Molar-Based Targeted Metabolic Profiling of Cyanobacterial Strains with Potential for Biological Production
Abstract
:1. Introduction
2. Results and Discussion
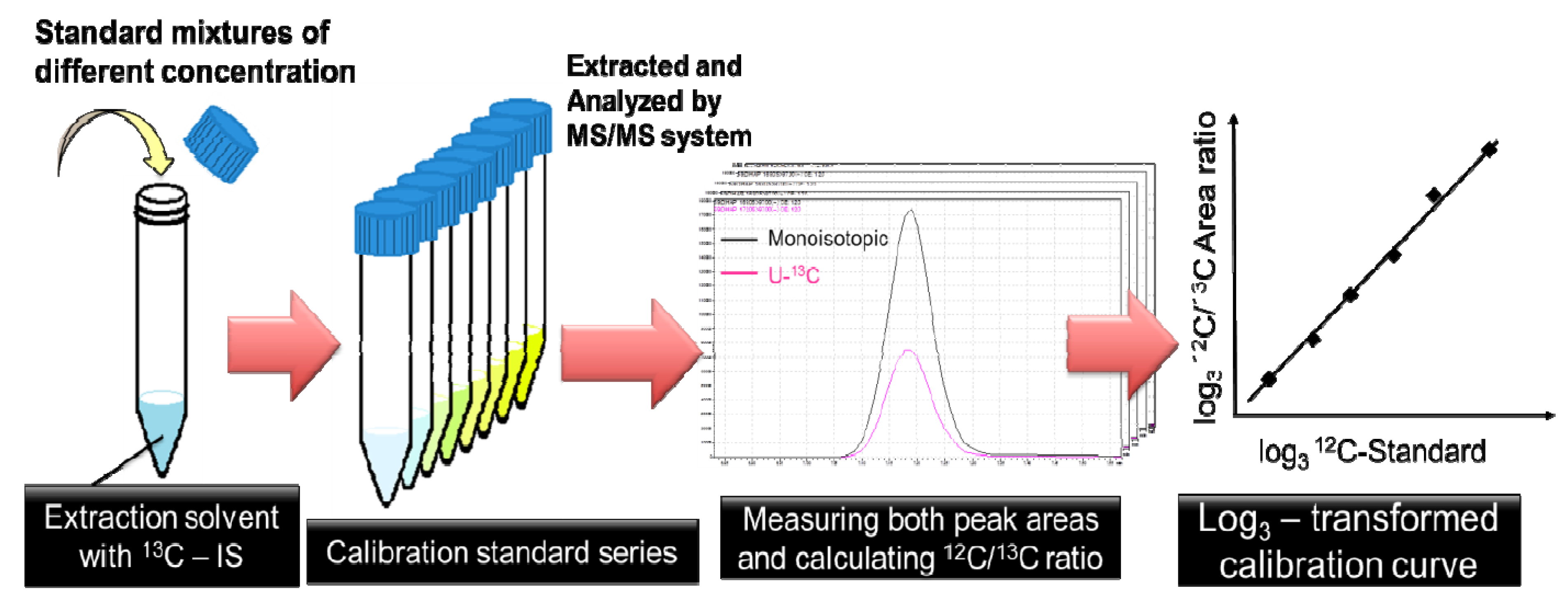
2.1. Optimization of 13C-internal Standard Amount for Quantitation


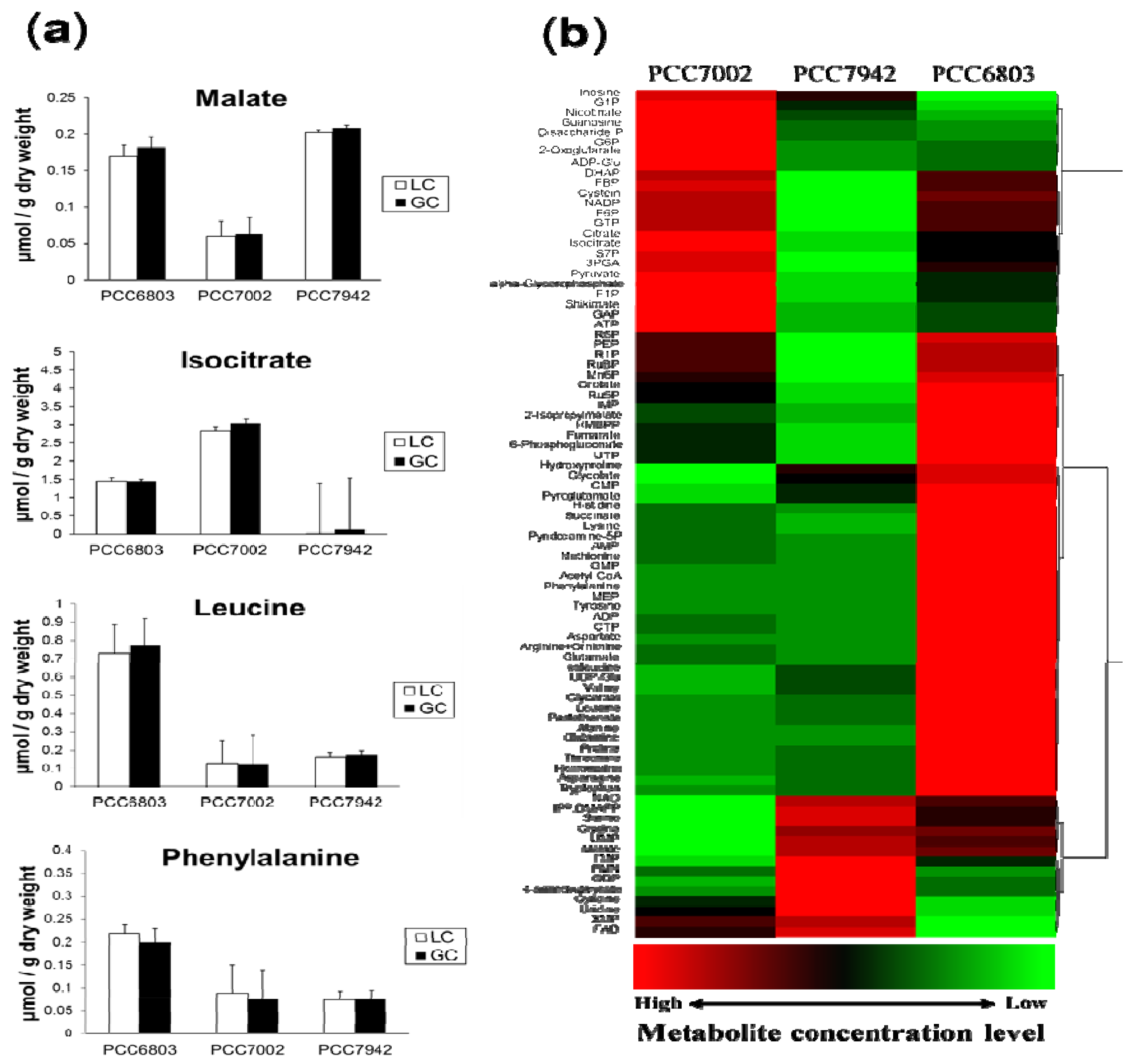
2.2. Overview of the Metabolic Differences between Type Strains
| Metabolite | PCC6803 | PCC7002 | PCC7942 | |||
|---|---|---|---|---|---|---|
| Average | S.D. | Average | S.D. | Average | S.D. | |
| 2-Isopropylmalate | 4.11 × 10 −1 | 7.75 × 10 −2 | 1.34 × 10 −1 | 2.01 × 10 −2 | 3.22 × 10 –2 | 1.18 × 10 −2 |
| 2-Oxoglutarate | 2.94 × 10 −1 | 7.06 × 10 −2 | 1.77 × 100 | 7.23 × 10 −1 | 1.77 × 10 −1 | 5.14 × 10 −2 |
| 3PGA | 1.48 × 101 | 1.86 × 100 | 2.02 × 101 | 4.87 × 100 | 7.13 × 100 | 1.18 × 100 |
| 4-Aminobutanoate | 9.67 × 10 −3 | 2.09 × 10 −3 | 9.23 × 10 −3 | 2.40 × 10 −3 | 1.42 × 10 −2 | 1.01 × 10 −2 |
| 6-Phosphogluconate | 4.66 × 100 | 8.35 × 10 −1 | 1.99 × 100 | 1.11 × 100 | 1.87 × 10 −1 | 4.76 × 10 −2 |
| Acetyl CoA | 4.43 × 10 −1 | 4.29 × 10 −2 | 6.22 × 10 −2 | 8.02 × 10 −3 | 6.54 × 10 −2 | 9.12 × 10 −3 |
| ADP | 6.51 × 100 | 7.58 × 10 −1 | 2.68 × 100 | 1.46 × 10 −2 | 2.38 × 100 | 6.44 × 10 −1 |
| ADP-Glc | 7.00 × 10 −2 | 4.13 × 10 −2 | 6.13 × 10 −1 | 2.66 × 10 −1 | 3.88 × 10 −2 | 1.76 × 10 −2 |
| Alanine | 4.53 × 100 | 1.16 × 100 | 6.50 × 10 −1 | 1.48 × 10 −1 | 8.12 × 10 −1 | 6.74 × 10 −1 |
| α-Glycerophosphate | 4.77 × 10 −1 | 7.50 × 10 −2 | 9.83 × 10 −1 | 1.94 × 10 −1 | 2.22 × 10 −1 | 3.74 × 10 −2 |
| AMP | 1.07 × 101 | 1.48 × 100 | 4.56 × 100 | 4.09 × 10 −1 | 3.92 × 100 | 1.09 × 100 |
| Arginine + Ornithine | 2.12 × 10 1 | 5.02 × 100 | 4.31 × 100 | 6.72 × 10 −1 | 3.52 × 100 | 9.79 × 10 −1 |
| Asparagine | 4.34 × 10 −1 | 8.01 × 10 −2 | 2.66 × 10 −2 | 3.97 × 10 −3 | 8.68 × 10 −2 | 3.49 × 10 −2 |
| Aspartate | 7.14 × 100 | 1.20 × 100 | 2.69 × 100 | 6.09 × 10 -1 | 2.53 × 100 | 1.07 × 100 |
| ATP | 2.14 × 100 | 5.58 × 10 −1 | 3.09 × 100 | 3.48 × 10 −1 | 1.90 × 100 | 8.72 × 10 −1 |
| Citrate | 2.16 × 100 | 7.83 × 10 −2 | 4.22 × 100 | 1.43 × 100 | 3.57 × 10 −1 | 8.69 × 10 −2 |
| CMP | 3.43 × 10 −1 | 7.71 × 10 −2 | 2.17 × 10 −1 | 4.07 × 10 −2 | 2.71 × 10 −1 | 3.67 × 10 −2 |
| CTP | 5.97 × 10 −1 | 1.08 × 10 -1 | 6.52 × 10 −2 | 9.81 × 10 −3 | 2.97 × 10 −2 | 1.13 × 10 −2 |
