MALDI-TOF MS Biomarkers for Methicillin-Resistant Staphylococcus aureus Detection: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Eligibility Criteria
2.3. Search Strategy and Database Search
2.4. Study Selection and Data Extraction
2.5. Risk of Bias Assessment
2.6. Statistical Analysis
3. Results
3.1. Selected Studies
3.2. PSM-Mec and Delta Toxin
3.3. Additional MRSA and MSSA Biomarkers
4. Discussion
4.1. Identification of PSM-Mec and Delta Toxin
4.2. Additional Biomarkers and the Contribution of AI Tools
4.3. Alternative Protocols for MRSA Detection
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1
| No. | Question |
|---|---|
| 1 | Were the criteria for inclusion in the sample clearly defined? |
| 2 | Were the study subjects and the setting described in detail? |
| 3 | Was the exposure measured in a valid and reliable way? |
| 4 | Were objective, standard criteria used for measurement of the condition? |
| 5 | Were confounding factors identified? |
| 6 | Were strategies to deal with confounding factors stated? |
| 7 | Were the outcomes measured in a valid and reliable way? |
| 8 | Was appropriate statistical analysis used? |
Appendix A.2
| Study | 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. |
|---|---|---|---|---|---|---|---|---|
| Abalkhail [21] | Y | Y | Y | Y | Y | NA | Y | Y |
| Alksne [24] | Y | Y | Y | Y | Y | NA | Y | Y |
| Du [29] | Y | Y | Y | Y | Y | NA | Y | Y |
| Edwards-Jones [30] | Y | Y | UC | Y | Y | NA | Y | Y |
| Elbehiry [22] | Y | Y | N | Y | Y | NA | Y | Y |
| Fan [35] | Y | Y | N | Y | Y | NA | Y | Y |
| Gao [36] | Y | Y | Y | Y | Y | NA | Y | Y |
| Hu [25] | Y | Y | N | Y | Y | NA | Y | Y |
| Josten [26] | Y | Y | Y | Y | Y | NA | Y | Y |
