Flux Balance Analysis Inspired Bioprocess Upgrading for Lycopene Production by a Metabolically Engineered Strain of Yarrowia lipolytica
Abstract
:1. Introduction
2. Results
2.1. Media Designing for Bioprocess Improvement
FBA-Independent Screening of Medium Components
| Std | Run | A:CuCl2·2H2O | B:NiSO4·6H2O | C:CoCl2·6H2O | D:MgSO4·7H2O | E:NiCl2·6H2O | F:H3BO3 | G:ZnCl2 | H:MnSO4·7H20 | J:KI | K:MnCl2·4H2O | L:FeSO4·7H2O | M:Na2MoO4·2H2O | N:FeCl3·6H2O | O:KNO3 | P:NaCl | Q:Glucose | R:Fructose | S:Yeast extract | T:(NH4)2SO4 | U:KH2PO4 | V:K2HPO4 | W:Vitamine B1 | X:BaCl2·2H2O | Biomass | Lycopene | Lycopene content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | mg/L | mg/g | |||
| 10 | 1 | 0.03 | 1 | 0.4 | 1.5 | 0.1 | 0.01 | 0.01 | 0.02 | 0.01 | 0.2 | 2.5 | 0.1 | 3 | 0.2 | 0.01 | 20 | 0 | 10 | 10 | 0.01 | 0 | 0.01 | 1 | 14.26 | 1.76 | 0.12 |
| 22 | 2 | 0.3 | 3 | 0.4 | 1.5 | 0.1 | 0.01 | 0.15 | 0.4 | 0.01 | 0.01 | 2.5 | 0.1 | 0.02 | 0.01 | 1 | 10 | 20 | 0.5 | 0.5 | 0.01 | 0 | 0.01 | 1 | 1.52 | 0 | 0 |
| 12 | 3 | 0.3 | 3 | 0.02 | 1.5 | 0.1 | 0.01 | 0.15 | 0.02 | 0.01 | 0.01 | 0.1 | 0.1 | 3 | 0.2 | 1 | 20 | 0 | 10 | 0.5 | 10 | 3.5 | 0 | 0.01 | 3.38 | 0 | 0 |
| 2 | 4 | 0.03 | 3 | 0.4 | 15 | 0.1 | 0.1 | 0.01 | 0.4 | 0.01 | 0.2 | 2.5 | 0.01 | 0.02 | 0.2 | 1 | 10 | 0 | 10 | 0.5 | 10 | 0 | 0 | 0.01 | 11.06 | 1.82 | 0.16 |
| 7 | 5 | 0.03 | 3 | 0.02 | 1.5 | 0.05 | 0.01 | 0.15 | 0.4 | 0.1 | 0.2 | 2.5 | 0.01 | 3 | 0.01 | 1 | 20 | 0 | 0.5 | 10 | 10 | 0 | 0 | 1 | 2.36 | 0.6 | 0.26 |
| 13 | 6 | 0.03 | 3 | 0.4 | 1.5 | 0.05 | 0.1 | 0.01 | 0.4 | 0.01 | 0.01 | 0.1 | 0.01 | 3 | 0.2 | 1 | 20 | 20 | 0.5 | 10 | 0.01 | 3.5 | 0.01 | 0.01 | 2.66 | 0.37 | 0.14 |
| 1 | 7 | 0.3 | 3 | 0.4 | 15 | 0.1 | 0.01 | 0.15 | 0.02 | 0.1 | 0.2 | 0.1 | 0.01 | 3 | 0.2 | 0.01 | 10 | 20 | 0.5 | 10 | 0.01 | 0 | 0 | 0.01 | 3.46 | 1.17 | 0.34 |
| 16 | 8 | 0.3 | 3 | 0.02 | 1.5 | 0.1 | 0.1 | 0.01 | 0.02 | 0.1 | 0.01 | 2.5 | 0.01 | 0.02 | 0.01 | 0.01 | 20 | 20 | 10 | 10 | 10 | 0 | 0.01 | 0.01 | 2.38 | 0 | 0 |
| 3 | 9 | 0.03 | 1 | 0.4 | 15 | 0.1 | 0.1 | 0.15 | 0.02 | 0.1 | 0.01 | 2.5 | 0.1 | 0.02 | 0.01 | 1 | 20 | 0 | 0.5 | 10 | 0.01 | 3.5 | 0 | 0.01 | 2.74 | 0.04 | 0.02 |
| 15 | 10 | 0.3 | 1 | 0.02 | 15 | 0.1 | 0.01 | 0.01 | 0.4 | 0.01 | 0.2 | 0.1 | 0.01 | 0.02 | 0.01 | 1 | 20 | 20 | 10 | 10 | 0.01 | 3.5 | 0 | 1 | 2.82 | 0 | 0 |