| Cystein | 4.79 × 10 −1 | 2.30 × 10 −1 | 5.28 × 10 −1 | 1.73 × 10 −1 | 2.32 × 10 −1 | 1.89 × 10 −1 |
| Cytidine | 1.28 × 10 −2 | 4.16 × 10 −3 | 2.11 × 10 −2 | 5.56 × 10 −3 | 3.40 × 10 −2 | 7.35 × 10 −3 |
| DHAP | 4.30 × 10 −1 | 7.71 × 10 −2 | 5.48 × 10 −1 | 2.91 × 10 −1 | 1.15 × 10 −1 | 1.67 × 10 −2 |
| Disaccharide-P | 6.03 × 10 −4 | 3.27 × 10 −4 | 1.21 × 10 −2 | 9.63 × 10 −3 | 1.56 × 10 −3 | 1.05 × 10 −3 |
| F1P | 1.12 × 10 −2 | 3.71 × 10 −3 | 1.90 × 10 −2 | 4.37 × 10 −3 | 6.99 × 10 −3 | 1.04 × 10 −3 |
| F6P | 4.61 × 10 −1 | 9.81 × 10 −2 | 5.19 × 10 −1 | 2.13 × 10 −2 | 2.75 × 10 −1 | 7.97 × 10 −2 |
| FAD | 9.98 × 10 −2 | 3.19 × 10 −2 | 1.60 × 10 −1 | 3.38 × 10 −2 | 1.98 × 10 −1 | 2.80 × 10 −2 |
| FBP | 2.40 × 10 −1 | 1.07 × 10 −1 | 3.39 × 10 −1 | 1.44 × 10 −2 | 2.08 × 10 −2 | 1.37 × 10 −3 |
| FMN | 4.77 × 10 −2 | 1.95 × 10 −2 | 4.81 × 10 −2 | 7.94 × 10 −3 | 5.26 × 10 −2 | 2.18 × 10 −2 |
| Fumarate | 1.63 × 10 −1 | 2.27 × 10 −2 | 9.08 × 10 −2 | 9.45 × 10 −3 | 5.11 × 10 −2 | 9.58 × 10 −3 |
| G1P | 6.91 × 10 −2 | 1.04 × 10 −2 | 1.41 × 10 −1 | 1.45 × 10 −2 | 9.23 × 10 −2 | 4.99 × 10 −2 |
| G6P | 1.84 × 100 | 3.10 × 10 −1 | 4.19 × 100 | 7.29 × 10 −1 | 1.57 × 100 | 3.43 × 10 −1 |
| GAP | 2.63 × 10 −1 | 7.81 × 10 −2 | 5.59 × 10 −1 | 2.24 × 10 −1 | 1.88 × 10 −1 | 4.91 × 10 −2 |
| GDP | 1.38 × 100 | 1.73 × 10 −1 | 9.38 × 10 −1 | 1.77 × 10 −1 | 3.46 × 100 | 4.42 × 100 |
| Glutamate | 2.13 × 102 | 4.79 × 101 | 4.25 × 101 | 6.52 × 100 | 3.39 × 101 | 8.11 × 100 |
| Glutamine | 3.47 × 100 | 4.61 × 10 −1 | 5.14 × 10 −1 | 1.37 × 10 −2 | 6.34 × 10 −1 | 2.12 × 10 −1 |
| Glycerate | 2.53 × 10 −2 | 9.28 × 10 −3 | 2.06 × 10 −2 | 5.33 × 10 −4 | 2.09 × 10 −2 | 1.93 × 10 −2 |
| Glycine | 1.67 × 10 1 | 4.51 × 100 | 2.17 × 100 | 8.78 × 10 −1 | 1.78 × 101 | 2.66 × 101 |
| Glycolate | 7.51 × 10 −2 | 2.03 × 10 −2 | 3.18 × 10 −2 | 1.47 × 10 −2 | 5.44 × 10 −2 | 1.49 × 10 −2 |
| GMP | 7.30 × 10 −1 | 2.05 × 10 −1 | 3.75 × 10 −1 | 3.47 × 10 −2 | 3.77 × 10 −1 | 6.40 × 10 −2 |
| GTP | 8.31 × 10 −1 | 2.35 × 10 −1 | 9.61 × 10 −1 | 1.93 × 10 −1 | 4.11 × 10 −1 | 1.31 × 10 −1 |
| Guanosine | 7.26 × 10 −2 | 3.51 × 10 −2 | 2.91 × 10 −1 | 1.92 × 10 −1 | 9.43 × 10 −2 | 3.71 × 10 −2 |
| Histidine | 1.10 × 10 −1 | 3.46 × 10 −2 | 7.03 × 10 −2 | 2.20 × 10 −2 | 6.45 × 10 −2 | 2.58 × 10 −2 |