| Kim [31] | Y | Y | Y | Y | Y | NA | Y | Y |
| Paskova [27] | Y | Y | UC | Y | Y | NA | Y | Y |
| Rhoads [28] | Y | Y | Y | Y | Y | NA | Y | Y |
| Sogawa [32] | Y | Y | Y | Y | Y | NA | Y | Y |
| Wang [33] | Y | Y | Y | Y | Y | NA | Y | UC |
| Yu [34] | Y | Y | N | Y | Y | NA | Y | Y |
Appendix A.3

Appendix B
Appendix B.1
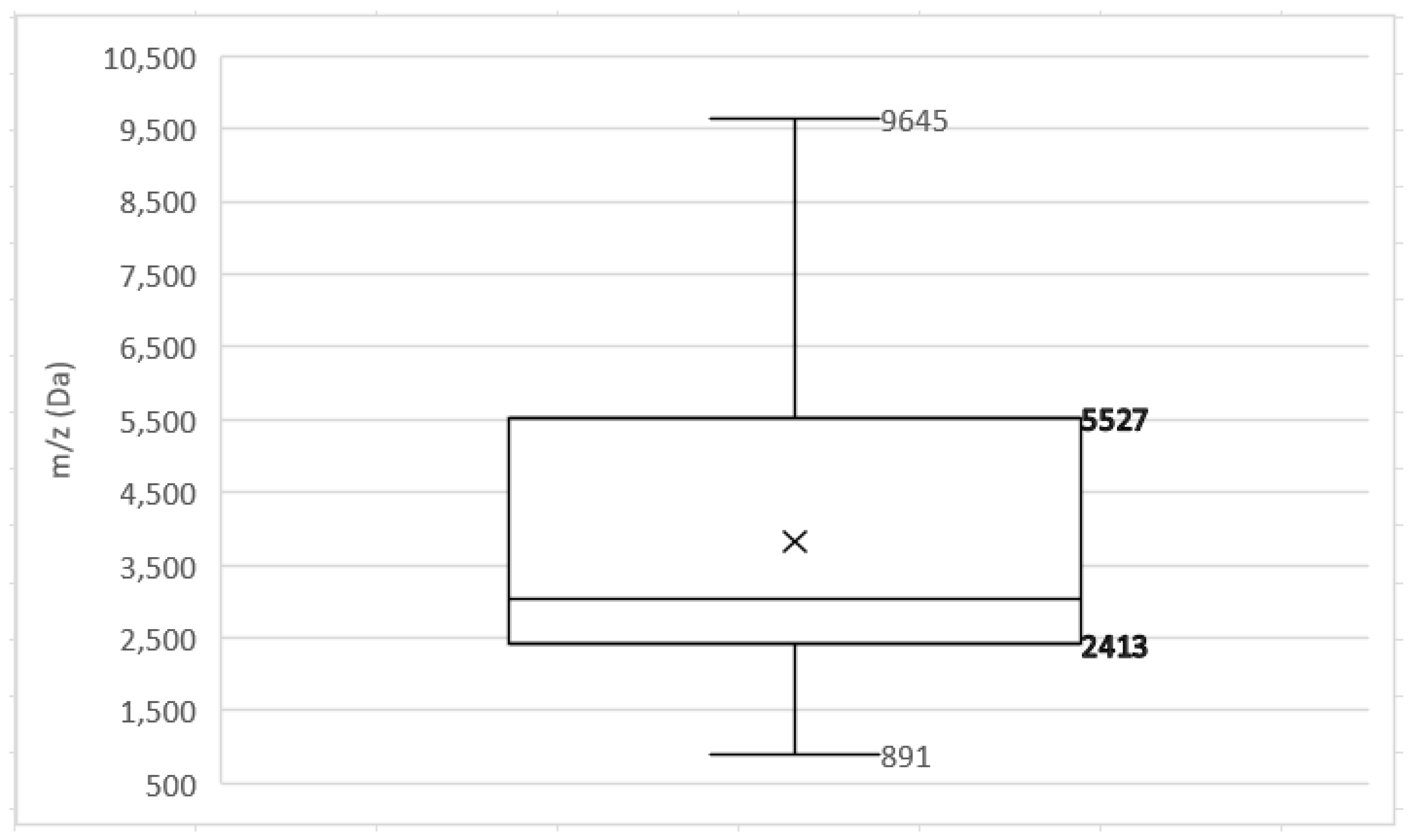
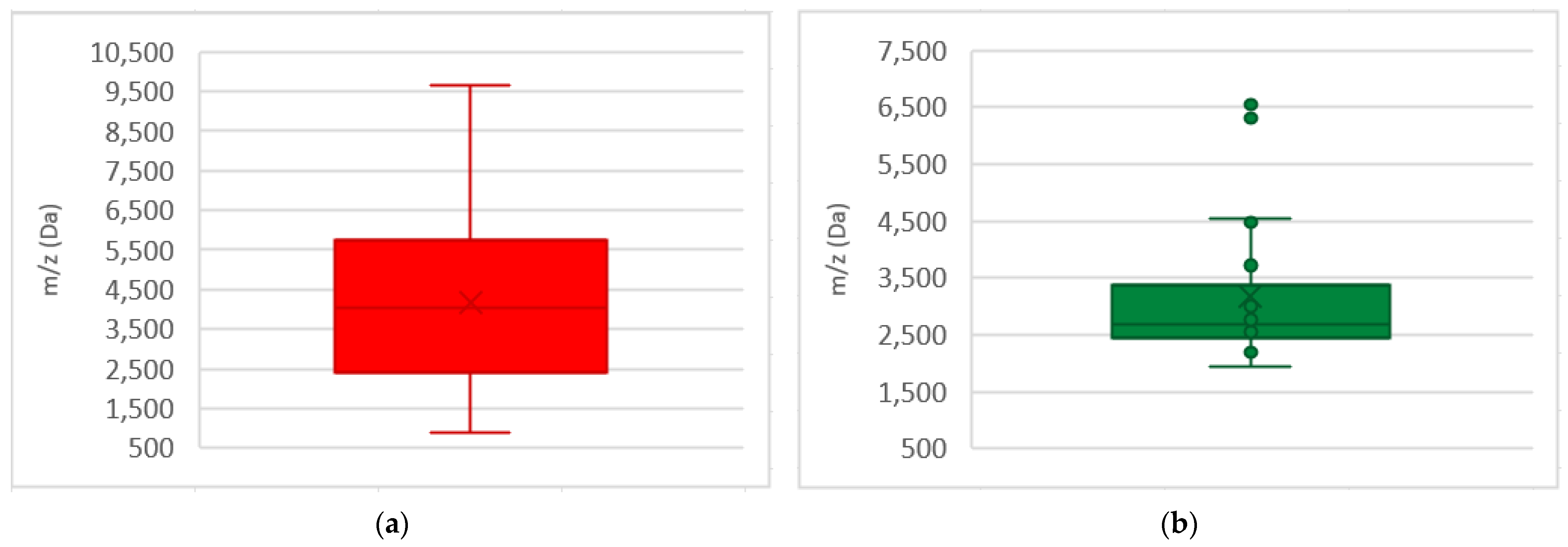
References
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, 00020-18. [Google Scholar] [CrossRef]
- Ji, Y. (Ed.) Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2007. [Google Scholar]
- Brown, D.F.; Edwards, D.I.; Hawkey, P.M.; Morrison, D.; Ridgway, G.L.; Towner, K.J.; Wren, M.W.; Joint Working Party of the British Society for Antimicrobial. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J. Antimicrob. Chemother. 2005, 56, 1000–1018. [Google Scholar] [CrossRef]
- Maugeri, G.; Lychko, I.; Sobral, R.; Roque, A.C.A. Identification and Antibiotic-Susceptibility Profiling of Infectious Bacterial Agents: A Review of Current and Future Trends. Biotechnol. J. 2019, 14, e1700750. [Google Scholar] [CrossRef]
- CLSI M100; Performance Standards for Antimicrobial Susceptibility Testing (M100). CLSI: Wayne, PA, USA, 2020.
- Calderaro, A.; Chezzi, C. MALDI-TOF MS: A Reliable Tool in the Real Life of the Clinical Microbiology Laboratory. Microorganisms 2024, 12, 322. [Google Scholar] [CrossRef]
- Ligozzi, M.; Bernini, C.; Bonora, M.G.; De Fatima, M.; Zuliani, J.; Fontana, R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 2002, 40, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Yarbrough, M.L.; Warren, D.K.; Allen, K.; Burkholder, D.; Daum, R.; Donskey, C.; Knaack, D.; LaMarca, A.; May, L.; Miller, L.G.; et al. Multicenter Evaluation of the Xpert MRSA NxG Assay for Detection of Methicillin-Resistant Staphylococcus aureus in Nasal Swabs. Clin. Microbiol. 2018, 52, 01381-17. [Google Scholar] [CrossRef]