| 14 | 11 | 0.03 | 1 | 0.4 | 15 | 0.05 | 0.01 | 0.15 | 0.02 | 0.1 | 0.01 | 0.1 | 0.01 | 0.02 | 0.2 | 1 | 20 | 20 | 10 | 0.5 | 10 | 0 | 0.01 | 1 | 12.84 | 3.82 | 0.3 |
| 21 | 12 | 0.3 | 3 | 0.02 | 15 | 0.05 | 0.1 | 0.15 | 0.02 | 0.01 | 0.2 | 2.5 | 0.01 | 0.02 | 0.2 | 0.01 | 20 | 0 | 0.5 | 0.5 | 0.01 | 3.5 | 0.01 | 1 | 3.76 | 0.04 | 0.01 |
| 24 | 13 | 0.03 | 1 | 0.02 | 1.5 | 0.05 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.1 | 0.01 | 0.02 | 0.01 | 0.01 | 10 | 0 | 0.5 | 0.5 | 0.01 | 0 | 0 | 0.01 | 0.64 | 0 | 0 |
| 18 | 14 | 0.3 | 1 | 0.4 | 15 | 0.05 | 0.01 | 0.15 | 0.4 | 0.01 | 0.01 | 2.5 | 0.01 | 3 | 0.01 | 0.01 | 10 | 0 | 10 | 10 | 10 | 3.5 | 0.01 | 0.01 | 3.52 | 0 | 0 |
| 19 | 15 | 0.03 | 3 | 0.02 | 15 | 0.1 | 0.01 | 0.01 | 0.4 | 0.1 | 0.01 | 0.1 | 0.1 | 0.02 | 0.2 | 0.01 | 10 | 0 | 0.5 | 10 | 10 | 3.5 | 0.01 | 1 | 7.36 | 2.82 | 0.38 |
| 6 | 16 | 0.3 | 1 | 0.02 | 1.5 | 0.05 | 0.1 | 0.15 | 0.4 | 0.1 | 0.2 | 0.1 | 0.1 | 0.02 | 0.2 | 1 | 10 | 0 | 10 | 10 | 0.01 | 0 | 0.01 | 0.01 | 1.38 | 0 | 0 |
| 23 | 17 | 0.3 | 3 | 0.4 | 15 | 0.05 | 0.1 | 0.01 | 0.4 | 0.1 | 0.01 | 0.1 | 0.1 | 3 | 0.01 | 0.01 | 20 | 0 | 10 | 0.5 | 0.01 | 0 | 0 | 1 | 2.4 | 0 | 0 |
| 5 | 18 | 0.03 | 1 | 0.02 | 1.5 | 0.1 | 0.1 | 0.15 | 0.4 | 0.1 | 0.01 | 2.5 | 0.01 | 3 | 0.2 | 0.01 | 10 | 20 | 10 | 0.5 | 0.01 | 3.5 | 0 | 1 | 16.14 | 5.76 | 0.36 |
| 8 | 19 | 0.3 | 1 | 0.4 | 1.5 | 0.05 | 0.01 | 0.01 | 0.4 | 0.1 | 0.2 | 2.5 | 0.1 | 0.02 | 0.2 | 0.01 | 20 | 20 | 0.5 | 0.5 | 10 | 3.5 | 0 | 0.01 | 4.44 | 0 | 0 |
| 17 | 20 | 0.03 | 3 | 0.4 | 1.5 | 0.05 | 0.1 | 0.15 | 0.02 | 0.01 | 0.2 | 0.1 | 0.1 | 0.02 | 0.01 | 0.01 | 10 | 20 | 10 | 10 | 10 | 3.5 | 0 | 1 | 2.76 | 0 | 0 |
| 4 | 21 | 0.03 | 1 | 0.02 | 15 | 0.1 | 0.1 | 0.15 | 0.4 | 0.01 | 0.2 | 0.1 | 0.1 | 3 | 0.01 | 0.01 | 20 | 20 | 0.5 | 0.5 | 10 | 0 | 0.01 | 0.01 | 8.3 | 9.18 | 1.11 |
| 11 | 22 | 0.3 | 1 | 0.02 | 15 | 0.05 | 0.1 | 0.01 | 0.02 | 0.01 | 0.01 | 2.5 | 0.1 | 3 | 0.2 | 1 | 10 | 20 | 0.5 | 10 | 10 | 0 | 0 | 1 | 11.26 | 2.17 | 0.19 |
| 9 | 23 | 0.03 | 3 | 0.02 | 15 | 0.05 | 0.01 | 0.01 | 0.02 | 0.1 | 0.2 | 2.5 | 0.1 | 3 | 0.01 | 1 | 10 | 20 | 10 | 0.5 | 0.01 | 3.5 | 0.01 | 0.01 | 15.16 | 3.27 | 0.22 |
| 20 | 24 | 0.3 | 1 | 0.4 | 1.5 | 0.1 | 0.1 | 0.01 | 0.02 | 0.1 | 0.2 | 0.1 | 0.01 | 3 | 0.01 | 1 | 10 | 0 | 0.5 | 0.5 | 10 | 3.5 | 0.01 | 1 | 4.36 | 0 | 0 |
2.2. Combination of Flux Balance Analysis and Plackett-Burman Design for Media Designing