| HMBPP | 9.27 × 10 −2 | 2.66 × 10 −2 | 5.40 × 10 −2 | 1.90 × 10 −2 | 3.32 × 10 −2 | 4.33 × 10 −3 |
| Homoserine | 3.05 × 10 −2 | 1.16 × 10 −2 | 1.32 × 10 −2 | 1.33 × 10 −3 | 1.51 × 10 −2 | 3.85 × 10 −3 |
| Hydroxyproline | 9.27 × 10 −1 | 3.05 × 10 −1 | 4.44 × 10 −1 | 1.72 × 10 −1 | 7.65 × 10 −1 | 5.11 × 10 −1 |
| IMP | 4.64 × 10 −1 | 3.84 × 10 −2 | 2.48 × 10 −1 | 9.97 × 10 −2 | 1.72 × 10 −1 | 6.00 × 10 −2 |
| Inosine | 1.46 × 10 −2 | 5.54 × 10 −3 | 9.41 × 10 −2 | 8.20 × 10 −2 | 6.80 × 10 −2 | 2.51 × 10 −2 |
| IPP + DMAPP | 1.40 × 10 −2 | 1.60 × 10 −3 | 9.64 × 10 − 3 | 1.79 × 10 −3 | 1.63 × 10 −2 | 1.28 × 10 −3 |
| Isocitrate | 1.45 × 100 | 1.25 × 10 −1 | 3.04 × 100 | 1.43 × 100 | 1.09 × 10 −1 | 4.96 × 10 −3 |
| Isoleucine | 3.70 × 10 −1 | 9.37 × 10 −2 | 7.14 × 10 −2 | 1.37 × 10 −2 | 1.50 × 10 −1 | 1.16 × 10 −1 |
| Leucine | 7.72 × 10 −1 | 1.55 × 10 −1 | 1.26 × 10 −1 | 2.32 × 10 −2 | 1.76 × 10 −1 | 1.33 × 10 −1 |
| Lysine | 1.57 × 10 −1 | 4.40 × 10 −2 | 1.02 × 10 −1 | 1.11 × 10 −2 | 9.32 × 10 −2 | 3.62 × 10 −2 |
| Malate | 1.82 × 10 −1 | 2.24 × 10 −2 | 6.36 × 10 −2 | 4.72 × 10 −3 | 2.08 × 10 −1 | 7.92 × 10 −2 |
| MEP | 8.38 × 10 −2 | 1.64 × 10 −2 | 3.94 × 10 −2 | 9.17 × 10 −3 | 3.83 × 10 −2 | 8.08 × 10 −3 |
| Methionine | 6.13 × 10 −1 | 1.10 × 10 −1 | 2.67 × 10 −1 | 4.40 × 10 −2 | 2.30 × 10 −1 | 4.51 × 10 −2 |
| Mn6P | 2.92 × 10 −1 | 4.19 × 10 −2 | 2.53 × 10 −1 | 7.48 × 10 −2 | 2.02 × 10 −1 | 3.06 × 10 −2 |
| NAD | 5.14 × 10 −1 | 1.26 × 10 −1 | 3.84 × 10 −1 | 2.97 × 10 −2 | 5.64 × 10 −1 | 1.04 × 10 −1 |
| NADP | 6.14 × 10 −1 | 1.26 × 10 −1 | 6.82 × 10 −1 | 1.38 × 10 −1 | 3.79 × 10 −1 | 8.28 × 10 −2 |
| Nicotinate | 3.56 × 10 −4 | 7.66 × 10 −5 | 7.72 × 10 −4 | 3.27 × 10 −4 | 4.62 × 10 −4 | 1.86 × 10 −4 |
| Orotate | 1.03 × 10 −2 | 1.58 × 10 −3 | 5.97 × 10 −3 | 2.34 × 10 −3 | 1.75 × 10 −3 | 7.23 × 10 −4 |
| Pantothenate | 4.91 × 10 −3 | 1.02 × 10 −3 | 2.13 × 10 −4 | 3.22 × 10 −5 | 4.56 × 10 −4 | 2.59 × 10 −4 |
| PEP | 2.51 × 100 | 7.59 × 10 −1 | 2.27 × 10 0 | 7.18 × 10 −1 | 1.47 × 100 | 2.45 × 10 −1 |
| Phenylalanine | 1.99 × 10 −1 | 6.09 × 10 −2 | 7.73 × 10 −2 | 1.86 × 10 −2 | 7.63 × 10 −2 | 4.77 × 10 −2 |
| Proline | 9.14 × 10 −1 | 1.76 × 10 −1 | 5.06 × 10 −2 | 9.05 × 10 −3 | 1.38 × 10 −1 | 1.23 × 10 −1 |
