Untargeted Metabolomics of Extracts from Faecal Samples Demonstrates Distinct Differences between Paediatric Crohn’s Disease Patients and Healthy Controls but No Significant Changes Resulting from Exclusive Enteral Nutrition Treatment
Abstract
1. Introduction
2. Results
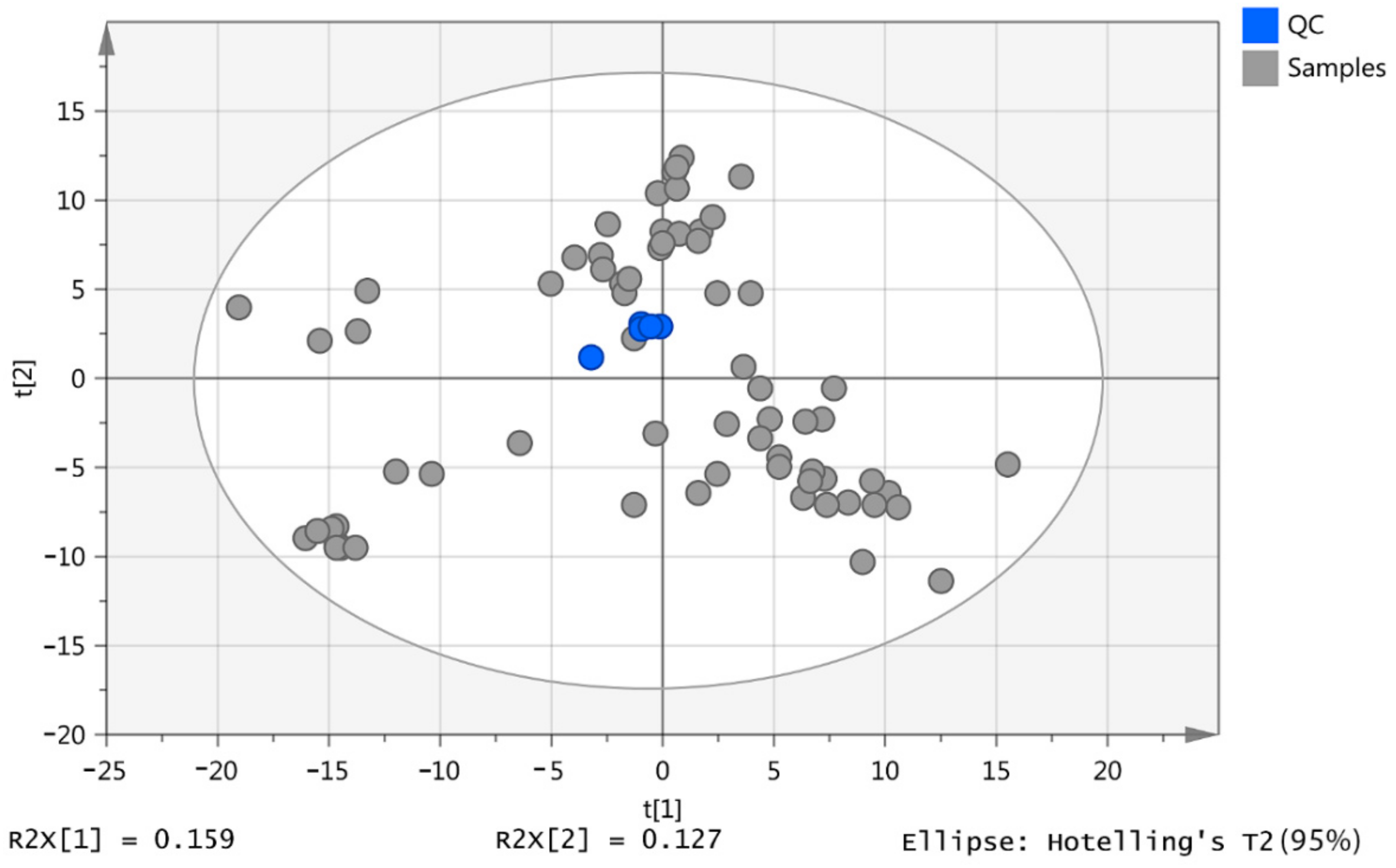
2.1. Pooled Samples
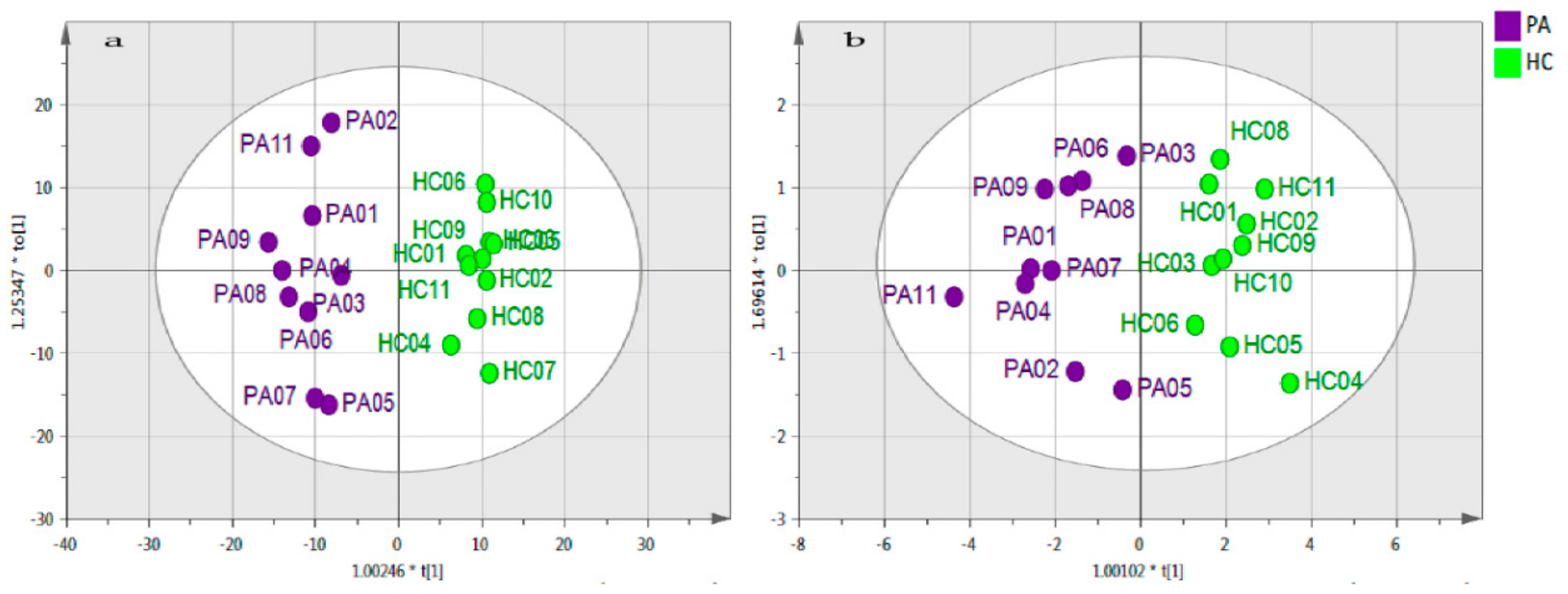
2.2. Data Visualisation
3. Discussion
4. Materials and Methods
4.1. Chemicals and Solvents
4.2. Samples and Sample Preparation
4.3. LC-MS Analysis
4.4. Data Pre-processing and Modelling
4.6. Model Validation
4.7. Data Filtration
4.8. Ranking, Grouping and Confirmation of Significant Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jansson, J.; Willing, B.; Lucio, M.; Fekete, A.; Dicksved, J.; Halfvarson, J.; Tysk, C.; Schmitt-Kopplin, P. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS ONE 2009, 4, e6386. [Google Scholar] [CrossRef] [PubMed]
- Day, A.S.; Lopez, R.N. Exclusive enteral nutrition in children with Crohn’s disease. World J. Gastroenterol. 2015, 21, 6809–6816. [Google Scholar] [CrossRef] [PubMed]
- Ruemmele, F.M.; Veres, G.; Kolho, K.L.; Griffiths, A.; Levine, A.; Escher, J.C.; Amil Dias, J.; Barabino, A.; Braegger, C.P.; Bronsky, J.; et al. Consensus guidelines of ecco/espghan on the medical management of pediatric crohn’s disease. J. Crohn Colitis 2014, 8, 1179–1207. [Google Scholar] [CrossRef] [PubMed]
- Cameron, F.L.; Gerasimidis, K.; Papangelou, A.; Missiou, D.; Garrick, V.; Cardigan, T.; Buchanan, E.; Barclay, A.R.; McGrogan, P.; Russell, R.K. Clinical progress in the two years following a course of exclusive enteral nutrition in 109 paediatric patients with crohn’s disease. Aliment. Pharmacol. Ther. 2013, 37, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, O.; Cordischi, L.; Cirulli, M.; Paganelli, M.; Labalestra, V.; Uccini, S.; Russo, P.M.; Cucchiara, S. Polymeric diet alone versus corticosteroids in the treatment of active pediatric crohn’s disease: A randomized controlled open-label trial. Clin. Gastroenterol 2006, 4, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Ijaz, U.Z.; Loman, N.; Eren, A.M.; Saulnier, D.; Russell, J.; Haig, S.J.; Calus, S.T.; Quick, J.; Barclay, A.; Bertz, M.; Blaut, M.; Hansen, R.; McGrogan, P.; Russell, R.K.; Edwards, C.A.; Gerasimidis, K. Extensive Modulation of the Fecal Metagenome in Children With Crohn’s Disease During Exclusive Enteral Nutrition. Am. J. Gastroenterol. 2015, 110, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Gerasimidis, K.; Bertz, M.; Hanske, L.; Junick, J.; Biskou, O.; Aguilera, M.; Garrick, V.; Russell, R.K.; Blaut, M.; McGrogan, P.; Edwards, C.A. