Enzymatic Evolution and Longitudinal Recovery in Biotinidase Deficiency: Genotypic and Clinical Insights from the Follow-Up of a Newborn-Screened Cohort in Emilia-Romagna, Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening Protocol and Diagnostic Confirmation Methods
2.2. Statistical Analysis
3. Results
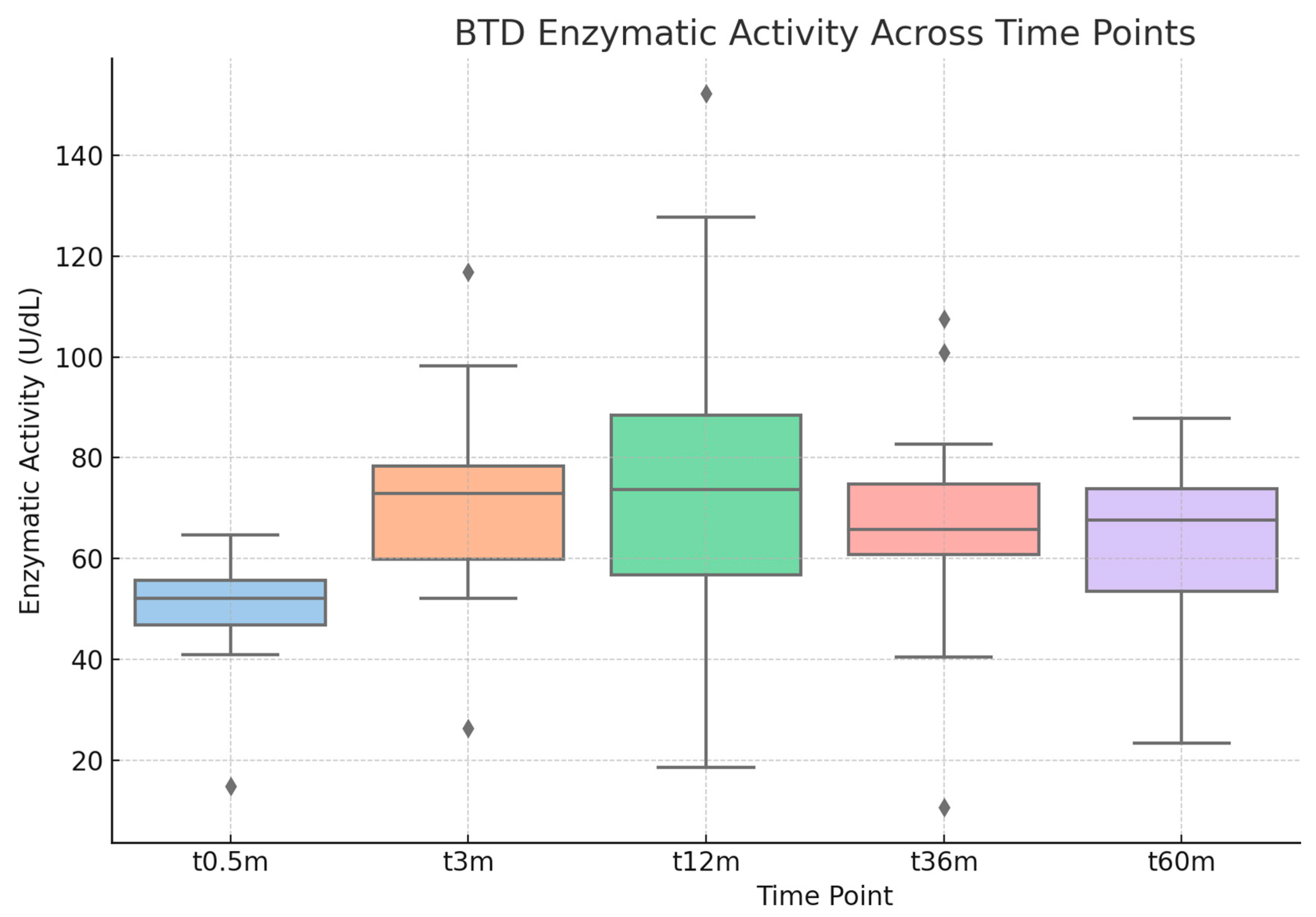
Follow-Up
- Red group: Patients whose BTD enzymatic activity remained persistently below 30% at all time points. These individuals showed no evidence of biochemical recovery during follow-up.