| Pyridoxamine-5P | 4.58 × 10 −2 | 5.05 × 10 −3 | 3.32 × 10 −2 | 2.92 × 10 −3 | 3.17 × 10 −2 | 3.82 × 10 −3 |
| Pyroglutamate | 2.81 × 100 | 7.22 × 10 −1 | 5.95 × 10 −1 | 8.06 × 10 −2 | 1.51 × 100 | 1.41 × 100 |
| Pyruvate | 1.05 × 101 | 1.38 × 100 | 1.81 × 10 1 | 4.20 × 10 0 | 5.76 × 100 | 1.12 × 100 |
| R1P | 3.62 × 10 −3 | 7.87 × 10 −4 | 2.90 × 10 −3 | 1.36 × 10 −3 | 9.68 × 10 −4 | 2.08 × 10 −4 |
| R5P | 1.95 × 10 −1 | 6.88 × 10 −2 | 1.52 × 10 −1 | 6.33 × 10 −2 | 5.82 × 10 −2 | 8.74 × 10 −3 |
| Ru5P | 4.04 × 10 −1 | 6.01 × 10 −2 | 2.19 × 10 −1 | 4.25 × 10 −2 | 4.48 × 10 −2 | 5.98 × 10 −3 |
| RuBP | 2.92 × 10 −1 | 5.35 × 10 −2 | 2.37 × 10 −1 | 5.29 × 10 −2 | 8.39 × 10 −2 | 2.74 × 10 −2 |
| S7P | 3.57 × 100 | 1.10 × 100 | 5.60 × 10 0 | 2.50 × 10 0 | 1.28 × 100 | 4.41 × 10 −1 |
| Serine | 7.12 × 10 −1 | 2.63 × 10 −1 | 3.11 × 10 −1 | 9.71 × 10 −2 | 9.12 × 10 −1 | 1.01 × 100 |
| Shikimate | 1.32 × 10 −3 | 5.41 × 10 −4 | 1.66 × 10 −3 | 1.13 × 10 −3 | 1.19 × 10 −3 | 7.67 × 10 −4 |
| Succinate | 3.23 × 10 −1 | 7.42 × 10 −2 | 2.15 × 10 −1 | 6.81 × 10 −2 | 1.97 × 10 −1 | 4.21 × 10 −2 |
| Threonine | 8.39 × 10 −1 | 1.99 × 10 −1 | 2.95 × 10 −1 | 8.09 × 10 −2 | 3.52 × 10 –1 | 2.68 × 10 −1 |
| TMP | 2.71 × 10 −2 | 6.61 × 10 −3 | 2.23 × 10 −2 | 6.80 × 10 −3 | 3.57 × 10 −2 | 2.71 × 10 −3 |
| Tryptophan | 2.19 × 10 −1 | 4.26 × 10 −2 | 6.95 × 10 −2 | 7.88 × 10 −3 | 8.85 × 10 −2 | 1.18 × 10 −2 |
| Tyrosine | 3.04 × 100 | 6.83 × 10 −1 | 6.09 × 10 −1 | 6.31 × 10 −2 | 5.62 × 10 −1 | 2.60 × 10 −1 |
| UDP-Glc | 2.24 × 100 | 2.22 × 10 −1 | 6.98 × 10 −1 | 7.48 × 10 −2 | 1.05 × 100 | 1.97 × 10 −1 |
| UMP | 7.44 × 10 −1 | 1.52 × 10 −1 | 6.05 × 10 −1 | 2.43 × 10 −1 | 7.79 × 10 −1 | 1.58 × 10 −1 |
| Uridine | 2.16 × 10 −2 | 9.04 × 10 −3 | 5.21 × 10 −2 | 3.01 × 10 −2 | 8.93 × 10 −2 | 2.92 × 10 −2 |
| UTP | 5.54 × 10 −1 | 1.49 × 10 −1 | 3.19 × 10 −1 | 4.17 × 10 −2 | 1.75 × 10 −1 | 4.96 × 10 −2 |
| Valine | 9.81 × 10 −1 | 2.00 × 10 −1 | 2.09 × 10 −1 | 3.94 × 10 −2 | 3.60 × 10 −1 | 2.13 × 10 −1 |
| XMP | 4.73 × 10 −2 | 1.37 × 10 −2 | 8.66 × 10 −2 | 3.06 × 10 −2 | 1.01 × 10 −1 | 2.73 × 10 −2 |
| Strain | Growth Rate (h−1) 1 | Genetic Modification | Utilizable Organic Substrate |
|---|---|---|---|
| Synechocystis sp. PCC6803 | 0.107 ± 0.003 | [30] | Glucose 2 |
| Synechococcus sp. PCC7002 | 0.165 ± 0.002 | [6] | Glucose, Glycerol |