2.2.1. Flux Balance Analysis and Prediction of Media Components
| ID | Reaction Name | Biomass | FPP | Flux |
|---|---|---|---|---|
| R1202 | l-isoleucine exchange | 0.0315 # | 0.4342 | −0.0042 |
| R1203 | l-leucine exchange | 0.0315 | 0.4352 | −0.0085 |
| R1205 | l-valine exchange | 0.0315 | 0.4350 | −0.0104 |
| R1226 | l-glutamate exchange | 0.0315 | 0.4763 | −2 $ |
| R1241 | l-alanine exchange | 0.0315 | 0.5871 | −2 |
| R1244 | l-arginine exchange | 0.0315 | 0.5159 | −2 |
| R1245 | l-asparagine exchange | 0.0315 | 0.4344 | −0.0052 |
| R1248 | l-cysteine exchange | 0.0315 | 0.5878 | −2 |
| R1268 | l-histidine exchange | 0.0315 | 0.4347 | −0.0030 |
| R1273 | l-methionine exchange | 0.0315 | 0.4352 | −0.0072 |
| R1275 | l-lysine exchange | 0.0315 | 0.4353 | −0.0067 |
| R1280 | l-serine exchange | 0.0315 | 0.6640 | −2 |
| R1283 | l-threonine exchange | 0.0315 | 0.7025 | −2 |
| R1285 | l-tryptophan exchange | 0.0315 | 0.4339 | −0.0010 |
| R1298 | l-aspartate exchange | 0.0315 | 0.6640 | −2 |
| R1300 | l-phenylalanine exchange | 0.0315 | 0.6215 | −2 |
| R1301 | l-proline exchange | 0.0315 | 0.5150 | −2 |
| R1303 | l-tyrosine exchange | 0.0315 | 0.5650 | −1.6980 |
| ID | Reaction name | Biomass | FPP | Flux |
|---|---|---|---|---|
| R1207 | Thiamine diphosphate exchange | 0.0315 # | 0.4332 | 0.0000 |
| R1239 | 4-aminobutanoate exchange | 0.0315 | 0.4525 | −1 $ |
| R1251 | Ergosterol exchange | 0.0315 | 0.4333 | −0.0002 |
| R1258 | Ethanolamine exchange | 0.0315 | 0.4392 | −0.0769 |
| R1267 | Guanine exchange | 0.0315 | 0.4345 | −1 |
| R1278 | (R)Pantothenate exchange | 0.0315 | 0.4332 | 0.0000 |
| R1281 | Sulfite exchange | 0.0315 | 0.4332 | 0 |
| R1287 | Urea exchange | 0.0315 | 0.4332 | 0 |
| R1296 | Hypoxanthine exchange | 0.0315 | 0.4353 | −0.0049 |
| R1306 | Pyridoxine exchange | 0.0315 | 0.4332 | 0 |
2.2.2. Plackett-Burman Design Screening of FBA-Predicted Components
| Std | Run | A:Thiamine pyrophosphate | B:Pyridoxine, hydrocloride | C:Succinate | D:(R) Pantothenate | E:4-Aminobutyric acid | F:KH2PO4 | G:(NH4)2SO4 | H:Urea | J:Sulfite Na2SO3 | K:Ergosterol | L:Ethanolamine | M:Hypoxanthine | N:Guanine | O:l-histidine | P:l-methionine | Q:l-lysine | R:l-tryptophan | S:l-valine | T:l-asparagine | U:l-isoleucine | V:l-leucine | W:d-glucose | X:d-fructose | Lycopene | Biomass | Lycopene content | Lipid | Lipid content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | g/L | mg/L | g/L | mg/g | g/L | % | ||