- Yellow group: Patients with fluctuating enzymatic activity, characterized by intermittent values above and below the 30% threshold. The timing of the first observed increase above 30% is documented in the column “Age at BTD Enzyme Activity Recovery.”
- Green group: Patients whose enzymatic activity increased above 30% and remained consistently above this threshold from a given follow-up point onward, indicating a sustained biochemical recovery.
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BD | Biotinidase deficiency |
| BTD | Biotinidase |
| NBS | Newborn screening |
| AER | Residual enzymatic activity |
| DBS | Dried blood spots |
| VUS | Variant of uncertain significance |
| ACMG | American College of Medical Genetics and Genomics |
| HGMD | Human Genetic Mutation Database |
| WT | Wild Type |
| RCTs | Randomized controlled trials |
References
- Wolf, B.; Hsia, Y.E.; Sweetman, L.; Feldman, G.; Boychuk, R.B.; Bart, R.D.; Crowell, D.H.; Di Mauro, R.M.; Nyhan, W.L. Multiple Carboxylase Deficiency: Clinical and Biochemical Improvement Following Neonatal Biotin Treatment. Pediatrics 1981, 68, 113–118. [Google Scholar] [CrossRef]
- Wolf, B.; Heard, G.S.; Jefferson, L.G.; Proud, V.K.; Nance, W.E.; Weissbecker, K.A. Clinical Findings in Four Children with Biotinidase Deficiency Detected through a Statewide Neonatal Screening Program. N. Engl. J. Med. 1985, 313, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Cowan, T.M.; Blitzer, M.G.; Wolf, B. Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee Technical Standards and Guidelines for the Diagnosis of Biotinidase Deficiency. Genet. Med. 2010, 12, 464–470. [Google Scholar] [CrossRef]
- Wolf, B.; Heard, G.S.; McVoy, J.R.; Raetz, H.M. Biotinidase Deficiency: The Possible Role of Biotinidase in the Processing of Dietary Protein-Bound Biotin. J. Inherit. Metab. Dis. 1984, 7 (Suppl. 2), 121–122. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase Deficiency: “If You Have to Have an Inherited Metabolic Disease, This Is the One to Have”. Genet. Med. 2012, 14, 565–575. [Google Scholar] [CrossRef]
- Strovel, E.T.; Cowan, T.M.; Scott, A.I.; Wolf, B. Laboratory Diagnosis of Biotinidase Deficiency, 2017 Update: A Technical Standard and Guideline of the American College of Medical Genetics and Genomics. Genet. Med. 2017, 19, 1–10. [Google Scholar] [CrossRef]
- Canda, E.; Kalkan Uçar, S.; Çoker, M. Biotinidase Deficiency: Prevalence, Impact and Management Strategies. Pediatr. Health Med. Ther. 2020, 11, 127–133. [Google Scholar] [CrossRef]
- Wolf, B. Clinical Issues and Frequent Questions about Biotinidase Deficiency. Mol. Genet. Metab. 2010, 100, 6–13. [Google Scholar] [CrossRef]
- Bracci, B.; Mala, D.; Pavanello, E.; Sauro, P.; Mussa, A.; Guaraldo, V.; Spada, M. Biotinidase Deficiency: Outcomes of 37 Years-Experience of Newborn Screening in Turin, Italy. JIM 2024, 1, e458. [Google Scholar]