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm. Bowel Dis. 2014, 20, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Wine, E. Effects of enteral nutrition on crohn’s disease: Clues to the impact of diet on disease pathogenesis. Inflamm. Bowel Dis. 2013, 19, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Leiss, K.A.; Choi, Y.H.; Verpoorte, R.; Klinkhamer, P.G. An overview of NMR-based metabolomics to identify secondary plant compounds involved in host plant resistance. Phytochem. Rev. 2011, 10, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. BioSyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys. Biol. 2008, 5, 011001. [Google Scholar] [CrossRef] [PubMed]
- Boccard, J.; Veuthey, J.L.; Rudaz, S. Knowledge discovery in metabolomics: An overview of MS data handling. J. Sep. Sci. 2010, 33, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Kolho, K.L.; Pessia, A.; Jaakkola, T.; de Vos, W.M.; Velagapudi, V. Faecal and serum metabolomics in paediatric inflammatory bowel disease. J. Crohns. Colitis. 2017, 11, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, J.T.; Wang, Y.; Hao, F.; Coskun, M.; Ludwig, C.; Gunther, U.; Nielsen, O.H. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics 2015, 11, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Schicho, R.; Shaykhutdinov, R.; Ngo, J.; Nazyrova, A.; Schneider, C.; Panaccione, R.; Kaplan, G.G.; Vogel, H.J.; Storr, M. Quantitative metabolomic profiling of serum, plasma, and urine by 1H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 2012, 11, 3344–3357. [Google Scholar] [CrossRef] [PubMed]
- De Preter, V. Metabolomics in the clinical diagnosis of inflammatory bowel disease. Digestive Dis. 2015, 33, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Karu, N.; Deng, L.; Slae, M.; Guo, A.C.; Sajed, T.; Huynh, H.; Wine, E.; Wishart, D.S. A Review on Human Fecal Metabolomics: Methods, Applications and the Human Fecal Metabolome Database. Analytica Chimica Acta 2018. [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Howe, C.C.; Alshehri, A.; Muggeridge, D.; Mullen, A.B.; Boyd, M.; Spendiff, O.; Moir, H.J.; Watson, D.G. Untargeted metabolomics profiling of an 80.5 km simulated treadmill ultramarathon. Metabolites 2018, 8, 14. [Google Scholar]
- Schuller-Levis, G.B.; Park, E. Taurine: New implications for an old amino acid. FEMS Microbiol. Lett. 2003, 226, 195–202. [Google Scholar] [CrossRef]
- Efron, B.; Gong, G. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am. Stat. 1983, 37, 36–48. [Google Scholar] [CrossRef]
- O’Brian, M.R.; Fabiano, E. Mechanisms and regulation of iron homeostasis in the Rhizobia. Iron Uptake Homeost. Microorg. 2010, 37–63. [Google Scholar]
