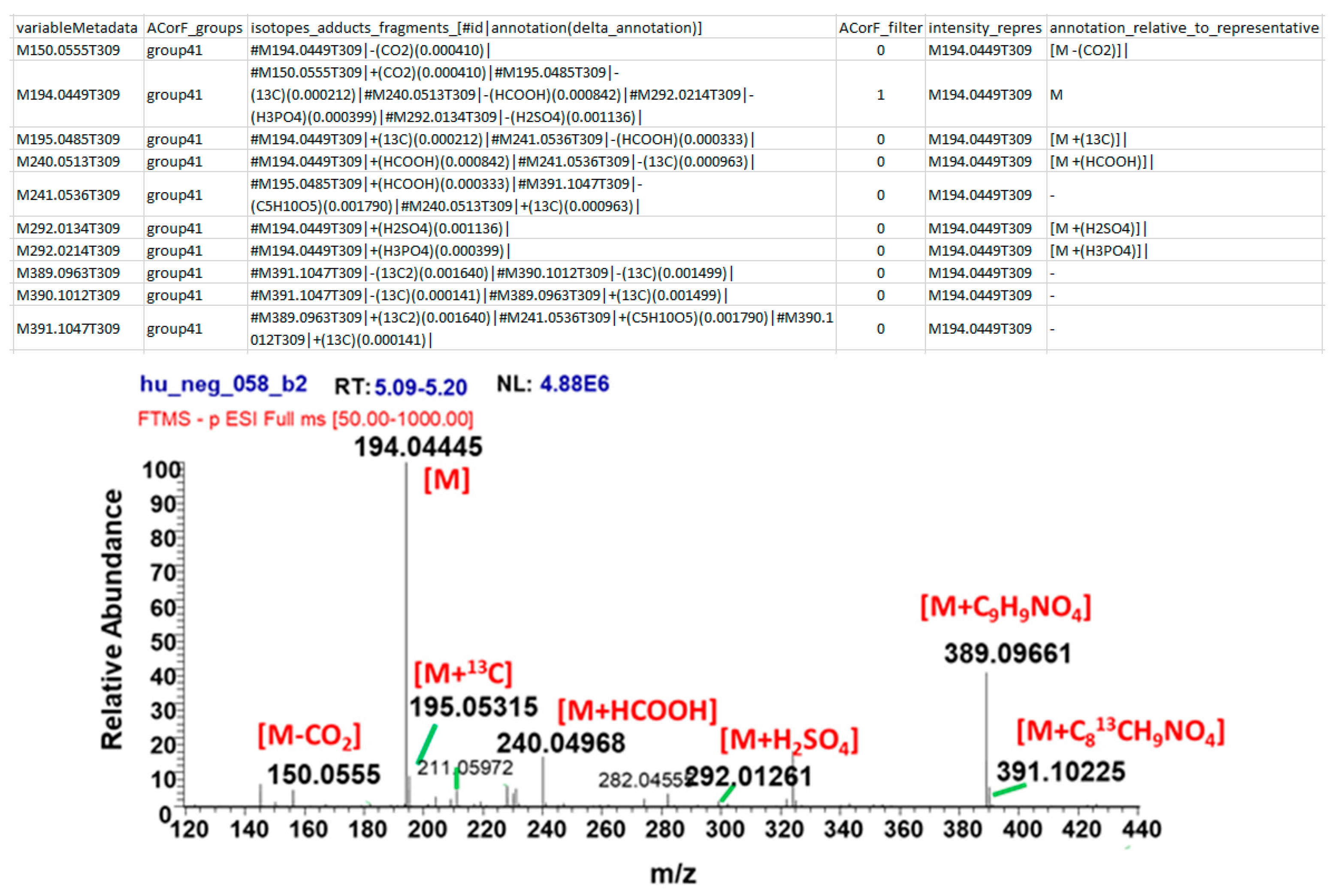
2.1.1. Input Files
2.1.2. Processing
- (1)
- Retaining the ion with the highest intensity
- (2)
- Retaining the ion with the highest mass
- (3)
- Retaining the ion with the highest ‘mass² × average intensity’
- (4)
- Retaining the highest mass among the top highest average intensities of the group. For this last option, the user determines the number of ions considered in the top list (top 5, top 3, top 10, etc.).