| Synechococcus elongatus PCC7942 | 0.123 ± 0.002 | [31] | None |
2.3. Characterization of Each Strain by Molar-Based Metabolite Distribution and AEC Values

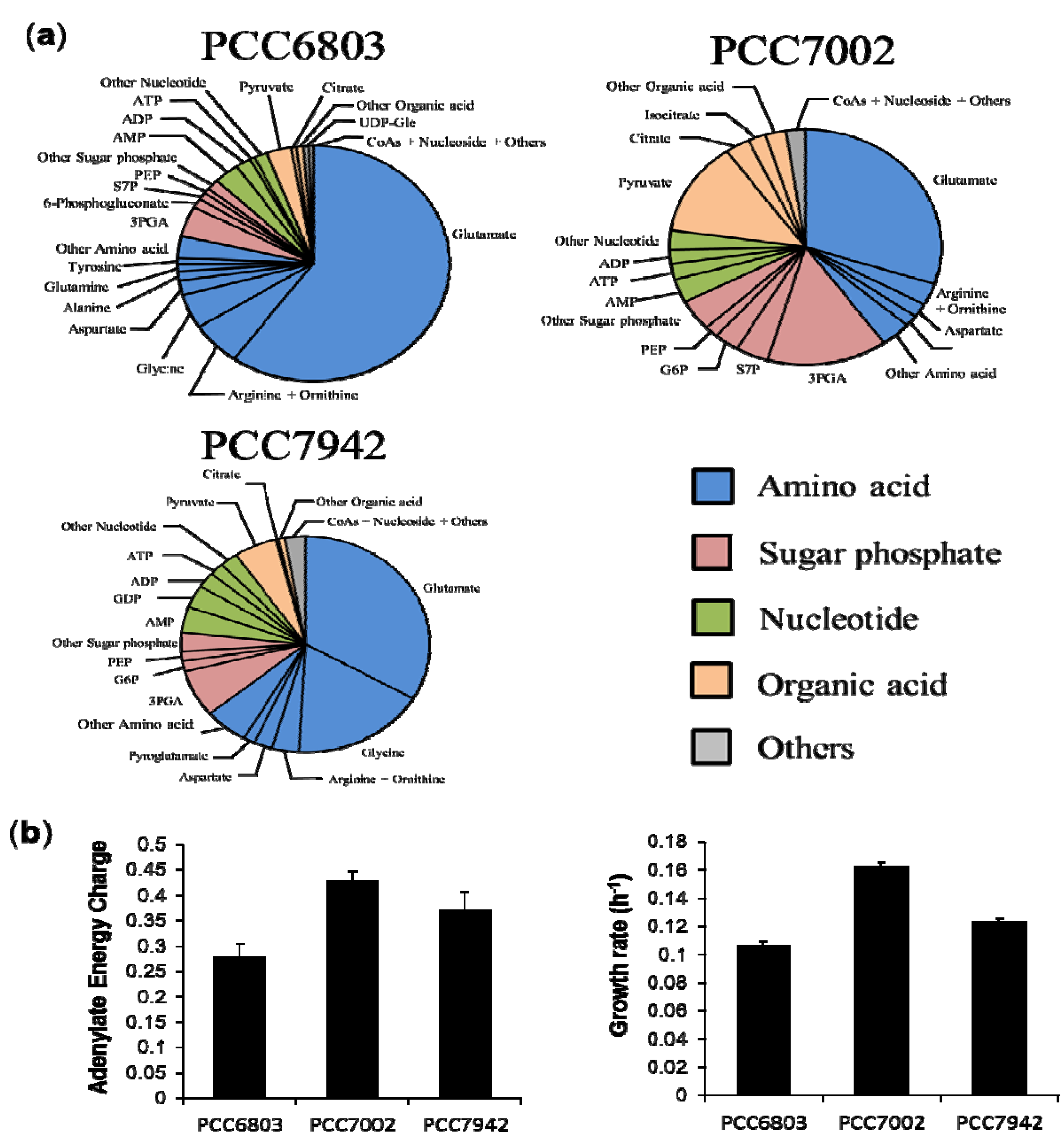
2.4. Overview and Prospect for Cyanobacterial Biological Production
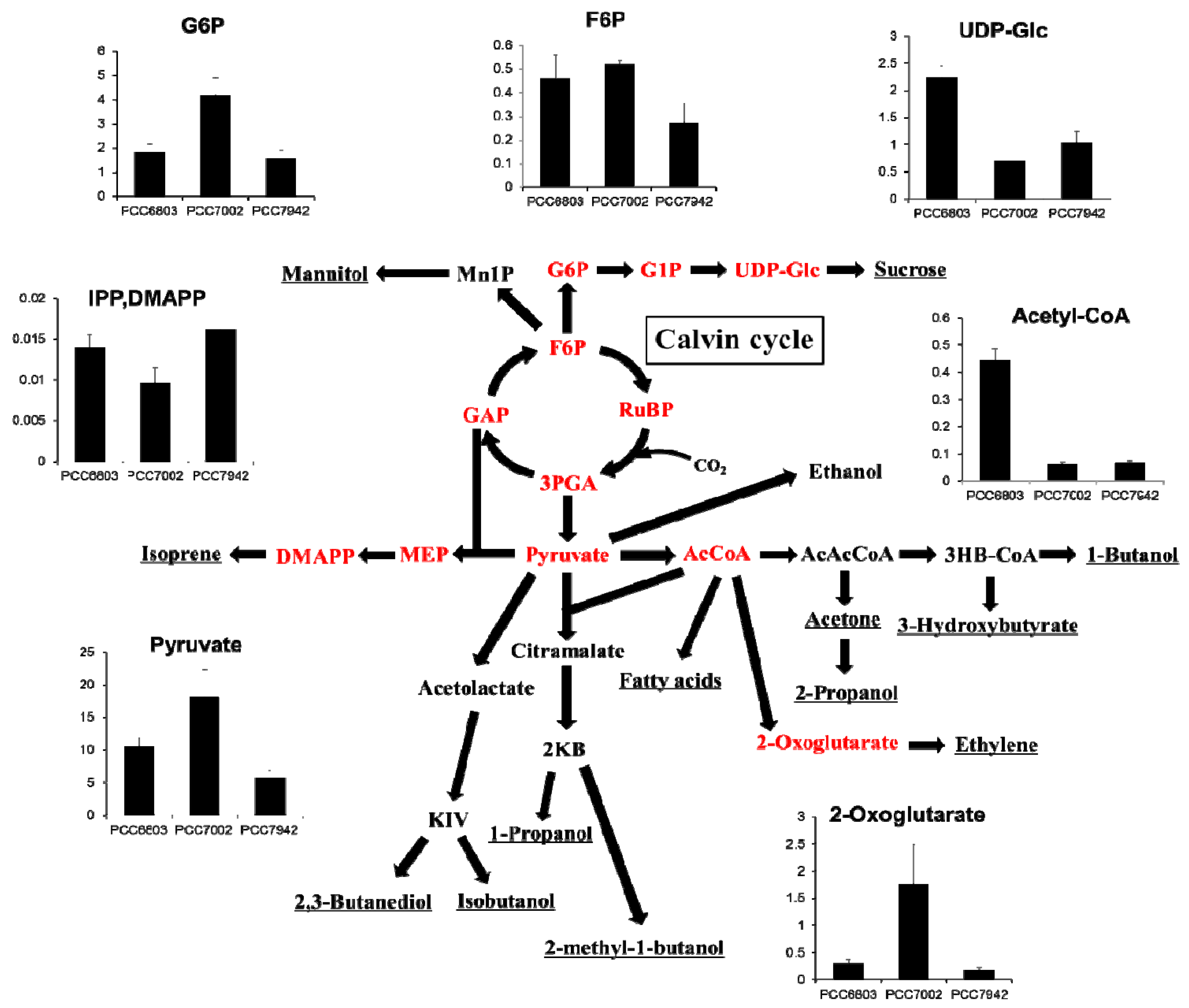
3. Experimental Section
3.1. Strain and Cultivation
3.2. Sampling Procedure for Metabolic Profiling Analysis
3.3. Metabolite Extraction and Sample Preparation
3.4. Preparation of 13C-Internal Standard
3.5. GC/QqQ-MS and IP-LC/QqQ-MS Analysis
3.6. Determine the Absolute Concentration
4. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, X.; Sheng, J.; Curtiss, R. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar] [CrossRef]
- Xu, Y.; Guerra, L.T.; Li, Z.; Ludwig, M.; Dismukes, G.C.; Bryant, D.A. Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC 7002: Cell factories for soluble sugars. Metab. Eng. 2013, 16, 56–67. [Google Scholar] [CrossRef]
- Suzuki, T.; Miyake, M.; Tokiwa, Y.; Saegusa, H.; Saito, T.; Asada, Y. A recombinant cyanobacterium that accumulates poly-(hydroxybutyrate). Biotech. Lett. 1996, 18, 1047–1050. [Google Scholar] [CrossRef]
- Kumaraswamy, G.K.; Guerra, T.; Qian, X.; Zhang, S.; Bryant, D.A.; Dismukes, G.C. Reprogramming the glycolytic pathway for increased hydrogen production in cyanobacteria: Metabolic engineering of NAD+-dependent GAPDH. Energy Environ. Sci. 2013, 6, 3722–3731. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.; Zhang, Y.; Li, Y.; Ma, Y. Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 2012, 14, 394–400. [Google Scholar] [CrossRef]
- Jacobsen, J.H.; Frigaard, N.-U. Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 2014, 21, 60–70. [Google Scholar] [CrossRef]
- Takahama, K.; Matsuoka, M.; Nagahama, K.; Ogawa, T. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng. 2003, 95, 302–305. [Google Scholar] [CrossRef]
- Ungerer, J.; Tao, L.; Davis, M.; Ghirardi, M.; Maness, P.-C.; Yu, J. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ. Sci. 2012, 5, 8998–9006. [Google Scholar] [CrossRef]
- Gao, Z.; Zhao, H.; Li, Z.; Tan, X.; Lu, X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 2012, 5, 9857–9865. [Google Scholar] [CrossRef]