- Funghini, S.; Tonin, R.; Malvagia, S.; Caciotti, A.; Donati, M.A.; Morrone, A.; la Marca, G. High Frequency of Biotinidase Deficiency in Italian Population Identified by Newborn Screening. Mol. Genet. Metab. Rep. 2020, 25, 100689. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase Deficiency. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Tankeu, A.T.; Van Winckel, G.; Elmers, J.; Jaccard, E.; Superti-Furga, A.; Wolf, B.; Tran, C. Biotinidase Deficiency: What Have We Learned in Forty Years? Mol. Genet. Metab. 2023, 138, 107560. [Google Scholar] [CrossRef]
- Gannavarapu, S.; Prasad, C.; DiRaimo, J.; Napier, M.; Goobie, S.; Potter, M.; Chakraborty, P.; Karaceper, M.; Munoz, T.; Schulze, A.; et al. Biotinidase Deficiency: Spectrum of Molecular, Enzymatic and Clinical Information from Newborn Screening Ontario, Canada (2007–2014). Mol. Genet. Metab. 2015, 116, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B. Why Screen Newborns for Profound and Partial Biotinidase Deficiency? Mol. Genet. Metab. 2015, 114, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Forny, P.; Wicht, A.; Rüfenacht, V.; Cremonesi, A.; Häberle, J. Recovery of Enzyme Activity in Biotinidase Deficient Individuals during Early Childhood. J. Inherit. Metab. Dis. 2022, 45, 605–620. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The Human Genomic Variant Search Engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Fokkema, I.F.A.C.; Kroon, M.; López Hernández, J.A.; Asscheman, D.; Lugtenburg, I.; Hoogenboom, J.; den Dunnen, J.T. The LOVD3 Platform: Efficient Genome-Wide Sharing of Genetic Variants. Eur. J. Hum. Genet. 2021, 29, 1796–1803. [Google Scholar] [CrossRef]
- Norrgard, K.J.; Pomponio, R.J.; Swango, K.L.; Hymes, J.; Reynolds, T.; Buck, G.A.; Wolf, B. Double Mutation (A171T and D444H) Is a Common Cause of Profound Biotinidase Deficiency in Children Ascertained by Newborn Screening the the United States. Mutations in Brief No. 128. Online. Hum. Mutat. 1998, 11, 410. [Google Scholar] [CrossRef]
- Pomponio, R.J.; Hymes, J.; Reynolds, T.R.; Meyers, G.A.; Fleischhauer, K.; Buck, G.A.; Wolf, B. Mutations in the Human Biotinidase Gene That Cause Profound Biotinidase Deficiency in Symptomatic Children: Molecular, Biochemical, and Clinical Analysis. Pediatr. Res. 1997, 42, 840–848. [Google Scholar] [CrossRef][Green Version]
- Pomponio, R.J.; Reynolds, T.R.; Mandel, H.; Admoni, O.; Melone, P.D.; Buck, G.A.; Wolf, B. Profound Biotinidase Deficiency Caused by a Point Mutation That Creates a Downstream Cryptic 3′ Splice Acceptor Site within an Exon of the Human Biotinidase Gene. Hum. Mol. Genet. 1997, 6, 739–745. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mühl, A.; Möslinger, D.; Item, C.B.; Stöckler-Ipsiroglu, S. Molecular Characterisation of 34 Patients with Biotinidase Deficiency Ascertained by Newborn Screening and Family Investigation. Eur. J. Hum. Genet. 2001, 9, 237–243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wolf, B.; Jensen, K.P.; Barshop, B.; Blitzer, M.; Carlson, M.; Goudie, D.R.; Gokcay, G.H.; Demirkol, M.; Baykal, T.; Demir, F.; et al. Biotinidase Deficiency: Novel Mutations and Their Biochemical and Clinical Correlates. Hum. Mutat. 2005, 25, 413. [Google Scholar] [CrossRef] [PubMed]