2.1.3. Output Files
2.2. Examples of Use
3. Results and Discussion
3.1. Functionalities
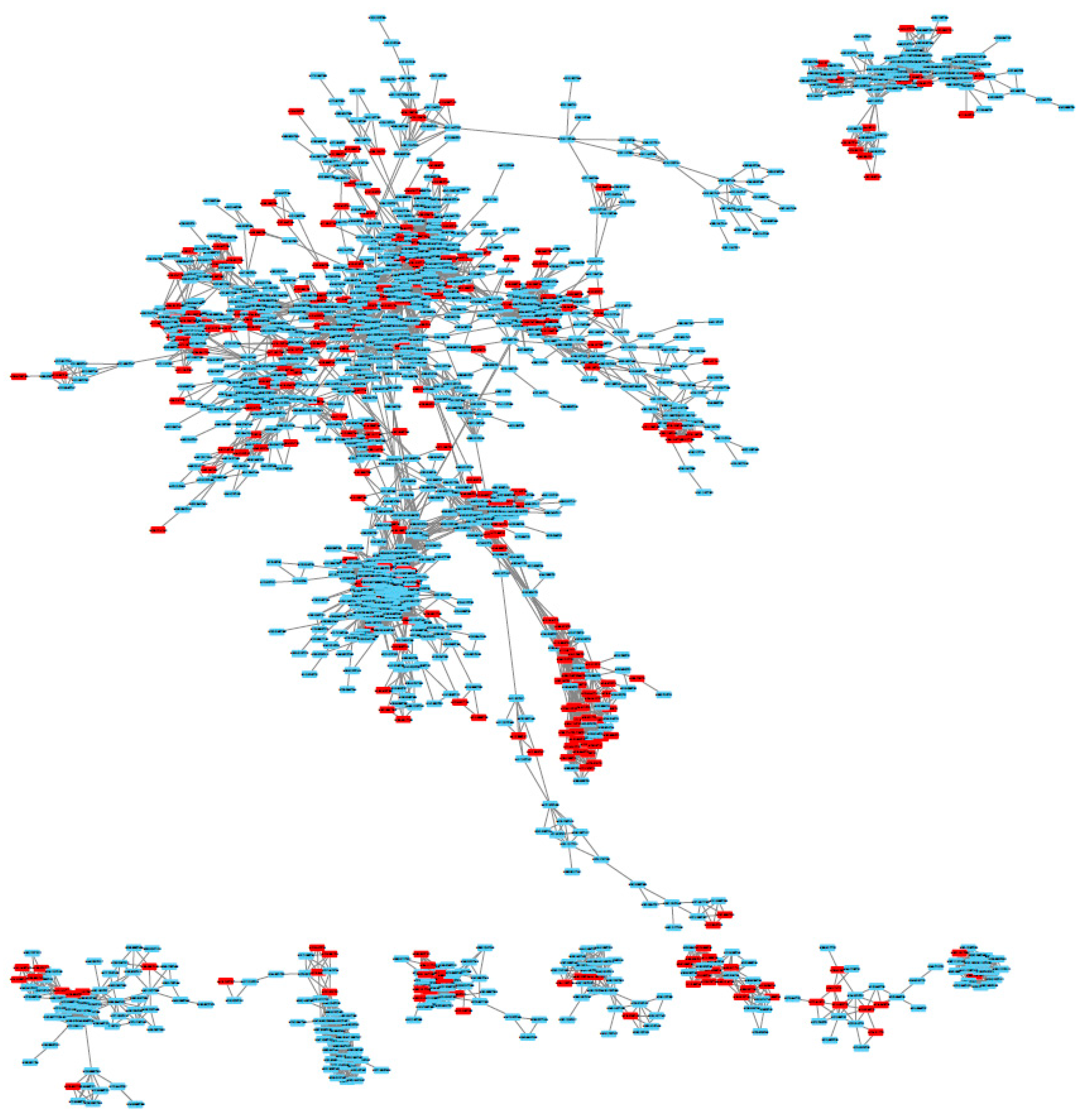
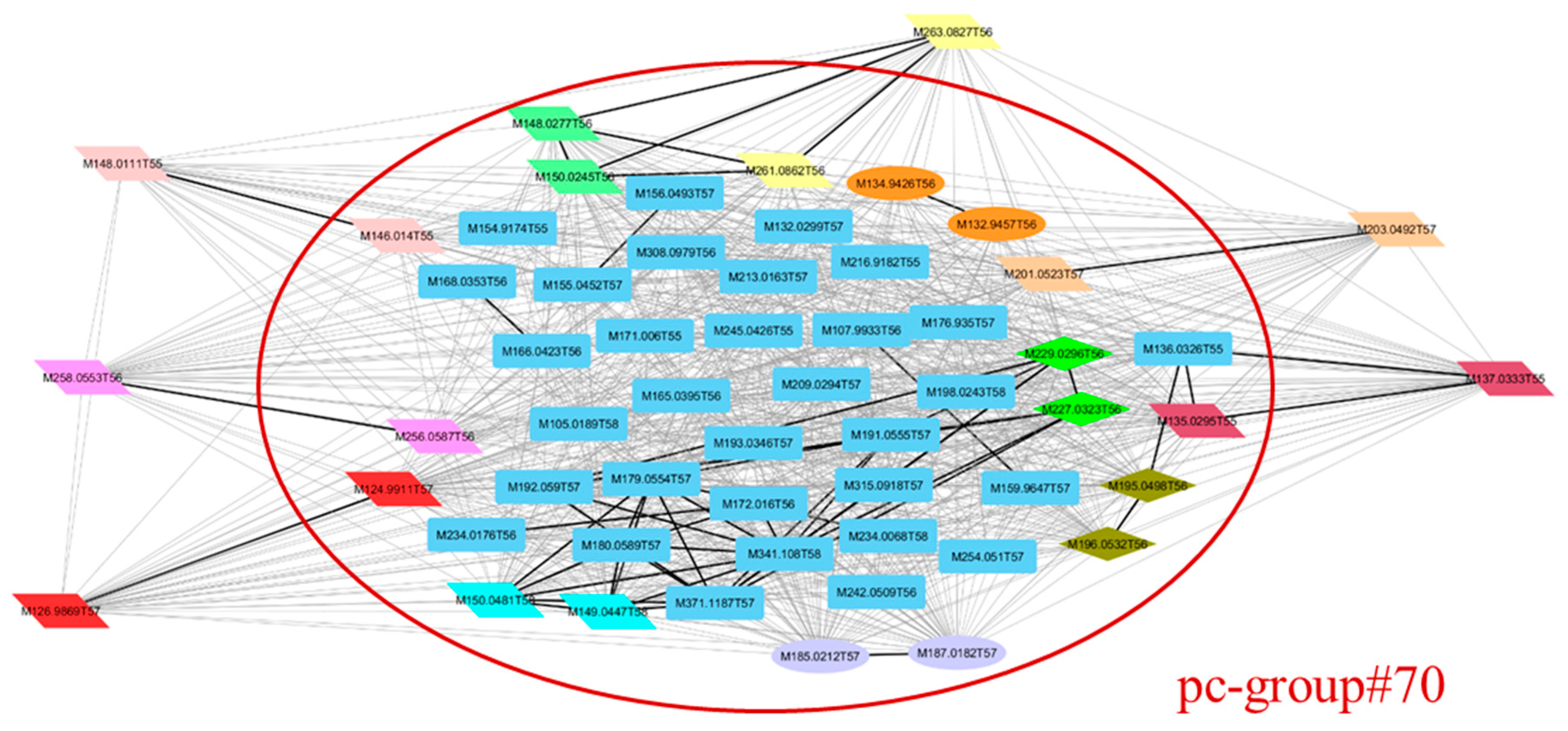
3.2. Example of Use: The Sacurine Dataset
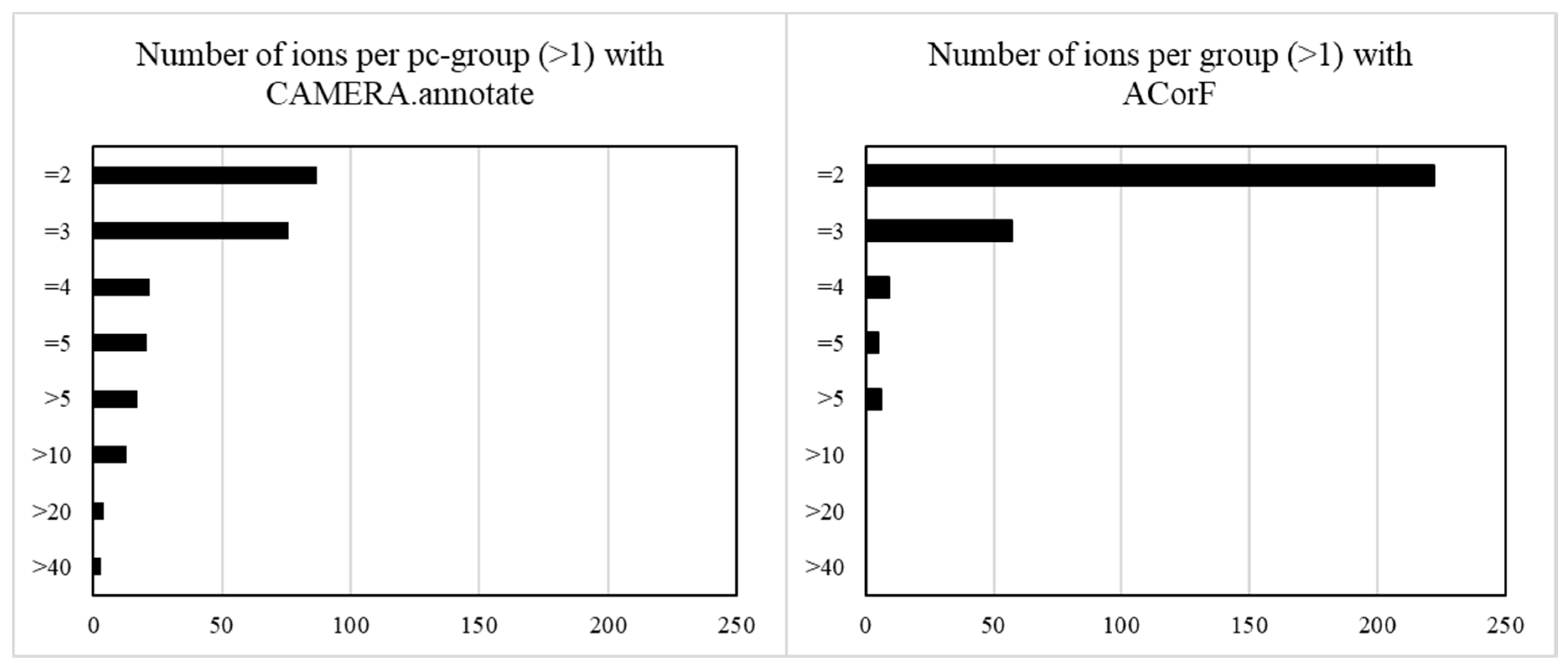
3.3. Result Comparisons
3.4. Use and Configuration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; Berger, R.; van der Greef, J.; Hankemeier, T. Human metabolomics: Strategies to understand biology. Curr. Opin. Chem. Biol. 2013, 17, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Forcisi, S.; Moritz, F.; Kanawati, B.; Tziotis, D.; Lehmann, R.; Schmitt-Kopplin, P. Liquid chromatography-mass spectrometry in metabolomics research: Mass analyzers in ultra-high pressure liquid chromatography coupling. J. Chromatogr. A 2013, 1292, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Kuehnbaum, N.L.; Britz-McKibbin, P. New advances in separation science for metabolomics: Resolving chemical diversity in a post-genomic era. Chem. Rev. 2013, 113, 2437–2468. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Marsal, S.; Julia, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Bottcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Julia, A.; Beltran, A.; Vinaixa, M.; Diaz, M.; Ibanez, L.; Correig, X.; Marsal, S. AStream: An R package for annotating LC/MS metabolomic data. Bioinformatics 2011, 27, 1339–1340. [Google Scholar] [CrossRef] [PubMed]