| 12 | 1 | 0.006 | 0.006 | 1 | 1 | 5 | 6 | 2 | 0.5 | 0.1 | 0.01 | 0.05 | 5 | 5 | 5 | 5 | 5 | 1 | 5 | 1 | 10 | 10 | 20 | 20 | 9.99 | 13.35 | 0.75 | 1.27 | 9.51 |
| 16 | 2 | 0.006 | 0.006 | 1 | 1 | 5 | 12 | 0.5 | 0.5 | 2 | 0.01 | 0.3 | 1 | 1 | 1 | 1 | 5 | 5 | 5 | 10 | 10 | 1 | 60 | 20 | 34.51 | 26.73 | 1.29 | 2.41 | 9.02 |
| 4 | 3 | 0.0006 | 0.0006 | 1 | 5 | 5 | 12 | 2 | 2 | 0.1 | 0.1 | 0.05 | 5 | 5 | 1 | 1 | 5 | 5 | 1 | 1 | 10 | 1 | 60 | 20 | 27.87 | 19.48 | 1.43 | 2.2 | 11.3 |
| 6 | 4 | 0.006 | 0.0006 | 1 | 1 | 1 | 12 | 2 | 2 | 2 | 0.1 | 0.05 | 5 | 1 | 5 | 5 | 1 | 1 | 5 | 10 | 1 | 1 | 60 | 20 | 9.85 | 27.4 | 0.36 | 1.57 | 5.73 |
| 20 | 5 | 0.006 | 0.0006 | 5 | 1 | 5 | 12 | 0.5 | 0.5 | 2 | 0.1 | 0.05 | 1 | 5 | 1 | 5 | 1 | 1 | 1 | 1 | 10 | 10 | 60 | 60 | 6.69 | 30.88 | 0.22 | 1.6 | 5.18 |
| 22 | 6 | 0.006 | 0.006 | 5 | 1 | 5 | 6 | 2 | 2 | 0.1 | 0.01 | 0.3 | 5 | 1 | 1 | 5 | 1 | 5 | 1 | 1 | 1 | 1 | 60 | 60 | 18.47 | 31.45 | 0.59 | 1.02 | 3.24 |
| 8 | 7 | 0.006 | 0.0006 | 5 | 1 | 1 | 6 | 0.5 | 2 | 2 | 0.1 | 0.3 | 5 | 1 | 5 | 1 | 5 | 5 | 1 | 1 | 10 | 10 | 20 | 20 | 41.23 | 14.23 | 2.9 | 1.55 | 10.9 |
| 24 | 8 | 0.0006 | 0.0006 | 1 | 1 | 1 | 6 | 0.5 | 0.5 | 0.1 | 0.01 | 0.05 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 20 | 20 | 40.54 | 16.53 | 2.45 | 1.53 | 9.23 |
| 18 | 9 | 0.006 | 0.0006 | 5 | 5 | 1 | 6 | 2 | 2 | 0.1 | 0.01 | 0.3 | 1 | 5 | 1 | 1 | 1 | 1 | 5 | 10 | 10 | 10 | 60 | 20 | 19.95 | 29.05 | 0.69 | 1.63 | 5.61 |
| 13 | 10 | 0.0006 | 0.006 | 5 | 1 | 1 | 12 | 0.5 | 2 | 0.1 | 0.01 | 0.05 | 1 | 5 | 5 | 5 | 5 | 5 | 1 | 10 | 1 | 10 | 60 | 20 | 12.41 | 30.58 | 0.41 | 0.82 | 2.68 |
| 3 | 11 | 0.0006 | 0.0006 | 5 | 5 | 5 | 12 | 2 | 0.5 | 2 | 0.01 | 0.3 | 5 | 1 | 1 | 5 | 5 | 1 | 1 | 10 | 1 | 10 | 20 | 20 | 2.07 | 21.05 | 0.1 | 1.45 | 6.86 |
| 5 | 12 | 0.0006 | 0.0006 | 1 | 1 | 5 | 12 | 2 | 2 | 2 | 0.01 | 0.3 | 1 | 5 | 5 | 1 | 1 | 5 | 5 | 1 | 1 | 10 | 20 | 60 | 9.8 | 33.23 | 0.3 | 1.4 | 4.21 |
| 7 | 13 | 0.0006 | 0.006 | 1 | 1 | 1 | 6 | 2 | 2 | 2 | 0.1 | 0.3 | 1 | 5 | 1 | 5 | 5 | 1 | 1 | 10 | 10 | 1 | 20 | 60 | 23.58 | 23.65 | 1 | 1.62 | 6.85 |
| 2 | 14 | 0.0006 | 0.006 | 5 | 5 | 5 | 12 | 0.5 | 2 | 0.1 | 0.1 | 0.3 | 1 | 1 | 5 | 5 | 1 | 1 | 5 | 1 | 10 | 1 | 20 | 20 | 16.44 | 21.33 | 0.77 | 0.88 | 4.13 |