- Musso, G.; Gambino, R.; Cassader, M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annual. Rev. Med. 2011, 62, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Odahara, S.; Nakamura, M.; Koido, S.; Katahira, K.; Shiraishi, H.; Ohkusa, T.; Fujise, K.; Tajiri, H. The fatty acid profile of the erythrocyte membrane in initial-onset inflammatory bowel disease patients. Dig. Dis. Sci. 2013, 58, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- Kaliannan, K.; Wang, B.; Li, X.Y.; Kim, K.J.; Kang, J.X. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Rep. 2015, 5, 11276. [Google Scholar] [CrossRef] [PubMed]
- Angulo, S.; Morales, A.; Danese, S.; Llacuna, L.; Masamunt, M.C.; Pultz, N.; Cifone, M.G.; de Simone, C.; Delgado, S.; Vila, J.; Panés, J.; Donskey, C.; Fernández-Checa, J.C.; Fiocchi, C.; Sans, M. Probiotic sonicates selectively induce mucosal immune cells apoptosis through ceramide generation via neutral sphingomyelinase. PLoS ONE 2011, 6, e16953. [Google Scholar] [CrossRef] [PubMed]
- Baur, P.; Martin, F.O.-P.; Gruber, L.; Bosco, N.; Brahmbhatt, V.; Collino, S.; Guy, P.; Montoliu, I.; Rozman, J.; Klingenspor, M. Metabolic phenotyping of the Crohn’s disease-like IBD etiopathology in the TNFΔARE/WT mouse model. J. Proteome Res. 2011, 10, 5523–5535. [Google Scholar] [CrossRef] [PubMed]
- Sewell, G.W.; Hannun, Y.A.; Han, X.; Koster, G.; Bielawski, J.; Goss, V.; Smith, P.J.; Rahman, F.Z.; Vega, R.; Bloom, S.L.; Walker, A.P.; Postle, A.D.; Segal, A.W. Lipidomic profiling in Crohn’s disease: Abnormalities in phosphatidylinositols, with preservation of ceramide, phosphatidylcholine and phosphatidylserine composition. Int. J. Biochem. Cell Biol. 2012, 44, 1839–1846. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Watson, D.G.; Wang, L.; Westrop, G.D.; Coombs, G.H.; Zhang, T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A 1362, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Gerasimidis, K.; Nikolaou, C.K.; Edwards, C.A.; McGrogan, P. Serial fecal calprotectin changes in children with Crohn’s disease on treatment with exclusive enteral nutrition: Associations with disease activity, treatment response, and prediction of a clinical relapse. J. Clin. Gastroenterol. 2011, 45, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Scheltema, R.A.; Jankevics, A.; Jansen, R.C.; Swertz, M.A.; Breitling, R. PeakML/mzMatch: A file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 2011, 83, 2786–2793. [Google Scholar] [CrossRef] [PubMed]
- Creek, D.J.; Jankevics, A.; Burgess, K.E.; Breitling, R.; Barrett, M.P. IDEOM: An Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 2012, 28, 1048–1049. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Blasco, H.; Błaszczyński, J.; Billaut, J.C.; Nadal-Desbarats, L.; Pradat, P.F.; Devos, D.; Moreau, C.; Andres, C.R.; Emond, P.; Corcia, P.; Słowiński, R. Comparative analysis. J. Biomed. Inform. 