- Senan, O.; Aguilar-Mogas, A.; Navarro, M.; Capellades, J.; Noon, L.; Burks, D.; Yanes, O.; Guimerà, R.; Sales-Pardo, M. CliqueMS: A computational tool for annotating in-source metabolite ions from LC-MS untargeted metabolomics data based on a coelution similarity network. Bioinformatics 2019, 35, 4089–4097. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: An R package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; Bourne, P.E.; Bouwman, J.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Guitton, Y.; Tremblay-Franco, M.; le Corguillé, G.; Martin, J.F.; Pétéra, M.; Roger-Mele, P.; Delabrière, A.; Goulitquer, S.; Monsoor, M.; Canlet, C.; et al. Create, run, share, publish, and reference your LC-MS, FIA-MS, GC-MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics. Int. J. Biochem. Cell Biol. 2017, 93, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Giacomoni, F.; le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-E.; Goulitquer, S.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed]
- Carusi, A.; Reimer, T. Virtual Research Environment Collaborative Landscape Study; UK’s Joint Information Systems Committee (JISC): Bristol, UK, 2010. [Google Scholar]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; Galaxy, T. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pujos-Guillot, E.; Brandolini, M.; Pétéra, M.; Grissa, D.; Joly, C.; Lyan, B.; Herquelot, É.; Czernichow, S.; Zins, M.; Comte, B.; et al. Systems metabolomics for prediction of metabolic syndrome. J. Proteome Res. 2017, 16, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