- Lindberg, P.; Park, S.; Melis, A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 2010, 12, 70–79. [Google Scholar] [CrossRef]
- Wang, B.; Pugh, S.; Nielsen, D.R.; Zhang, W.; Meldrum, D.R. Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab. Eng. 2013, 16, 68–77. [Google Scholar] [CrossRef]
- Oliver, J.W.K.; Machado, I.M.P.; Yoneda, H.; Atsumi, S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. USA. 2013, 110, 1249–1254. [Google Scholar] [CrossRef]
- Atsumi, S.; Higashide, W.; Liao, J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009, 27, 1177–1180. [Google Scholar] [CrossRef]
- Kusakabe, T.; Tatsuke, T.; Tsuruno, K.; Hirokawa, Y.; Atsumi, S.; Liao, J.C.; Hanai, T. Engineering a synthetic pathway in cyanobacteria for isopropanol production directly from carbon dioxide and light. Metab. Eng. 2013, 20, 101–108. [Google Scholar] [CrossRef]
- Lan, E.I.; Liao, J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2012, 109, 6018–6023. [Google Scholar] [CrossRef]
- Shen, C.R.; Liao, J.C. Photosynthetic production of 2-methyl-1-butanol from CO2 in cyanobacterium Synechococcus elongatus PCC7942 and characterization of the native acetohydroxyacid synthase. Energy Environ. Sci. 2012, 5, 9574–9583. [Google Scholar] [CrossRef]
- Putri, S.P.; Nakayama, Y.; Matsuda, F.; Uchikata, T.; Kobayashi, S.; Matsubara, A.; Fukusaki, E. Current metabolomics: practical applications. J. Biosci. Bioeng. 2013, 115, 579–589. [Google Scholar] [CrossRef]
- Putri, S.P.; Yamamoto, S.; Tsugawa, H.; Fukusaki, E. Current metabolomics: Technological advances. J. Biosci. Bioeng. 2013, 116, 9–16. [Google Scholar] [CrossRef]
- Bennett, B.D.; Kimball, E.H.; Gao, M.; Osterhout, R.; Van Dien, S.J.; Rabinowitz, J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5, 593–599. [Google Scholar] [CrossRef]
- Trethewey, N.R. Metabolite profiling as an aid to metabolic engineering in plants. Curr. Opin. Plant Biol. 2004, 7, 196–201. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, S.Y.; Cho, J.; Kim, T.Y.; Lee, J.W.; Park, J.H.; Han, M.-J. Global physiological understanding and metabolic engineering of microorganisms based on omics studies. Appl. Microbiol. Biotechnol. 2005, 68, 567–579. [Google Scholar] [CrossRef]
