A Major Update and Improved Validation Functionality in the mwtab Python Library and the Metabolomics Workbench File Status Website
Abstract
1. Introduction
2. Materials and Methods
2.1. Updates to the mwTab Format
2.2. mwtab Package Implementation
2.3. mwFileStatusWebsite Updates
2.4. Evaluation of the Metabolomics Workbench Repository
3. Results
3.1. Analysis IDs with Files Missing from the Metabolomics Workbench
3.2. Analysis Files Which Could Not Be Parsed
3.3. Comparing Parsability Between mwtab Version 1.2.5 and Version 2.0.0
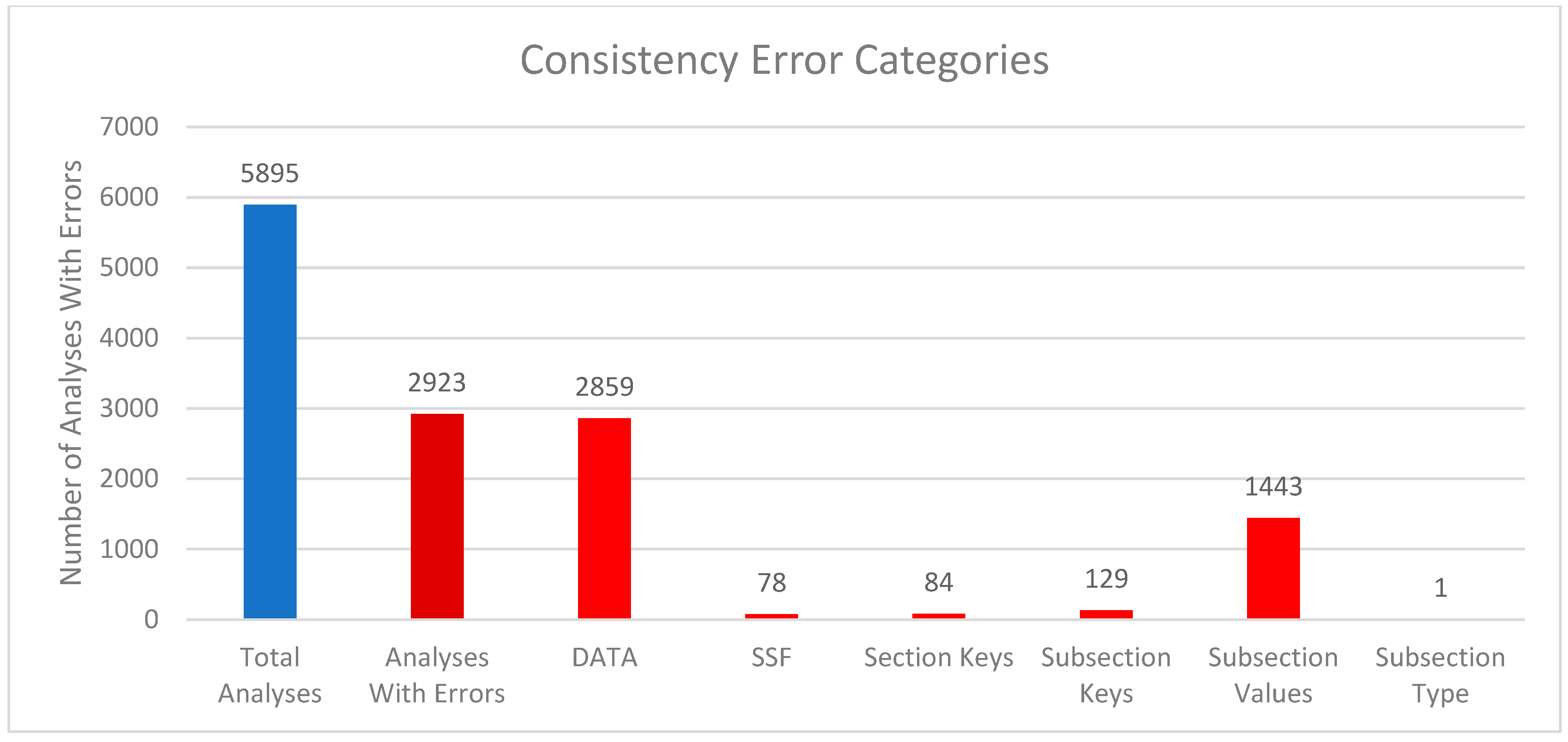
3.4. Consistency Errors Between mwTab- and JSON-Formatted Files
3.5. Validation Issues
3.6. Conversions and Roundtripping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MW | Metabolomics Workbench |
| MS | Mass Spectroscopy |
| NMR | Nuclear Magnetic Resonance |
| FAIR | Findable Accessible Interoperable Reusable |
| BSD | Berkeley Software Distribution |
| REST | Representational State Transfer |
| API | Application Programming Interface |
| CLI | Command Line Interface |
| JSON | JavaScript Object Notation |
| CSS | Cascading Style Sheets |
| PYPI | Python Package Index |
References
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
- Bray, T. The JavaScript Object Notation (JSON) Data Interchange Format; RFC Editor: Marina Del Rey, CA, USA, 2017. [Google Scholar]
- Crockford, D. The Application/Json Media Type for JavaScript Object Notation (JSON); RFC Editor: Marina Del Rey, CA, USA, 2006. [Google Scholar]
- Fielding, R.T. Architectural Styles and the Design of Network-based Software Architectures. Ph.D. Thesis, University of California, Irvine, CA, USA, 2000. [Google Scholar]
- Howell, A.; Yaros, C. Downloading and Analysis of Metabolomic and Lipidomic Data from Metabolomics Workbench Using MetaboAnalyst 5.0. In Lipidomics: Methods and Protocols; Springer: New York, NY, USA, 2023; pp. 313–321. [Google Scholar]