- Norrgard, K.J.; Pomponio, R.J.; Hymes, J.; Wolf, B. Mutations Causing Profound Biotinidase Deficiency in Children Ascertained by Newborn Screening in the United States Occur at Different Frequencies than in Symptomatic Children. Pediatr. Res. 1999, 46, 20–27. [Google Scholar] [CrossRef]
- Yılmaz, B.; Ceylan, A.C.; Gündüz, M.; Ünal Uzun, Ö.; Küçükcongar Yavaş, A.; Bilginer Gürbüz, B.; Öncül, Ü.; Güleç Ceylan, G.; Kasapkara, Ç.S. Evaluation of Clinical, Laboratory, and Molecular Genetic Features of Patients with Biotinidase Deficiency. Eur. J. Pediatr. 2024, 183, 1341–1351. [Google Scholar] [CrossRef]
- Sharma, R.; Kucera, C.R.; Nery, C.R.; Lacbawan, F.L.; Salazar, D.; Tanpaiboon, P. Biotinidase Biochemical and Molecular Analyses: Experience at a Large Reference Laboratory. Pediatr. Int. 2024, 66, e15726. [Google Scholar] [CrossRef]
- Swango, K.L.; Demirkol, M.; Hüner, G.; Pronicka, E.; Sykut-Cegielska, J.; Schulze, A.; Mayatepek, E.; Wolf, B. Partial Biotinidase Deficiency Is Usually Due to the D444H Mutation in the Biotinidase Gene. Hum. Genet. 1998, 102, 571–575. [Google Scholar] [CrossRef]
- Sarafoglou, K.; Bentler, K.; Gaviglio, A.; Redlinger-Grosse, K.; Anderson, C.; McCann, M.; Bloom, B.; Babovic-Vuksanovic, D.; Gavrilov, D.; Berry, S.A. High Incidence of Profound Biotinidase Deficiency Detected in Newborn Screening Blood Spots in the Somalian Population in Minnesota. J. Inherit. Metab. Dis. 2009, 32, 169–173. [Google Scholar] [CrossRef]
- Maguolo, A.; Rodella, G.; Dianin, A.; Monge, I.; Messina, M.; Rigotti, E.; Pellegrini, F.; Molinaro, G.; Lupi, F.; Pasini, A.; et al. Newborn Screening for Biotinidase Deficiency. The Experience of a Regional Center in Italy. Front. Pediatr. 2021, 9, 661416. [Google Scholar] [CrossRef]
- Semeraro, D.; Verrocchio, S.; Di Dalmazi, G.; Rossi, C.; Pieragostino, D.; Cicalini, I.; Ferrante, R.; Di Michele, S.; Stuppia, L.; Rizzo, C.; et al. High Incidence of Partial Biotinidase Deficiency in the First 3 Years of a Regional Newborn Screening Program in Italy. Int. J. Environ. Res. Public Health 2022, 19, 8141. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.; Heard, G.S. Screening for Biotinidase Deficiency in Newborns: Worldwide Experience. Pediatrics 1990, 85, 512–517. [Google Scholar] [CrossRef] [PubMed]
- McVoy, J.R.; Levy, H.L.; Lawler, M.; Schmidt, M.A.; Ebers, D.D.; Hart, P.S.; Pettit, D.D.; Blitzer, M.G.; Wolf, B. Partial Biotinidase Deficiency: Clinical and Biochemical Features. J. Pediatr. 1990, 116, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Procter, M.; Wolf, B.; Mao, R. Forty-Eight Novel Mutations Causing Biotinidase Deficiency. Mol. Genet. Metab. 2016, 117, 369–372. [Google Scholar] [CrossRef]
- Norrgard, K.J.; Pomponio, R.J.; Swango, K.L.; Hymes, J.; Reynolds, T.R.; Buck, G.A.; Wolf, B. Mutation (Q456H) Is the Most Common Cause of Profound Biotinidase Deficiency in Children Ascertained by Newborn Screening in the United States. Biochem. Mol. Med. 1997, 61, 22–27. [Google Scholar] [CrossRef]