2015, 53, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi- and Megavariate Data Analysis Basic Principles and Applications, 3rd ed.; MKS Umetrics AB: Malmo, Sweden, 2013; ISBN 978-9197373050. [Google Scholar]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metabolomics. 2013, 1, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Wheelock, Å.M.; Wheelock, C.E. Trials and tribulations of ’omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chen, J.Q.; Huang, W.Q.; Li, W.; Huang, Y.; Zhang, Z.J.; Xu, F.G. Renal medulla is more sensitive to cisplatin than cortex revealed by untargeted mass spectrometry-based metabolomics in rats. Sci. Rep. 2017, 7, 44804. [Google Scholar] [CrossRef] [PubMed]
| Model | R2X (Cum) | R2 | Q2 | Permutation (999 times) | R2 − Q2 | Valid | p CV-ANOVA | Significance |
|---|---|---|---|---|---|---|---|---|
| PA vs. HC | 0.63 | 0.95 | 0.71 | yes | 0.24 | yes | 1.83 × 10−3 | yes |
| PA vs. PB | 0.60 | 0.88 | 0.51 | yes | 0.37 | no | 1.37 × 10−1 | no |
| PA vs. PC | 0.65 | 0.88 | 0.66 | yes | 0.22 | yes | 1.00 × 10−2 | yes |
| PA vs. PD | 0.67 | 0.89 | 0.43 | yes | 0.46 | no | 2.42 × 10−1 | no |
| PA vs. PE | 0.47 | 0.67 | 0.33 | yes | 0.34 | no | 1.56 × 10−1 | no |
| HC vs. PB | 0.68 | 0.99 | 0.91 | yes | 0.08 | yes | 2.03 × 10−6 | yes |
| HC vs. PC | 0.67 | 0.99 | 0.91 | yes | 0.08 | yes | 4.81 × 10−7 | yes |
| HC vs. PD | 0.72 | 0.99 | 0.86 | yes | 0.13 | yes | 6.69 × 10−4 | yes |
| HC vs. PE | 0.54 | 0.99 | 0.72 | yes | 0.27 | yes | 1.19 × 10−2 | yes |
| PB vs. PC | 0.68 | 0.97 | 0.08 | yes | 0.89 | no | 9.97 × 10−1 | no |
| PB vs. PD | 0.61 | 0.76 | 0.12 | yes | 0.64 | no | 7.16 × 10−1 | no |
| PB vs. PE | 0.63 | 0.99 | 0.93 | yes | 0.06 | yes | 3.31 × 10−7 | yes |
| PC vs. PD | 0.58 | 0.68 | 0.24 | yes | 0.44 | no | 2.98 × 10-1 | no |
| PC vs. PE | 0.60 | 0.98 | 0.89 | yes | 0.09 | yes | 1.90 × 10−7 | yes |
| PD vs. PE | 0.57 | 0.84 | 0.69 | yes | 0.15 | yes | 3.43 × 10−4 | yes |
| Mass | RT | Putative Metabolite | p-Value HCPA | PA/HC | p-Value HCPB | PB/HC | p-Value HCPC | PC/HC | p-Value HCPD | D/HC | p-Value HCPE | E/HC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 254.2246 | 3.6 | Hexadecenoic acid | 0.005 | 2.334 | 0.004 | 3.028 | 0.001 | 3.219 | 0.027 | 2.287 | 0.035 | 1.623 |
| 256.2401 | 3.6 | Hexadecanoic acid | 0.839 | 1.038 | 0.860 | 1.038 | 0.957 | 1.011 | 0.522 | 0.866 | 0.249 | 1.212 |
| 258.1829 | 3.6 | Tetradecanedioic acid | 0.016 | 0.362 | 0.010 | 0.332 | 0.007 | 0.300 | 0.068 | 0.507 | 0.637 | 1.200 |
| 258.2198 | 3.5 | Hydroxypentadecanoic acid | 0.984 | 1.005 | 0.713 | 1.143 | 0.401 | 1.379 | 0.266 | 1.448 | 0.716 | 1.095 |
| 260.1988 | 3.3 | Dihydroxytetradecanoic acid | 0.891 | 0.935 | 0.051 | 0.240 | 0.037 | 0.180 | 0.034 | 0.161 | 0.241 | 1.512 |
| 266.1882 | 3.4 | Hydroxyhexadecatrienoic acid | 0.537 | 0.839 | 0.007 | 0.391 | 0.015 | 0.459 | 0.029 | 0.506 | 0.192 | 1.387 |