- Fahy, E.; Subramaniam, S. RefMet: A reference nomenclature for metabolomics. Nat. Methods 2020, 17, 1173–1174. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.T.; Moseley, H.N. MESSES: Software for transforming messy research datasets into clean submissions to metabolomics workbench for public sharing. Metabolites 2023, 13, 842. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Salek, R.M.; Steinbeck, C. Global open data management in metabolomics. Curr. Opin. Chem. Biol. 2017, 36, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Smelter, A.; Moseley, H.N.B. A Python library for FAIRer access and deposition to the Metabolomics Workbench Data Repository. Metabolomics 2018, 14, 64. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.D.; Moseley, H.N.B. The mwtab Python Library for RESTful Access and Enhanced Quality Control, Deposition, and Curation of the Metabolomics Workbench Data Repository. Metabolites 2021, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Boeckhout, M.; Zielhuis, G.A.; Bredenoord, A.L. The FAIR guiding principles for data stewardship: Fair enough? Eur. J. Hum. Genet. 2018, 26, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.D.; Moseley, H.N.B. The metabolomics workbench file status website: A metadata repository promoting FAIR principles of metabolomics data. BMC Bioinform. 2023, 24, 299. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E. mwTab File Format Specification. Available online: https://www.metabolomicsworkbench.org/data/mwTab_specification.pdf (accessed on 17 December 2025).
- Droettboom, M. Understanding JSON Schema. Available online: https://json-schema.org/UnderstandingJSONSchema.pdf (accessed on 17 December 2025).
- Pezoa, F.; Reutter, J.L.; Suarez, F.; Ugarte, M.; Vrgoč, D. Foundations of JSON schema. In Proceedings of the 25th International Conference on World Wide Web, Montreal, QC, Canada, 11–15 April 2016; International World Wide Web Conferences Steering Committee: Geneva, Switzerland, 2016; pp. 263–273. [Google Scholar]
- Writing Your Pyproject.toml. Available online: https://packaging.python.org/en/latest/guides/writing-pyproject-toml/ (accessed on 17 December 2025).
- Setuptools-scm. Available online: https://pypi.org/project/setuptools-scm/ (accessed on 17 December 2025).