- Tsugawa, H.; Tsujimoto, Y.; Sugitate, K.; Sakui, N.; Nishiumi, S.; Bamba, T.; Fukusaki, E. Highly sensitive and selective analysis of widely targeted metabolomics using gas chromatography/triple-quadrupole mass spectrometry. J. Biosci. Bioeng. 2014, 117, 122–128. [Google Scholar] [CrossRef]
- Kato, H.; Izumi, Y.; Hasunuma, T.; Matsuda, F.; Kondo, A. Widely targeted metabolic profiling analysis of yeast central metabolites. J. Biosci. Bioeng. 2012, 113, 665–673. [Google Scholar]
- Bennett, B.D.; Yuan, J.; Kimball, E.H.; Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008, 3, 1299–1311. [Google Scholar] [CrossRef]
- Ellis, D.I.; Goodacre, R. Metabolomics-assisted synthetic biology. Curr. Opin. Biotechnol. 2012, 23, 22–28. [Google Scholar] [CrossRef]
- Kopf, M.; Klähn, S.; Pade, N.; Weingärtner, C.; Hagemann, M.; Voß, B.; Hess, W.R. Comparative genome analysis of the closely related Synechocystis strains PCC 6714 and PCC 6803. DNA Res. 2014. [Google Scholar] [CrossRef]
- Blank, L.M.; Sauer, U. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology 2004, 150, 1085–1093. [Google Scholar] [CrossRef]
- Steinhauser, D.; Fernie, A.R.; Araújo, W.L. Unusual cyanobacterial TCA cycles: Not broken just different. Trends Plant Sci. 2012, 17, 503–509. [Google Scholar] [CrossRef]
- Zhang, S.; Bryant, D.A. The tricarboxylic acid cycle in cyanobacteria. Science. 2011, 334, 1551–1553. [Google Scholar] [CrossRef]
- Grigorieva, G.; Shestakov, S. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol. Lett. 1982, 13, 367–370. [Google Scholar] [CrossRef]
- Golden, S.; Brusslan, J.; Haselkom, R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987, 153, 215–231. [Google Scholar] [CrossRef]
- Li, H.; Sherman, D.M.; Bao, S.; Sherman, L.A. Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 2001, 176, 9–18. [Google Scholar] [CrossRef]
- Sakamoto, T.; Bryant, D.A. Growth at low temperature causes nitrogen limitation in the cyanobacterium Synechococcus sp. PCC7002. Arch. Microbiol. 1998, 169, 10–19. [Google Scholar] [CrossRef]
- Atkinson, D.E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 1968, 7, 4030–4034. [Google Scholar]
- Hardie, D.G.; Hawley, S.A. AMP-activated protein kinase: The energy charge hypothesis revisited. BioEssays 2001, 23, 1112–1119. [Google Scholar] [CrossRef]
- Chapman, A.G.; Fall, L.; Atkinson, D.E. Adenylate energy charge in Escherichia coli during growth and starvation. J. Bacteriol. 1971, 108, 1072–1086. [Google Scholar]