| 268.2036 | 3.1 | Hydroxyhexadecadienoic acid | 0.016 | 0.376 | 0.001 | 0.118 | 0.001 | 0.113 | 0.002 | 0.176 | 0.552 | 0.852 |
| 270.2195 | 3.5 | Hydroxyhexadecenoic acid | 0.940 | 1.024 | 0.174 | 0.702 | 0.846 | 1.081 | 0.902 | 0.963 | 0.095 | 1.472 |
| 272.2351 | 3.5 | Hydroxyhexadecanoic acid | 0.905 | 0.947 | 0.228 | 0.578 | 0.389 | 0.690 | 0.538 | 0.785 | 0.449 | 1.287 |
| 278.2245 | 3.5 | Octadecatrienoic acid | 0.004 | 0.115 | 0.005 | 0.143 | 0.004 | 0.135 | 0.004 | 0.128 | 0.014 | 0.293 |
| 280.2401 | 3.6 | Octadecadienoic acid | 0.212 | 0.513 | 0.220 | 0.517 | 0.211 | 0.508 | 0.098 | 0.342 | 0.195 | 0.500 |
| 282.2559 | 3.6 | Octadecenoic acid | 0.826 | 1.093 | 0.374 | 0.666 | 0.839 | 1.102 | 0.407 | 0.688 | 0.065 | 1.871 |
| 284.2713 | 3.5 | Octadecanoic acid | 0.399 | 0.756 | 0.022 | 0.400 | 0.020 | 0.394 | 0.018 | 0.374 | 0.928 | 0.976 |
| 288.23 | 3.2 | Dihydroxyhexadecanoic acid | 0.810 | 0.893 | 0.047 | 0.252 | 0.048 | 0.256 | 0.052 | 0.269 | 0.296 | 1.436 |
| 296.2349 | 3.5 | Hydroxyoctadecadienenoic acid | 0.004 | 0.285 | 0.004 | 0.271 | 0.004 | 0.269 | 0.004 | 0.279 | 0.235 | 0.700 |
| 298.2506 | 3.6 | Hydroxyoctadecenoic acid | 0.797 | 1.105 | 0.509 | 0.784 | 0.921 | 1.038 | 0.950 | 1.023 | 0.019 | 2.064 |
| 304.2401 | 3.5 | Eicosatetraenoic acid | 0.088 | 18.052 | 0.100 | 4.904 | 0.049 | 2.915 | 0.228 | 1.968 | 0.008 | 3.448 |
| 306.2558 | 3.5 | Eicosatrienoic acid | 0.002 | 16.182 | 0.008 | 13.854 | 0.014 | 9.671 | 0.049 | 8.821 | 0.003 | 8.751 |
| 308.2715 | 3.5 | Eicosadienoic acid | 0.028 | 8.716 | 0.041 | 6.338 | 0.017 | 6.119 | 0.008 | 5.658 | 0.000 | 3.709 |
| 310.2145 | 3.5 | Dihydroxyoctadecatrienoic acid | 0.042 | 0.593 | 0.691 | 0.890 | 0.368 | 0.783 | 0.017 | 0.496 | 0.676 | 1.147 |
| 310.2871 | 3.5 | Eicosenoic acid | 0.045 | 1.793 | 0.308 | 1.315 | 0.422 | 1.181 | 0.772 | 1.076 | 0.020 | 1.588 |
| 312.2301 | 3.6 | Dihydroxyoctadecadienoic acid | 0.871 | 1.053 | 0.868 | 1.053 | 0.860 | 0.947 | 0.191 | 0.647 | 0.350 | 1.244 |
| 312.2663 | 3.5 | Hydroxynonadecenoic acid | 0.232 | 1.695 | 0.013 | 2.360 | 0.018 | 2.560 | 0.045 | 2.148 | 0.733 | 1.149 |
| 312.3028 | 3.5 | Eicosanoic acid | 0.851 | 1.071 | 0.203 | 1.595 | 0.082 | 2.166 | 0.304 | 1.614 | 0.766 | 1.085 |
| 330.2405 | 3.7 | Trihydroxyoctadecenoic acid | 0.175 | 0.508 | 0.373 | 1.590 | 0.646 | 1.220 | 0.878 | 0.938 | 0.583 | 1.261 |
| 332.2716 | 3.5 | Docosatetraenoic acid | 0.006 | 20.326 | 0.004 | 9.694 | 0.003 | 6.946 | 0.006 | 6.532 | <0.001 | 8.116 |
| 334.2144 | 3.7 | Dihydroxyeicosapentaenoic acid | 0.964 | 1.024 | 0.938 | 1.043 | 0.661 | 0.775 | 0.605 | 0.743 | 0.233 | 0.414 |
| 334.2871 | 3.5 | Docosatrienoic acid | 0.127 | 1.701 | 0.491 | 1.457 | 0.856 | 1.098 | 0.405 | 1.491 | 0.201 | 1.664 |