- Huckvale, E.D.; Thompson, P.T.; Flight, R.M.; Moseley, H.N.B. High-Quality Predicted Pathway Annotations Greatly Improve Pathway Enrichment Analysis of Metabolomics Datasets. bioRxiv 2025. [Google Scholar] [CrossRef]
- El Abiead, Y.; Strobel, M.; Payne, T.; Fahy, E.; O’Donovan, C.; Subramamiam, S.; Vizcaíno, J.A.; Yurekten, O.; Deleray, V.; Zuffa, S. Enabling pan-repository reanalysis for big data science of public metabolomics data. Nat. Commun. 2025, 16, 4838. [Google Scholar] [CrossRef] [PubMed]


| Modules | Changes |
|---|---|
| cli | Refactored for DRY (Do not Repeat Yourself) purposes. Added error recovery for many commands. |
| converter | Added error recovery when converting multiple files. ixed write-out errors due to non-ASCII characters. |
| fileio | Refactored for DRY purposes. dded error recovery when reading and writing multiple files. ixed write-out errors due to non-ASCII characters. ile paths that do not exist will be created instead of raising an error. |
| mwextract | Added error recovery when operating on multiple files. ile paths that do not exist will be created instead of raising an error. |
| mwrest | Added error recovery when operating on multiple files. |
| mwschema | Completely refactored to use the jsonschema package instead of the schema package. dded more validations, particularly on the values of some attributes. |
| tokenizer | Improved parsing, making some formerly unreadable files readable. |
| mwtab | Fixed round-trip errors by adding support for duplicate keys. ey/block order is now always the same when writing out. dded convenience properties for attributes in the METABOLOMICS WORKBENCH header. dded convenience methods to convert the tabular sections of the mwTab format to pandas DataFrames and vice versa. mproved file parsing by taking into account some common errors in the mwTab files. |
| validator | Added many new validations. mproved issue printing by standardizing formatting and greatly reducing the printing of spurious issues. dded a new JSON output with all validation messages tagged with ‘format’, ‘value’, or ‘consistency’. |
| duplicates_dict | A new module that contains the DuplicatesDict class used to support reading and writing duplicate keys. |
| metadata_column_matching | A new module for matching metadata columns by names and values. ontains regular expressions, functions, and classes used for matching metadata columns. ontains the “column_finders” dictionary of prebuilt ColumnFinder objects used to match the most common metadata columns in the MW repository. |
| File Format | Analysis ID |
|---|---|
| mwTab | AN002312, AN005082, AN005098, AN005099, AN005557, AN006051, AN006148, AN006593 |
| JSON | AN005098, AN005099, AN005557, AN006051, AN006148, AN006593 |
| Issue Category | mwTab | JSON |
|---|---|---|
| Passing | 5497 | 4907 |
| Missing | 8 | 6 |
| Parsing Error | 162 | 69 |
| Validation Bug | 10 | 24 |
| Errors | 456 | 1125 |
| Issue Category | mwTab | JSON |
|---|---|---|
| Passing | 16 | 23 |
| Missing | 8 | 6 |
| Parsing Error | 51 | 124 |
| Validation Bug | 0 | 0 |
| Warnings Only | 17 | 21 |
| Errors | 6033 | 5951 |
| Consistency Errors | 538 | 457 |
| Value Errors | 5328 | 5198 |
| Format Errors | 3660 | 3405 |
| Only New Errors | 2634 | 2548 |
| Only New Errors and Warnings | 5717 | 5640 |
| Previous Standard Name Errors | 2926 | 2883 |
| Issue Category | Description |
|---|---|
| Parsing Error | File could not be parsed. |
| Validation Bug | A coding error was encountered during validation. The file did not necessarily have something wrong with it. |
| Passing | The file had no validation issues. |
| Missing | The file could not be downloaded. |
| Warnings Only | All of the validation issues were only warnings. |
| Errors | There was at least 1 validation issue with the file that was not a warning. |
| Consistency Errors | There was at least 1 validation issue due to inconsistencies between data in the file. |
| Value Errors | There was at least 1 validation issue due to incorrect value(s) in the file. |
| Format Errors | There was at least 1 validation issue due to the file not meeting the mwTab specification. |
| Only New Errors | Every issue found is one that has been added in this latest version of the mwtab package. |
| Only New Errors and Warnings | Every issue found is one that has been added in this latest version of the mwtab package or is a warning. |
| Previous Standard Name Errors | If the previous version of the mwtab package had validated while using its code for standard name matching, then the file would have errored instead of passing. |
| ID | Validation Short Name | mwTab | JSON |
|---|---|---|---|
| 1 | Duplicate Sub-section | 30 | 0 |
| 2 | Bad Headers | 44 | 0 |
| 3 | Factor Mismatch | 27 | 0 |