- Montanari, G.; Candela, E.; Baronio, F.; Ferrari, V.; Biasucci, G.; Lanari, M.; Ortolano, R. Early-Onset Inherited Metabolic Diseases: When Clinical Symptoms Precede Newborn Screening-Insights from Emilia-Romagna (Italy). Children 2025, 12, 464. [Google Scholar] [CrossRef]
- Candela, E.; Zagariello, M.; Di Natale, V.; Ortolano, R.; Righetti, F.; Assirelli, V.; Biasucci, G.; Cassio, A.; Pession, A.; Baronio, F. Cystathionine Beta-Synthase Deficiency: Three Consecutive Cases Detected in 40 Days by Newborn Screening in Emilia Romagna (Italy) and a Comprehensive Review of the Literature. Children 2023, 10, 396. [Google Scholar] [CrossRef]
- Lampe, C.; Dionisi-Vici, C.; Bellettato, C.M.; Paneghetti, L.; van Lingen, C.; Bond, S.; Brown, C.; Finglas, A.; Francisco, R.; Sestini, S.; et al. The Impact of COVID-19 on Rare Metabolic Patients and Healthcare Providers: Results from Two MetabERN Surveys. Orphanet J. Rare Dis. 2020, 15, 341. [Google Scholar] [CrossRef]
- Sacket, S.J.; Chung, H.-Y.; Okajima, F.; Im, D.-S. Increase in Sphingolipid Catabolic Enzyme Activity during Aging. Acta Pharmacol. Sin. 2009, 30, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
| Patient ID | Allele 1 | Allele 2 | Expected Type of BD According to Genotype | Type of BD According to Enzyme Activity | BTD Activity U/dL at Diagnosis (%) | Group |
|---|---|---|---|---|---|---|
| 1 | p.Gln456His | p.Gln456His | Profound | Profound | 15.0 (6.9) | A |
| 2 | p.Asp444His | p.Gln456His | Partial | Partial | 54.6 (25.2) | B |
| 3 | p.Asp444His | p.Gln456His | Partial | Partial | 64.7 (29.8) | B |
| 4 | p.Asp444His | p.Gln456His | Partial | Partial | 41.0 (18.9) | B |
| 5 | p.Asp444His | p.Gln456His | Partial | Partial | 55.5 (25.6) | B |
| 6 | p.Asp444His | p.Gln456His | Partial | Partial | 53.5 (24.7) | B |
| 7 | p.Asp444His | p.Gln456His | Partial | Partial | 47.5 (21.9) | B |
| 8 | p.Asp444His | p.Gln456His | Partial | Partial | 47.2 (21.7) | B |
| 9 | p.Asp444His | p.Gln456His | Partial | Partial | 66.3 (19.85) | B |
| 10 | p.Asp444His | p.Gln456His | Partial | Partial | 45.4 (20.9) | B |
| 11 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Partial | 52.2 (24.0) | B |
| 12 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Partial | 55.8 (25.7) | B |
| 13 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Partial | 44.2 (20.4) | B |
| 14 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Partial | 53.6 (24.7) | B |
| 15 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Partial | 54.2 (25.0) | B |
| 16 | p.Asp444His | p.Cys33PhefsTer36 | Partial | Partial | 63.1 (29.1) | B |
| 17 | p.Asp444His | p.Cys33PhefsTer36 | Partial | Partial | 65.1 (30.0) | B |
| 18 | p.Asp444His | p.Cys33PhefsTer36 | Partial | Partial | 48.9 (22.5) | B |
| 19 | p.Asp444His | p.Cys33PhefsTer36 | Partial | Partial | 52.7 (24.3) | B |
| 20 | p.Asp444His | p.Cys33PhefsTer36 | Partial | Partial | 63.5 (29.3) | B |
| 21 | p.Asp444His | p.Asp252Gly | Partial | Partial | 59.6 (27.5) | B |
| 22 | p.Asp444His | p.Asp252Gly | Partial | Partial | 53.6 (24.7) | B |
| 23 | p.Asp444His | p.Asp444His;p.Thr532Met | Partial | Partial | 48.0 (22.1) | B |