| 19 | 15 | 0.0006 | 0.006 | 1 | 5 | 5 | 6 | 0.5 | 2 | 2 | 0.01 | 0.05 | 5 | 1 | 5 | 1 | 1 | 1 | 1 | 10 | 10 | 10 | 60 | 60 | 1.77 | 9.08 | 0.2 | 1.09 | 12.01 |
| 9 | 16 | 0.0006 | 0.006 | 1 | 5 | 1 | 6 | 0.5 | 0.5 | 2 | 0.1 | 0.3 | 5 | 5 | 1 | 5 | 1 | 5 | 5 | 1 | 1 | 10 | 60 | 20 | 4.09 | 21.3 | 0.19 | 1.56 | 7.3 |
| 15 | 17 | 0.006 | 0.0006 | 1 | 5 | 5 | 6 | 0.5 | 2 | 0.1 | 0.1 | 0.05 | 1 | 1 | 1 | 5 | 5 | 5 | 5 | 10 | 1 | 10 | 20 | 60 | 12.36 | 29.13 | 0.42 | 1.44 | 4.93 |
| 14 | 18 | 0.0006 | 0.0006 | 5 | 5 | 1 | 6 | 2 | 0.5 | 2 | 0.01 | 0.05 | 1 | 1 | 5 | 5 | 5 | 5 | 5 | 1 | 10 | 1 | 60 | 60 | 11.14 | 24.75 | 0.45 | 1.56 | 6.3 |
| 10 | 19 | 0.0006 | 0.0006 | 5 | 1 | 5 | 6 | 0.5 | 0.5 | 0.1 | 0.1 | 0.3 | 5 | 5 | 5 | 1 | 5 | 1 | 5 | 10 | 1 | 1 | 60 | 60 | 16.32 | 35.5 | 0.46 | 1.89 | 5.32 |
| 1 | 20 | 0.006 | 0.006 | 5 | 5 | 5 | 6 | 2 | 0.5 | 2 | 0.1 | 0.05 | 1 | 5 | 5 | 1 | 1 | 5 | 1 | 10 | 1 | 1 | 20 | 20 | 24.84 | 20.73 | 1.2 | 2 | 9.65 |
| 21 | 21 | 0.006 | 0.006 | 1 | 5 | 1 | 12 | 2 | 0.5 | 0.1 | 0.1 | 0.3 | 1 | 1 | 5 | 1 | 5 | 1 | 1 | 1 | 1 | 10 | 60 | 60 | 9.99 | 28.65 | 0.35 | 2.65 | 9.23 |
| 17 | 22 | 0.0006 | 0.006 | 5 | 1 | 1 | 12 | 2 | 0.5 | 0.1 | 0.1 | 0.05 | 5 | 1 | 1 | 1 | 1 | 5 | 5 | 10 | 10 | 10 | 20 | 60 | 24.79 | 32.63 | 0.76 | 2.22 | 6.8 |
| 11 | 23 | 0.006 | 0.0006 | 1 | 5 | 1 | 12 | 0.5 | 0.5 | 0.1 | 0.01 | 0.3 | 5 | 5 | 5 | 5 | 1 | 5 | 1 | 10 | 10 | 1 | 20 | 60 | 22.7 | 33.83 | 0.67 | 1.19 | 3.52 |
| 23 | 24 | 0.006 | 0.006 | 5 | 5 | 1 | 12 | 0.5 | 2 | 2 | 0.01 | 0.05 | 5 | 5 | 1 | 1 | 5 | 1 | 5 | 1 | 1 | 1 | 20 | 60 | 3.07 | 27.73 | 0.11 | 1.62 | 5.83 |
2.2.3. Analysis of the Correlation between Lipid Content and Lycopene Content
2.2.4. Analysis of the Effect of FBA Predicted Factors on the Expression of Genes of the Terpenoids Backbone
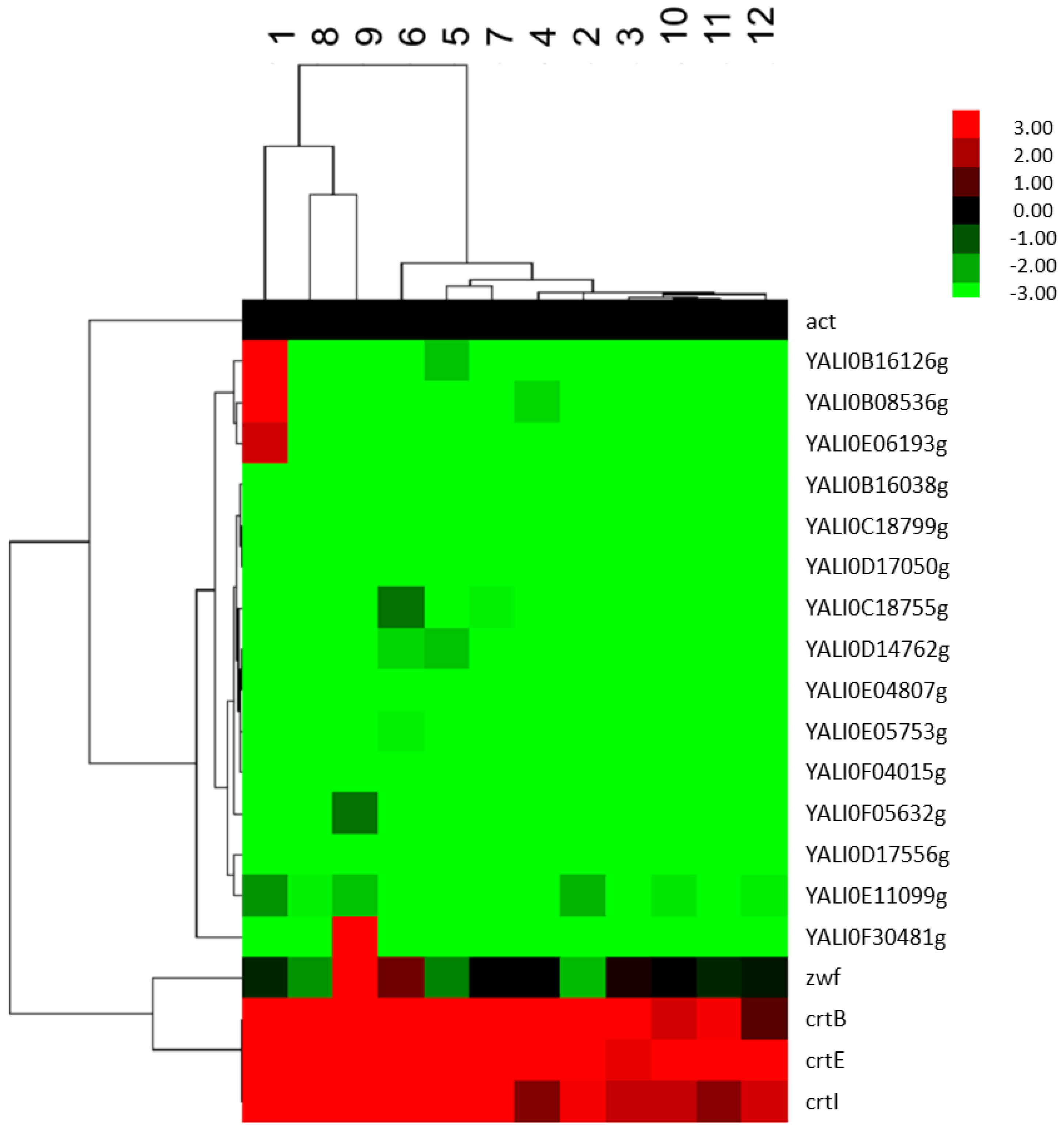
2.3. Fed-Batch Fermentation of Y. lipolytica for Lycopene Production on Designed Media
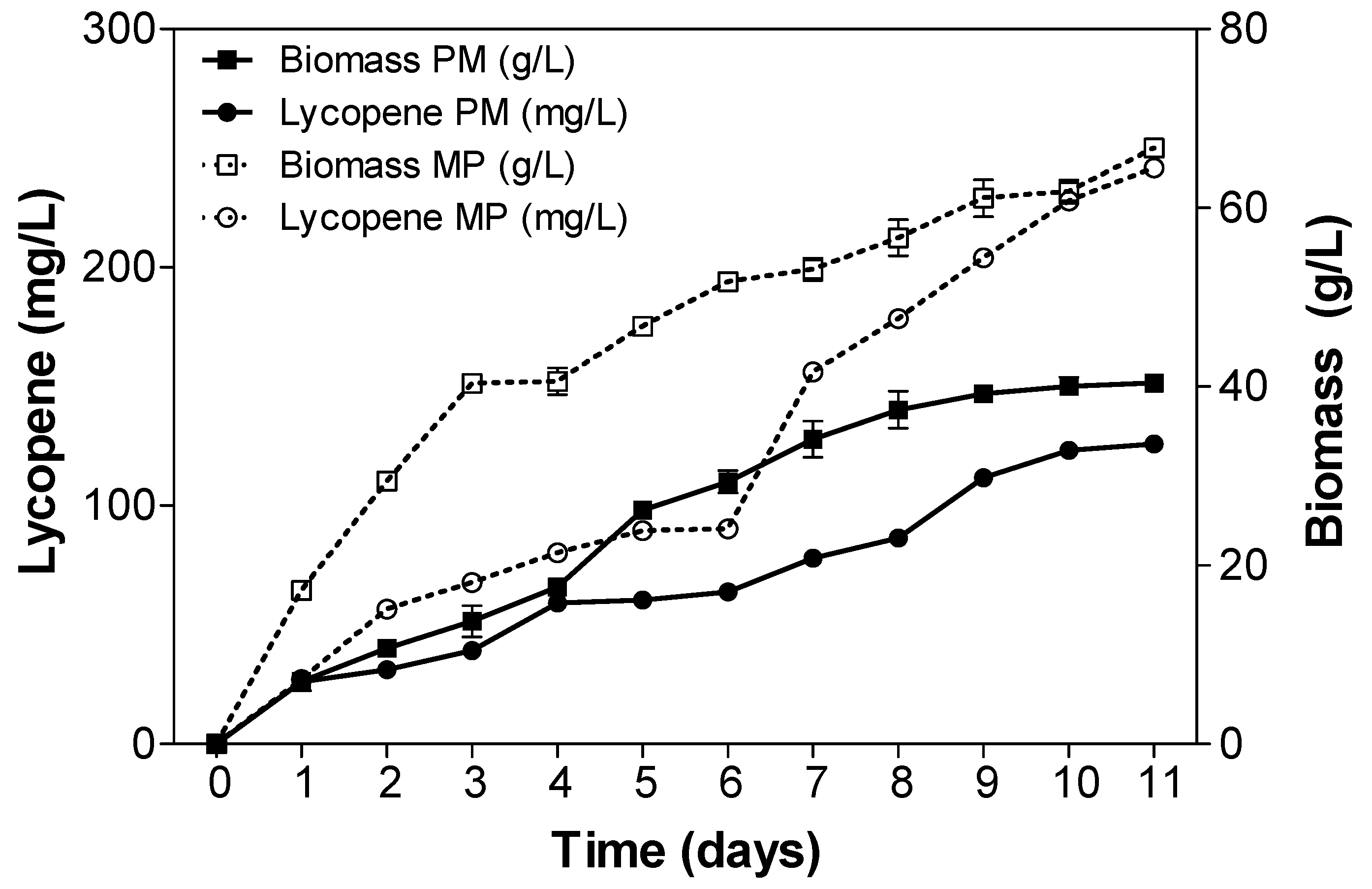
3. Discussion
4. Material and Methods
4.1. Description of the Engineered Lycopene Producing Y. Lipolytica Strain
4.2. Modeling Platform
4.3. Plackett-Burman Design and Shake Flasks Cultivation
4.4. Bioreactor Scale-up
4.5. Determination of Glucose Concentration, Dry Cell Weight, and Lipids
4.6. Lycopene Extraction and HPLC Analysis
4.7. Quantitative Reverse Transcription-PCR (qRT-PCR) Studies