| 336.3029 | 3.5 | Docosadienoic acid | 0.738 | 1.226 | 0.444 | 0.705 | 0.037 | 0.341 | 0.116 | 0.488 | 0.880 | 0.939 |
| 338.3186 | 3.5 | Docosenoic acid | 0.119 | 1.665 | 0.768 | 0.935 | 0.192 | 0.759 | 0.185 | 0.728 | 0.039 | 1.459 |
| 340.334 | 3.4 | Docosanoic acid | 0.610 | 1.264 | 0.017 | 0.320 | 0.019 | 0.342 | 0.026 | 0.356 | 0.262 | 1.570 |
| 342.2769 | 3.4 | Eicosanedioic acid | 0.021 | 0.311 | 0.061 | 0.454 | 0.069 | 0.468 | 0.088 | 0.504 | 0.331 | 0.718 |
| 346.2353 | 3.9 | Tetrahydroxyoctadecenoic acid | 0.046 | 0.447 | 0.301 | 0.691 | 0.168 | 0.618 | 0.244 | 0.670 | 0.978 | 0.992 |
| 352.3341 | 3.4 | Tricosenoic acid | 0.164 | 1.402 | 0.006 | 2.492 | 0.005 | 2.596 | 0.127 | 1.788 | 0.088 | 1.392 |
| 354.2408 | 3.7 | Trihydroxyeicosatetraenoic acid | 0.799 | 0.938 | 0.254 | 0.697 | 0.078 | 0.598 | 0.239 | 0.730 | 0.097 | 1.433 |
| 354.3134 | 3.4 | Hydroxydocosenoic acid | 0.209 | 0.563 | 0.022 | 0.361 | 0.036 | 0.419 | 0.088 | 0.529 | 0.602 | 1.160 |
| 354.3498 | 3.4 | Tricosanoic acid | 0.999 | 1.000 | 0.011 | 0.351 | 0.019 | 0.427 | 0.019 | 0.408 | 0.511 | 1.225 |
| 356.329 | 3.4 | Hydroxydocosanoic acid | 0.494 | 0.713 | 0.011 | 0.239 | 0.013 | 0.270 | 0.021 | 0.325 | 0.461 | 0.763 |
| 364.3342 | 3.4 | Tetracosadienoic acid | 0.037 | 4.794 | 0.214 | 2.076 | 0.651 | 1.220 | 0.312 | 2.154 | 0.048 | 3.661 |
| 370.2358 | 3.8 | Tetrahydroxyeicosatrienoic acid | 0.456 | 0.840 | 0.044 | 0.532 | 0.012 | 0.464 | 0.039 | 0.536 | 0.075 | 1.544 |
| 372.2509 | 3.8 | Tetrahydroxyeicosadienenoic acid | 0.086 | 0.574 | 0.030 | 0.537 | 0.004 | 0.402 | 0.016 | 0.497 | 0.080 | 1.459 |
| 382.2719 | 3.6 | Dihydroxydocosatrienoic acid | 0.039 | 0.268 | 0.101 | 0.379 | 0.041 | 0.278 | 0.140 | 0.459 | 0.131 | 0.485 |
| 382.3447 | 3.3 | Hydroxy tetracosanoic acid | 0.556 | 0.702 | 0.090 | 0.282 | 0.084 | 0.270 | 0.121 | 0.349 | 0.835 | 1.103 |
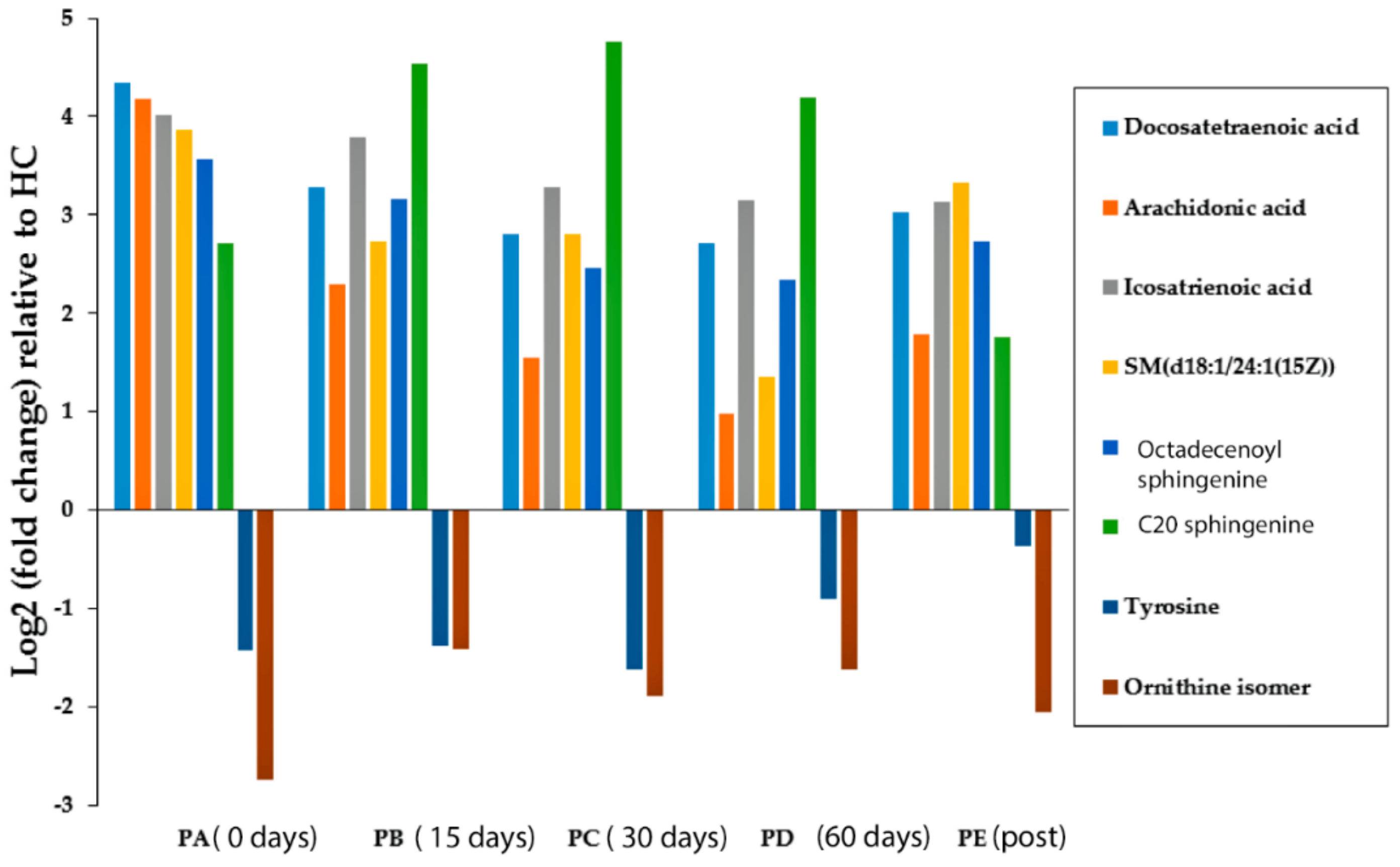
| Putative Metabolite | Pathway | (PA/HC) | p-Value | q-Value | AUC | VIP Total | VIP (Pred./Ortho.) |
|---|---|---|---|---|---|---|---|
| Ornithine isomer | unknown | 0.15 | 7.82 × 10−3 | 2.67 × 10−2 | 0.84 | 1.85 | 4.28 |
| C20 sphingenine | Sphingoid bases | 6.54 | 2.03 × 10−2 | 3.92 × 10−2 | 0.75 | 1.81 | 2.82 |
| Tyrosine | Tyrosine metabolism | 0.37 | 2.64 × 10−2 | 4.98 × 10−2 | 0.83 | 1.63 | 1.84 |
| SM (d18:1/24:1) | Ceramide phosphocholines (sphingomyelins) | 14.52 | 3.28 × 10−3 | 2.67 × 10−2 | 0.87 | 1.26 | 1.08 |
| Eicosatrienoic acid | Biosynthesis of unsaturated fatty acids | 16.18 | 3.48 × 10−4 | 4.67 × 10−3 | 0.88 | 1.07 | 1.53 |
| Docosatetraenoic acid | Biosynthesis of unsaturated fatty acids | 20.32 | 9.11 × 10−4 | 6.15 × 10−3 | 0.92 | 1.02 | 1.01 |
| Arachidonic acid | Fatty Acids and Conjugates | 18.05 | 4.79 × 10−3 | 1.94 × 10−2 | 0.88 | 0.99 | 2.91 |