| 24 | p.Asp444His | p.Arg538Cys | Partial | Partial | 64.1 (29.5) | B |
| 25 | p.Asp444His | p.Gly114Val | Partial | Partial | 44.5 (20.5) | B |
| 26 | p.Asp444His | p.Gly34Ser | Partial | Partial | 56.0 (25.8) | B |
| 27 | p.Asp444His | p.Thr532Met | Partial | Partial | 46.7 (21.5) | B |
| 28 | p.Asp444His | p.Val62Met | Partial | Partial | 57.4 (26.5) | B |
| 29 | p.Asp444His | p.Glu436AlafsTer8 * | Partial $ | Partial | 51.0 (23.5) | B |
| 30 | p.Asp444His | p.Gly238AlafsTer36 * | Partial $ | Partial | 51.7 (23.8) | B |
| 31 | p.Asp444His | p.Met98_Ala100delinsIle * | Partial $ | Partial | 49 (22.6) | B |
| 32 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 80.6 (37.2) | C |
| 33 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 68.4 (31.5) | C |
| 34 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 93.7 (43.2) | C |
| 35 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 89.5 (41.3) | C |
| 36 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 102 (47.0) | C |
| 37 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 101 (46.6) | C |
| 38 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 78.6 (36.2) | C |
| 39 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 84.1 (38.8) | C |
| 40 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 80.0 (37.0) | C |
| 41 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 89.8 (41.4) | C |
| 42 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 118.2 (54.5) | C |
| 43 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 100 (46.1) | C |
| 44 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 104 (48) | C |
| 45 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 72.8 (33.6) | C |
| 46 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 93.7 (43.2) | C |
| 47 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 80.6 (37.2) | C |
| 48 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 95.3 (43.9) | C |
| 49 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 104.8 (48.3) | C |
| 50 | p.Asp444His | p.Asp444His | Mild reduction | Mild reduction | 97.4 (44.9) | C |
| 51 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Mild reduction | 80.1 (36.9) | C |
| 52 | p.Asp444His | p.Asp444His;p.Ala171Thr | Partial | Mild reduction | 76.8 (35.4) | C |
| 53 | p.Asp444His | p.Arg157His | Partial | Mild reduction | 85.2 (39.3) | C |
| 54 | p.Asp444His | p.Pro497Ser | Partial | Mild reduction | 69.5 (32.1) | C |
| 55 | p.Asp444His | p.Cys33PhefsTer36 | Partial | Mild reduction | 69.0 (54.1) | C |
| 56 | p.Asp444His | p.His323Arg | Partial or Mild reduction | Mild reduction | 68.0 (31.3) | C |
| 57 | p.Asp444His | p.Pro133Leu * | Partial or Mild reduction | Mild reduction | 84.9 (39.2) | C |
| 58 | p.Asp444His | p.Pro133Leu * | Partial or Mild reduction | Mild reduction | 90.4 (41.7) | C |
| 59 | p.Asp444His | p.Pro133Leu * | Partial or Mild reduction | Mild reduction | 101 (46.6) | C |
| 60 | p.Asp444His | p.Val109Ala * | Partial or Mild reduction | Mild reduction | 67.7 (31.2) | C |
| 61 | p.Gly293Ser * | p.Phe361Ser * | Unknown | Mild reduction | 89.2 (41.1) | C |