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heider, S.E.; Peters-Wendisch, P.; Netzer, R.; Stafnes, M.; Brautaset, T.; Wendisch, V. Production and glucosylation of c50 and c40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014, 98, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Britton, G. Carotenoids. In Natural Food Colorants; Hendry, G.A.F., Houghton, J.D., Eds.; Springer: London, UK, 1996; pp. 197–243. [Google Scholar]
- Krinsky, N.I. Actions of carotenoids in biological systems. Annu. Rev. Nutr. 1993, 13, 561–587. [Google Scholar] [CrossRef] [PubMed]
- Amorim-Carrilho, K.T.; Cepeda, A.; Fente, C.; Regal, P. Review of methods for analysis of carotenoids. TrAC Trend Anal. Chem. 2014, 56, 49–73. [Google Scholar] [CrossRef]
- Gloria, N.F.; Soares, N.; Brand, C.; Oliveira, F.L.; Borojevic, R.; Teodoro, A.J. Lycopene and beta-carotene induce cell-cycle arrest and apoptosis in human breast cancer cell lines. Anticancer Res. 2014, 34, 1377–1386. [Google Scholar] [PubMed]
- Ilic, D. Lycopene for the prevention and treatment of prostate disease. Recent Results Cancer Res. 2014, 202, 109–114. [Google Scholar] [PubMed]
- Zhang, B.; Gu, Y. Low expression of erk signaling pathway affecting proliferation, cell cycle arrest and apoptosis of human gastric hgc-27 cells line. Mol. Biol. Rep. 2014, 41, 3659–3669. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, M.; Ono, M.; Higuchi, T.; Chen, C.; Hara, T.; Nakano, S. Anti-proliferative and apoptosis-inducing activity of lycopene against three subtypes of human breast cancer cell lines. Cancer Sci. 2014, 105, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Bilecova-Rabajdova, M.; Birkova, A.; Urban, P.; Gregova, K.; Durovcova, E.; Marekova, M. Naturally occurring substances and their role in chemo-protective effects. Cent. Eur. J. Public Health 2013, 21, 213–219. [Google Scholar] [PubMed]
- Khuda-Bukhsh, A.R.; Das, S.; Saha, S.K. Molecular approaches toward targeted cancer prevention with some food plants and their products: Inflammatory and other signal pathways. Nutr. Cancer 2014, 66, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, M.M.; Cortese, F.; Gesualdo, M.; Carbonara, S.; Zito, A.; Ricci, G.; de Pascalis, F.; Scicchitano, P.; Riccioni, G. Dietary intake of carotenoids and their antioxidant and anti-inflammatory effects in cardiovascular care. Mediat. Inflamm. 2013, 2013, 782137. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.W.; Sharpe, P.L.; Zhu, Q. Bioengineering of oleaginous yeast Yarrowia lipolytica for lycopene production. Methods Mol. Biol. 2012, 898, 153–159. [Google Scholar] [PubMed]