| Octadecenoylsphingenine | Ceramides | 11.88 | 5.31 × 10−5 | 1.08 × 10−3 | 0.94 | 0.89 | 1.21 |
| m/z | Rt min | Elemental Composition | Putative ID | Deviation ppm | MS2/MS3 | Comments |
|---|---|---|---|---|---|---|
| 133.0971 (+) | 87 | C5H13O2N2 | Ornithine isomer | −0.332 | MS2 115.085 (C5H11ON2), 98.060 (C5H8ON), 69.033 (C4H5NO) | Nearest alternative composition C3H11N5O (+9.8 ppm) Despite the MS2 fragments making sense (Figure S4), it is difficult to propose a definitive structure. |
| 328.3211 (+) | 3.4 | C20H42O2N | C20 sphinganine | +0499 | MS2 311.2943 (C20H39O2),310.30951 (C20H42ON) 228.1957(C13H26O2N) 188.1644 (C10H22O2N) | Proposed fragmentation scheme shown in Figure S5. (Spectrum Figure S6). |
| 813.6851 (+) | 3.3 | C47H94N2PO6 | Ceramide d18:1 24:1 | −1.145 | MS2 795.61, 553.53 MS3 (7956) 777.3, 614.6, 495.22, 264.1 | This marker remains unidentified since it is not possible to relate the fragments to the proposed structure. Nearest alternative composition. Nearest match C51H91NO6 (1.5 ppm). MS2 and MS3 spectra Figures S7 and S8. |
| 182.0810 (+) | 13.5 | C9H12NO3 | Tyrosine | −0.810 | - | Matches retention time of standard. Nearest alternative composition C7H10NO2 (+7.9 ppm) |
| 305.2484 (−) | 19.5 C4 | C20H33O2 | Eicosatrienoic acid | −0.438 | - | Matches retention time of standard. Nearest alternative composition C18H31ON3 (+3.9 ppm) |
| 329.2484 (−) | 191 C4 | C22H33O2 | Docosapentaenoic acid | −0.406 | - | No standard available but logically the retention time falls close to eicosatrienoic acid because number of hydrogens is the same. |
| 303.2329 (−) | 18.5 C4 | C20H31O2 | Arachidonic acid | −0.045 | - | Matches retention time of standard. Nearest alternative composition C18H29ON3 (+4.3 ppm) |
| 564.5361 (+) | 3.1 | C36H70NO3 | Octadecenoylsphinganine | +19.8 | MS2 546.5239(C36H68NO2) 528.5128 (C36H66NO) 282.2782 (C18H38NO) 264.2680 (C18H36N) | Proposed fragmentation scheme shown in Figure S9. MS2 spectrum Figure S10. |
| Healthy Controls | ||
| Sample | Arachidonic Acid | Cis-8, 11, 14-Eicosatrienoic acid |
| HC01 | 47.6 | 112.4 |
| HC02 | 13.6 | 10 |
| HC03 | 25.6 | 19.6 |
| HC04 | 7.6 | 3.6 |
| HC05 | 15.2 | 2 |
| HC06 | 63.6 | 210.4 |
| HC07 | 86.8 | 136.4 |
| HC08 | 144.8 | 127.6 |
| HC09 | 12.8 | 5.6 |
| HC10 | 13.2 | 16.4 |
| HC11 | 34.8 | 60 |
| Mean | 42.4 | 64 |
| SD | 42.4 | 71.6 |
| SEM | 12.8 | 21.6 |
| Crohn’s disease | ||
| Sample | Arachidonic Acid | Cis-8, 11, 14-Eicosatrienoic acid |
| PA01 | 4406 | 3854.4 |
| PA02 | 4365.2 | 1671.6 |
| PA03 | 432.4 | 492.4 |
| PA04 | 1644.8 | 9462.4 |
| PA05 | 92 | 514.4 |
| PA06 | 3510 | 3600.4 |
| PA07 | 98 | 196 |
| PA08 | 44.8 | 904.8 |
| PA09 | 27.6 | 181.6 |
| PA10 | 14.0 | 50.0 |
| PA11 | 4262.8 | 1029.2 |
| Mean | 1718 | 1996 |
| SD | 1985.2 | 2806.8 |