| 62 | p.His323Arg | p.His323Arg | Partial or Mild reduction | Mild reduction | 78.8 (36.3) | C |
| 63 | p.His323Arg | p.His323Arg | Partial or Mild reduction | Mild reduction | 69.0 (31.8) | C |
| 64 | p.Gln456His | WT | Mild reduction | Mild reduction | 88.7 (40.9) | C |
| Variants | Deficit Associated | Reference | |
|---|---|---|---|
| Nucleotide Change | Amino Acid Change | ||
| c.98_104delGCGGCTGinsTCC | p.Cys33PhefsTer36 | Profound | [21] |
| c.100G > A | p.Gly34Ser | Profound | [22] |
| c.184G > A | p.Val62Met | Profound | [23] |
| c.294_299del | p.Met98_Ale100delinsIle | Unknown | |
| c.326T > C | p.Val109Ala | Unknown | |
| c.341G > T | p.Gly114Val | Profound | [24] |
| c.398C > T | p.Pro133Leu | Unknown | |
| c.470G > A | p.Arg157His | Profound | [21] |
| c.511G > A | p.Ala171Thr | profound (allele combined with p.Asp444His) | [20] |
| c.713delG | p.Gly238AlafsTer36 | Unknown | |
| c.755A > G | p.Asp252Gly | Profound | [25] |
| c.877G > A | p. Gly293Ser | Unknown | |
| c.968A > G | p.His323Arg | mild reduction/partial | [15,26,27] |
| c.1082T > C | p.Phe361Ser | Unknown | |
| c.1307_1308delAG | p.Glu436AlafsTer8 | Unknown | |
| c.1330G > C | p.Asp444His | partial (in compound heterozygosity with a profound variant) | [28] |
| c.1368A > T | p.Gln456His | Profound | [28] |
| c.1489C > T | p.Pro497Ser | Partial | [29] |
| c.1595C > T | p.Thr532Met | Profound | [28] |
| c.1612C > T | p.Arg538Cys | Profound | [21] |
| Patient ID | Allele 1 | Allele 2 | BTD Enzyme Activity (%) at Confirmation | Age (Months) at BTD Enzyme Activity Recovery | BTD Value Stability | Biotin Treatment at Last Visit | Age at Last Visit (Year) | Outcome/Sequelae |
|---|---|---|---|---|---|---|---|---|
| 1 | p.Gln456His | p.Gln456His | 6.92 | No |  | 10 mg/day | 6 | Healthy |
| 12 | p.Asp444His | p.Asp444His; p.Ala171Thr | 25.75 | No |  | 5 mg/day | 6 | Mild dermatologic signs (hypopigmentation, café-au-lait macule, regressing hemangioma) |
| 27 | p.Asp444His | p.Thr532Met | 21.55 | No |  | 5 mg/day | 5.8 | Healthy |
| 29 | p.Asp444His | p.Glu436AlafsTer8 | 23.54 | 3 |  | 5 mg/day | 8.7 | Healthy |
| 25 | p.Asp444His | p.Gly114Val | 20.54 | 3 |  | 5 mg/day | 8.8 | Healthy |
| 23 | p.Asp444His | p.Asp444His; p.Thr532Met | 22.15 | 3 |  | N.A. | N.A. | Follow-up discontinued by patient |
| 24 | p.Asp444His | p.Arg538Cys | 29.58 | 3 |  | 5 mg/day | 7.5 | Healthy |
| 16 | p.Asp444His | p.Cys33PhefsTer36 | 29.12 | 12 |  | 5 mg/day | 9 | Healthy |
| 31 | p.Asp444His | p.Met98_Ala100 delinsIle | 22.62 | 3 |  | 5 mg/day | 8,4 | Healthy |
| 13 | p.Asp444His | p.Asp444His; p.Ala171Thr | 20.40 | 3 |  | 5 mg/day | 5.9 | Recurrent asthma-like bronchitis |
| 18 | p.Asp444His | p.Cys33PhefsTer36 | 22.57 | 3 |  | 5 mg/day | 5.8 | Healthy |
| 28 | p.Asp444His | p.Val62Met | 26.49 | 3 |  | 5 mg/day | 5.8 | Healthy |
| 7 | p.Asp444His | p.Gln456His | 21.92 | 3 |  | 5 mg/day | 5 | Alternating esotropia |
| 26 | p.Asp444His | p.Gly34Ser | 25.86 | 12 |  | 5 mg/day | 5.9 | Healthy |
| 15 | p.Asp444His | p.Asp444His; p.Ala171Thr | 25.02 | 3 |  | 5 mg/day | 5.5 | Speech therapy for phonetic disorder and language delay |