- Matthäus, F.; Ketelhot, M.; Gatter, M.; Barth, G. Production of lycopene in the non-carotenoid-producing yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2014, 80, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, P.L.; Ye, R.W.; Zhu, Q.Q. Carotenoid Production in a Recombinant Oleaginous Yeast. WO Patent 2,008,073,367 A1, 19 June 2008. [Google Scholar]
- Jin, Y.S.; Stephanopoulos, G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 2007, 9, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Alper, H.; Jin, Y.-S.; Moxley, J.F.; Stephanopoulos, G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 2005, 7, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, Ø.P.A. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar]
- Henry, C.S.; DeJongh, M.; Best, A.A.; Frybarger, P.M.; Linsay, B.; Stevens, R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010, 28, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Computational systems biology. Nature 2002, 420, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.E.; Hixson, K.K.; Conrad, T.M.; Lerman, J.A.; Charusanti, P.; Polpitiya, A.D.; Adkins, J.N.; Schramm, G.; Purvine, S.O.; Lopez-Ferrer, D.; et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Sports Med. Arthrosc. Rev. 2010. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Palsson, B.O. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli w3110. Appl. Environ. Microbiol. 1994, 60, 3724–3731. [Google Scholar] [PubMed]
- Schellenberger, J.; Que, R.; Fleming, R.M.; Thiele, I.; Orth, J.D.; Feist, A.M.; Zielinski, D.C.; Bordbar, A.; Lewis, N.E.; Rahmanian, S.; et al. Quantitative prediction of cellular metabolism with constraint-based models: The cobra toolbox v2.0. Nat. Protoc. 2011, 6, 1290–1307. [Google Scholar] [CrossRef] [PubMed]
- Ye, V.M.; Bhatia, S.K. Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol. Lett. 2012, 34, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G. Phytoene desaturase: Genes, enzymes and phylogenetic aspects. J. Plant Physiol. 1994, 143, 444–447. [Google Scholar] [CrossRef]