| - | CAMERA.annotate (W4M version) | “Analytic Correlation Filtration” (ACorF) Tool |
|---|---|---|
| Interface | Galaxy (W4M) | Galaxy (W4M) |
| Language | R | Perl |
| Version | Galaxy version 2.1.3 | - |
| Input files | .Rdata output from XCMS Galaxy pre-processing | DataMatrix, variableMetadata and similarity matrix |
| Parameters | - | - |
| Mandatory | - | Correlation rate Representative selection method |
| Optional | Correlation rate RT window determination variables | Mass difference list Retention time tolerance delta |
| Correlation information | Calculation of correlation is included in the tool | A correlation table has to be obtained before using the tool |
| Correlation type | Pearson correlation | Any type of correlation are possible |
| Possibility to set a Correlation threshold | Yes—only for the second step of grouping | Yes |
| Retention time (RT) window | Calculated for each peak | Defined by a threshold |
| Parameter settings | Two different parameters ([sigma] and [perfwhm]) are available | The user can set the RT tolerance delta value |
| Comparison to a mass defect list | Conditioned by obtained group but not used for grouping | When used, directly impact the group determination |
| Isotope identification | Yes—performed in a previous step | Yes—if the isotope mass difference is included in the list |
| Existing default list | Yes | Yes |
| Possibility to upload a personal list | Yes | Yes |