| 4 | Duplicate Sample ID in SSF | 6 | 6 |
| 5 | Duplicate Factors in SSF | 29 | 28 |
| 6 | Duplicate Additional Data | 346 | 341 |
| 7 | Missing Sample ID(s) in SSF | 45 | 75 |
| 8 | Duplicate Samples in DATA | 67 | 0 |
| 9 | Metabolite(s) in DATA not METABOLITES | 390 | 363 |
| 10 | Blank Metabolite(s) in DATA | 4 | 1 |
| 11 | Duplicate Metabolite(s) in DATA | 24 | 22 |
| 12 | Metabolite(s) in METABOLITES not DATA | 420 | 371 |
| 13 | Blank Metabolite(s) in METABOLITES | 1 | 1 |
| 14 | Duplicate Metabolite(s) in METABOLITES | 30 | 29 |
| 15 | Standard Column Name Match | 3800 | 3740 |
| 16 | METABOLITES Bad Standard Values | 658 | 628 |
| 17 | Missing Implied Column | 772 | 764 |
| 18 | Paired Columns Value Mismatch | 472 | 457 |
| 19 | “other_id” Column | 1263 | 1221 |
| 20 | Multiple Standard Name Match | 33 | 32 |
| 21 | Missing “sample_id” in EXTENDED | 0 | 0 |
| 22 | Missing Sample ID(s) in EXTENDED | 3 | 0 |
| 23 | Bad Metabolite Name | 4 | 0 |
| 24 | JSON Schema Error | 6032 | 5947 |
| 25 | Inconsistent Columns | 0 | 0 |
| 26 | Column With No Name | 55 | 526 |
| 27 | Null Column | 1388 | 1398 |
| 28 | Possible Bad Column Values | 504 | 458 |
| 29 | Duplicate Rows | 2 | 0 |
| 30 | Duplicate Column Names | 18 | 0 |
| 31 | Multiple Polarities | 11 | 11 |
| 32 | No MS or NM Section | 2 | 0 |
| 33 | Missing METABOLITES Section | 0 | 0 |
| 34 | Missing Header | 54 | 629 |
| 35 | Blank Sample ID(s) in EXTENDED | 3 | 0 |
| 36 | Blank Metabolite(s) in EXTENDED | 0 | 0 |
| ID | Validation Short Name | Description |
|---|---|---|
| 1 | Duplicate Sub-section | The same sub-section appears twice in a section. For example, COLLECTION_SUMMARY shows up twice in COLLECTION. Will only be detected in mwTab-formatted files and not JSON, since duplicate keys tend to overwrite in JSON. |
| 2 | Bad Headers | A table section has a mismatch between the number of columns in the data rows and the header row. Can only be found in mwTab-formatted files because tables are lists of dictionaries in JSON, so there is no header line. Each dictionary repeats the column names. |
| 3 | Factor Mismatch | The factors in the METABOLITE_DATA section and SUBJECT_SAMPLE_FACOTRS do not match. Can only be found in mwTab-formatted files since the JSON version only has the SUBJECT_SAMPLE_FACTORS section. |
| 4 | Duplicate Sample ID in SSF | There are at least two Sample IDs with the same name in the SUBJECT_SAMPLE_FACTORS section. |
| 5 | Duplicate Factors in SSF | There are at least two factors with the same name in the SUBJECT_SAMPLE_FACTORS section. |
| 6 | Duplicate Additional Data | There are at least two keys in the Additional sample data with the same name in the SUBJECT_SAMPLE_FACTORS section. |
| 7 | Missing Sample ID(s) in SSF | There is at least one Sample ID in the METABOLITE_DATA that is not in the SUBJECT_SAMPLE_FACTORS. |
| 8 | Duplicate Samples in DATA | There are at least two Sample IDs with the same name in the METABOLITE_DATA section. |
| 9 | Metabolite(s) in DATA not METABOLITES | There is at least one Metabolite in the METABOLITE_DATA section that is not in the METABOLITES section. |
| 10 | Blank Metabolite(s) in DATA | There is at least one Metabolite in the METABOLITE_DATA section that is blank or null valued. |
| 11 | Duplicate Metabolite(s) in DATA | There are at least two Metabolites in the METABOLITE_DATA section with the same name. |
| 12 | Metabolite(s) in METABOLITES, not DATA | There is at least one Metabolite in the METABOLITES section that is not in the METABOLITE_DATA section. |
| 13 | Blank Metabolite(s) in METABOLITES | There is at least one Metabolite in the METABOLITES section that is blank or null valued. |
| 14 | Duplicate Metabolite(s) in METABOLITES | There are at least two Metabolites in the METABOLITES section with the same name. |
| 15 | Standard Column Name Match | There is at least one column in the METABOLITES section with a name that matches a standard name. |
| 16 | METABOLITES Bad Standard Values | There is at least one column in the METABOLITES section with a name that matches a standard name, and at least one value in that column does not match what values are supposed to look like for that column. For example, a KEGG_ID column has a value that does not look like a KEGG ID. |
| 17 | Missing Implied Column | There is at least one column in the METABOLITES section that matches a column that should be paired with another column, but that column is not present. For example, a “retention_index” column should also have a “retention_index_type” column. |
| 18 | Paired Columns Value Mismatch | There is at least one column in the METABOLITES section that matches a column that should be paired with another column, but the values in each column do not align. For example, if there is a value in row 5 of a “retention_index” column, then there should also be a value in row 5 for the “retention_index_type” column. |