- Streusand, V.J.; Portis, A.R. Rubisco activase mediates ATP-dependent activation of ribulose bisphosphate carboxylase. Plant Physiol. 1987, 85, 152–154. [Google Scholar] [CrossRef]
- Li, X.; Hu, H.; Zhang, Y. Growth and lipid accumulation properties of a freshwater microalga Scenedesmus sp. under different cultivation temperature. Bioresour. Technol. 2011, 102, 3098–3102. [Google Scholar] [CrossRef]
- Pettit, F.H.; Pelley, J.W.; Reed, L.J. Regulation of pyruvate dehydrogenase kinase and phosphatase by acetyl-CoA/CoA and NADH/NAD ratios. Biochem. Biophys. Res. Commun. 1975, 65, 575–582. [Google Scholar] [CrossRef]
- Bogorad, I.W.; Lin, T.-S.; Liao, J.C. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 2013, 502, 693–697. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 110, 1–61. [Google Scholar] [CrossRef]
- Hasunuma, T.; Kikuyama, F.; Matsuda, M.; Aikawa, S.; Izumi, Y.; Kondo, A. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J. Exp. Bot. 2013, 64, 2943–2954. [Google Scholar]
- Lu, W.; Clasquin, M.F.; Melamud, E.; Amador-Noguez, D.; Caudy, A.A.; Rabinowitz, J.D. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 2010, 82, 3212–3221. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dempo, Y.; Ohta, E.; Nakayama, Y.; Bamba, T.; Fukusaki, E. Molar-Based Targeted Metabolic Profiling of Cyanobacterial Strains with Potential for Biological Production. Metabolites 2014, 4, 499-516. https://doi.org/10.3390/metabo4020499
Dempo Y, Ohta E, Nakayama Y, Bamba T, Fukusaki E. Molar-Based Targeted Metabolic Profiling of Cyanobacterial Strains with Potential for Biological Production. Metabolites. 2014; 4(2):499-516. https://doi.org/10.3390/metabo4020499
Chicago/Turabian StyleDempo, Yudai, Erika Ohta, Yasumune Nakayama, Takeshi Bamba, and Eiichiro Fukusaki. 2014. "Molar-Based Targeted Metabolic Profiling of Cyanobacterial Strains with Potential for Biological Production" Metabolites 4, no. 2: 499-516. https://doi.org/10.3390/metabo4020499
APA StyleDempo, Y., Ohta, E., Nakayama, Y., Bamba, T., & Fukusaki, E. (2014). Molar-Based Targeted Metabolic Profiling of Cyanobacterial Strains with Potential for Biological Production. Metabolites, 4(2), 499-516. https://doi.org/10.3390/metabo4020499