| SEM | 598.4 | 846.4 |
| * p-value | 0.019 | 0.046 |
| Group ID | Description | n |
|---|---|---|
| PA | CD children pre-EEN treatment | 11 |
| PB | CD children 15 days of EEN treatment | 10 |
| PC | CD children 30 days of EEN treatment | 11 |
| PD | CD children 60 days of EEN treatment | 11 |
| PE | CD children back to normal diet | 11 |
| HC | Healthy children control | 11 |
| Subjects | Sex | Age | BMI at Enrolment (kg/m2) | Weight (kg) at Enrolment (kg/m2) | BMI Z Score at Enrolment (kg/m2) | BMI (kg/m2) at 4 Weeks | Weight (kg) at 4 Weeks | BMI Z Score Increase 4 Weeks | BMI (kg/m2) at 8 Weeks | Weight (kg) at 8 Weeks | BMI Z Score Increase 8 Weeks | Treatment Naïve | Previously Treated | PCDAI at Start | PCDAI at End |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD Patients | 4 F 7 M | 11.5 ± 2.4 | 13.8 ± 1.4 | 28.9 ± 6.0 | −1.61 ± 0.27 | 15.7 ± 1.3 | 30.8 ± 6.3 | 1.6 ± 0.38 | 16.2 ± 1.5 | 33.3 ± 5.2 | 1.7 ± 0.35 | 7 | 4 | 11 > 10 | 7 < 10 |
| Healthy controls | 4 F 7 M | 10.2 ± 2.3 | na | na | na | nr | nr | nr | nr | nr | nr | nr | nr | nr | nr |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alghamdi, A.; Gerasimidis, K.; Blackburn, G.; Akinci, D.; Edwards, C.; Russell, R.K.; Watson, D.G. Untargeted Metabolomics of Extracts from Faecal Samples Demonstrates Distinct Differences between Paediatric Crohn’s Disease Patients and Healthy Controls but No Significant Changes Resulting from Exclusive Enteral Nutrition Treatment. Metabolites 2018, 8, 82. https://doi.org/10.3390/metabo8040082
Alghamdi A, Gerasimidis K, Blackburn G, Akinci D, Edwards C, Russell RK, Watson DG. Untargeted Metabolomics of Extracts from Faecal Samples Demonstrates Distinct Differences between Paediatric Crohn’s Disease Patients and Healthy Controls but No Significant Changes Resulting from Exclusive Enteral Nutrition Treatment. Metabolites. 2018; 8(4):82. https://doi.org/10.3390/metabo8040082
Chicago/Turabian StyleAlghamdi, Adel, Konstantinos Gerasimidis, Gavin Blackburn, Didem Akinci, Christine Edwards, Richard K. Russell, and David G. Watson. 2018. "Untargeted Metabolomics of Extracts from Faecal Samples Demonstrates Distinct Differences between Paediatric Crohn’s Disease Patients and Healthy Controls but No Significant Changes Resulting from Exclusive Enteral Nutrition Treatment" Metabolites 8, no. 4: 82. https://doi.org/10.3390/metabo8040082
APA StyleAlghamdi, A., Gerasimidis, K., Blackburn, G., Akinci, D., Edwards, C., Russell, R. K., & Watson, D. G. (2018). Untargeted Metabolomics of Extracts from Faecal Samples Demonstrates Distinct Differences between Paediatric Crohn’s Disease Patients and Healthy Controls but No Significant Changes Resulting from Exclusive Enteral Nutrition Treatment. Metabolites, 8(4), 82. https://doi.org/10.3390/metabo8040082