| 19 | “other_id” Column | There is at least one column in the METABOLITES section that matches the column name “other_id”. In general, this column should be avoided and the IDs given a more specific name, such as “kegg_id” or “lab_id”. |
| 20 | Multiple Standard Name Match | There is at least one column in the METABOLITES section that matches more than one standard name. Columns like this usually pull double duty and should be separated into multiple columns. |
| 21 | Missing “sample_id” in EXTENDED | The EXTENDED_DATA section is missing the required “sample_id” column. |
| 22 | Missing Sample ID(s) in EXTENDED | The EXTENDED_DATA section does not have entries for some Sample IDs in the SUBJECT_SAMPLE_FACTORS. |
| 23 | Bad Metabolite Name | There is at least one Metabolite name in one of the tables that is close to a name like “metabolite_name” or other names that are likely a mistake and are actually part of a header. |
| 24 | JSON Schema Error | One of the many possible validations performed through JSON Schema. Usually, a required subsection is missing or blank, or a value in a subsection does not look right. |
| 25 | Inconsistent Columns | One of the tables has at least one row with different columns from another one. Since tables are stored as lists of dictionaries in JSON and the internal MWTabFile object, there is the possibility of a dictionary missing or having extra columns compared to other dictionaries in the same list. |
| 26 | Column With No Name | There is at least one column in a table section with no name. |
| 27 | Null Column | There is at least one column in a table section where every value is a null value. |
| 28 | Possible Bad Column Values | There is at least one column in a table section where 90% of the values are the same, but 10% are different. This could be intentional, but many times it is a mistake. |
| 29 | Duplicate Rows | There are at least two rows in a table section that are exactly the same. |
| 30 | Duplicate Column Names | There are at least two columns in a table section with the same name. |
| 31 | Multiple Polarities | A column in the METABOLITES table was matched to be a “polarity” column, and that column has more than 1 value. It should only ever be positive or negative and not mixed because each analysis is supposed to be one mass spec run. A different mass spec polarity should be in its own analysis. |
| 32 | No MS or NM Section | The file does not have an “MS” or “NM” section. At least one or the other is required. |
| 33 | Missing METABOLITES Section | The file has a METABOLITE_DATA section but does not have a METABOLITES section. |
| 34 | Missing Header | A table is missing a required column, or it has the wrong name. For example, the METABOLITES and METABOLITE_DATA tables require a “Metabolite” column. |
| 35 | Blank Sample ID(s) in EXTENDED | The EXTENDED_DATA section has at least one Sample ID that has a blank or null value. |
| 36 | Blank Metabolite(s) in EXTENDED | The EXTENDED_DATA section has at least one Metabolite that has a blank or null value. |
| Section | mwTab | JSON |
|---|---|---|
| CHROMATOGRAPHY | 5679 | 5621 |
| MS_METABOLITE_DATA | 3770 | 4198 |
| MS | 1757 | 1724 |
| SUBJECT | 1730 | 1695 |
| TREATMENT | 1241 | 1225 |
| no section | 895 | 139 |
| COLLECTION | 806 | 805 |
| SAMPLEPREP | 704 | 695 |
| SUBJECT_SAMPLE_FACTORS | 457 | 471 |
| STUDY | 412 | 411 |
| METABOLOMICS WORKBENCH | 303 | 5 |
| PROJECT | 288 | 287 |
| NM | 223 | 233 |
| NMR_METABOLITE_DATA | 122 | 150 |
| ANALYSIS | 82 | 51 |
| NMR_BINNED_DATA | 3 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Thompson, P.T.; Moseley, H.N.B. A Major Update and Improved Validation Functionality in the mwtab Python Library and the Metabolomics Workbench File Status Website. Metabolites 2026, 16, 76. https://doi.org/10.3390/metabo16010076
Thompson PT, Moseley HNB. A Major Update and Improved Validation Functionality in the mwtab Python Library and the Metabolomics Workbench File Status Website. Metabolites. 2026; 16(1):76. https://doi.org/10.3390/metabo16010076
Chicago/Turabian StyleThompson, P. Travis, and Hunter N. B. Moseley. 2026. "A Major Update and Improved Validation Functionality in the mwtab Python Library and the Metabolomics Workbench File Status Website" Metabolites 16, no. 1: 76. https://doi.org/10.3390/metabo16010076
APA StyleThompson, P. T., & Moseley, H. N. B. (2026). A Major Update and Improved Validation Functionality in the mwtab Python Library and the Metabolomics Workbench File Status Website. Metabolites, 16(1), 76. https://doi.org/10.3390/metabo16010076