| 20 | p.Asp444His | p.Cys33PhefsTer36 | 29.31 | 3 |  | 5 mg/day | 5.1 | Healthy |
| 58 | p.Asp444His | p.Gln456His | 25.20 | 36 |  | 5 mg/day | 8 | Healthy |
| 21 | p.Aps444His | p.Asp252Gly | 27.28 | 3 |  | N.A. | N.A. | Follow-up discontinued by patient |
| 22 | p.Aps444His | p.Asp252Gly | 24.74 | 3 |  | N.A. | N.A. | Follow-up discontinued by patient |
| 11 | p.Asp444His | p.Asp444His; p.Ala171Thr | 24.0 | 36 |  | 5 mg/day | 8 | Mild language delay |
| 3 | p.Asp444His | p.Gln456His | 29.86 | 60 |  | 5 mg/day | 6.5 | Healthy |
| 4 | p.Asp444His | p.Gln456His | 18.92 | 3 |  | 5 mg/day | 5.9 | Hypertrichosis of the limbs |
| 17 | p.Asp444His | p.Cys33PhefsTer36 | 30 | 3 |  | 5 mg/day | 5.8 | Healthy |
| 30 | p.Asp444His | p.Gly238AlafsTer36 | 23.86 | 60 |  | 5 mg/day | 5.8 | Healthy |
| 5 | p.Asp444His | p.Gln456His | 25.62 | 3 |  | 5 mg/day | 5.7 | Healthy |
| 6 | p.Asp444His | p.Gln456His | 24.69 | 36 |  | 5 mg/day | 5 | Frequent URTIs; past acute lymphoblastic leukemia |
| 8 | p.Asp444His | p.Gln456His | 21.78 | 3 |  | 5 mg/day | 5 | Healthy |
| 14 | p.Asp444His | p.Asp444His; p.Ala171Thr | 24.74 | 3 |  | 5 mg/day | 5 | Healthy |
| 9 | p.Asp444His | p.Gln456His | 19.85 | 3 |  | 5 mg/day | 5.2 | Healthy |
| 19 | p.Asp444His | p.Cys33PhefsTer36 | 24.32 | 36 |  | 5 mg/day | 5.1 | Healthy |
| 10 | p.Asp444His | p.Gln456His | 20.9 | 3 |  | 5 mg/day | 5.3 | Healthy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortolano, R.; Menabò, S.; Candela, E.; Biasucci, G.; Bortolamedi, E.; Montanari, G.; Zuccotti, A.; Cattini, U.; Lanari, M.; Baronio, F. Enzymatic Evolution and Longitudinal Recovery in Biotinidase Deficiency: Genotypic and Clinical Insights from the Follow-Up of a Newborn-Screened Cohort in Emilia-Romagna, Italy. Metabolites 2025, 15, 605. https://doi.org/10.3390/metabo15090605
Ortolano R, Menabò S, Candela E, Biasucci G, Bortolamedi E, Montanari G, Zuccotti A, Cattini U, Lanari M, Baronio F. Enzymatic Evolution and Longitudinal Recovery in Biotinidase Deficiency: Genotypic and Clinical Insights from the Follow-Up of a Newborn-Screened Cohort in Emilia-Romagna, Italy. Metabolites. 2025; 15(9):605. https://doi.org/10.3390/metabo15090605
Chicago/Turabian StyleOrtolano, Rita, Soara Menabò, Egidio Candela, Giacomo Biasucci, Elisa Bortolamedi, Giulia Montanari, Alessandro Zuccotti, Umberto Cattini, Marcello Lanari, and Federico Baronio. 2025. "Enzymatic Evolution and Longitudinal Recovery in Biotinidase Deficiency: Genotypic and Clinical Insights from the Follow-Up of a Newborn-Screened Cohort in Emilia-Romagna, Italy" Metabolites 15, no. 9: 605. https://doi.org/10.3390/metabo15090605
APA StyleOrtolano, R., Menabò, S., Candela, E., Biasucci, G., Bortolamedi, E., Montanari, G., Zuccotti, A., Cattini, U., Lanari, M., & Baronio, F. (2025). Enzymatic Evolution and Longitudinal Recovery in Biotinidase Deficiency: Genotypic and Clinical Insights from the Follow-Up of a Newborn-Screened Cohort in Emilia-Romagna, Italy. Metabolites, 15(9), 605. https://doi.org/10.3390/metabo15090605