- Jin, Y.S.; Jeffries, T.W. Stoichiometric network constraints on xylose metabolism by recombinant Saccharomyces cerevisiae. Metab. Eng. 2004, 6, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Morgan, J.A. Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst. Biol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Tajparast, M.; Frigon, D. Genome-scale metabolic model of Rhodococcus jostii rha1 (imt1174) to study the accumulation of storage compounds during nitrogen-limited condition. BMC Syst. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.; Hua, Q. Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS ONE 2012, 7, e51535. [Google Scholar] [CrossRef] [PubMed]
- Nambou, K.; Zhao, C.; Wei, L.; Chen, J.; Imanaka, T.; Hua, Q. Designing of a “cheap to run” fermentation platform for an enhanced production of single cell oil from Yarrowia lipolytica dsm3286 as a potential feedstock for biodiesel. Bioresour. Technol. 2014, 173, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Verwaal, R.; Paalman, J.W.; Hogenkamp, A.; Verkleij, A.J.; Verrips, C.T.; Boonstra, J. Hxt5 expression is determined by growth rates in Saccharomyces cerevisiae. Yeast 2002, 19, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2003–2007. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nambou, K.; Jian, X.; Zhang, X.; Wei, L.; Lou, J.; Madzak, C.; Hua, Q. Flux Balance Analysis Inspired Bioprocess Upgrading for Lycopene Production by a Metabolically Engineered Strain of Yarrowia lipolytica. Metabolites 2015, 5, 794-813. https://doi.org/10.3390/metabo5040794
Nambou K, Jian X, Zhang X, Wei L, Lou J, Madzak C, Hua Q. Flux Balance Analysis Inspired Bioprocess Upgrading for Lycopene Production by a Metabolically Engineered Strain of Yarrowia lipolytica. Metabolites. 2015; 5(4):794-813. https://doi.org/10.3390/metabo5040794
Chicago/Turabian StyleNambou, Komi, Xingxing Jian, Xinkai Zhang, Liujing Wei, Jiajia Lou, Catherine Madzak, and Qiang Hua. 2015. "Flux Balance Analysis Inspired Bioprocess Upgrading for Lycopene Production by a Metabolically Engineered Strain of Yarrowia lipolytica" Metabolites 5, no. 4: 794-813. https://doi.org/10.3390/metabo5040794
APA StyleNambou, K., Jian, X., Zhang, X., Wei, L., Lou, J., Madzak, C., & Hua, Q. (2015). Flux Balance Analysis Inspired Bioprocess Upgrading for Lycopene Production by a Metabolically Engineered Strain of Yarrowia lipolytica. Metabolites, 5(4), 794-813. https://doi.org/10.3390/metabo5040794