| Possibility of setting a mass difference tolerance value | No | Yes |
| Possibility of selecting a representative ion for each group | No | Yes |
| Output files | variableMetadata with additional columns | variableMetadata with additional columns and a .sif file for network visualisation |
| Optional output files | EIC for main pc-group visualisation pdf file | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monnerie, S.; Petera, M.; Lyan, B.; Gaudreau, P.; Comte, B.; Pujos-Guillot, E. Analytic Correlation Filtration: A New Tool to Reduce Analytical Complexity of Metabolomic Datasets. Metabolites 2019, 9, 250. https://doi.org/10.3390/metabo9110250
Monnerie S, Petera M, Lyan B, Gaudreau P, Comte B, Pujos-Guillot E. Analytic Correlation Filtration: A New Tool to Reduce Analytical Complexity of Metabolomic Datasets. Metabolites. 2019; 9(11):250. https://doi.org/10.3390/metabo9110250
Chicago/Turabian StyleMonnerie, Stephanie, Melanie Petera, Bernard Lyan, Pierrette Gaudreau, Blandine Comte, and Estelle Pujos-Guillot. 2019. "Analytic Correlation Filtration: A New Tool to Reduce Analytical Complexity of Metabolomic Datasets" Metabolites 9, no. 11: 250. https://doi.org/10.3390/metabo9110250
APA StyleMonnerie, S., Petera, M., Lyan, B., Gaudreau, P., Comte, B., & Pujos-Guillot, E. (2019). Analytic Correlation Filtration: A New Tool to Reduce Analytical Complexity of Metabolomic Datasets. Metabolites, 9(11), 250. https://doi.org/10.3390/metabo9110250