- WHO. The Diagnostic Stewardship Concept Refers to the Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef]
- Elbehiry, A.; Aldubaib, M.; Abalkhail, A.; Marzouk, E.; Albeloushi, A.; Moussa, I.; Ibrahem, M.; Albazie, H.; Alqarni, A.; Anagreyyah, S.; et al. How MALDI-TOF Mass Spectrometry Technology Contributes to Microbial Infection Control in Healthcare Settings. Vaccines 2022, 10, 1881. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.Y.; Chiang-Ni, C.; Teng, S.H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Veloo, A.C.; de Vries, E.D.; Jean-Pierre, H.; Justesen, U.S.; Morris, T.; Urban, E.; Wybo, I.; van Winkelhoff, A.J. The optimization and validation of the Biotyper MALDI-TOF MS database for the identification of Gram-positive anaerobic cocci. Clin. Microbiol. Infect. 2016, 22, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Cai, K.; Yu, P.; Liu, Y.; Zhao, G.; Chen, R.; Xu, R.; Yu, M. Evaluation of three sample preparation methods for the identification of clinical strains by using two MALDI-TOF MS systems. J. Mass Spectrom. 2021, 56, e4696. [Google Scholar] [CrossRef]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front. Cell. Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef]
- Vrioni, G.; Tsiamis, C.; Oikonomidis, G.; Theodoridou, K.; Kapsimali, V.; Tsakris, A. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: Current achievements and future perspectives. Ann. Transl. Med. 2018, 6, 240. [Google Scholar] [CrossRef] [PubMed]
- Abalkhail, A.; Elbehiry, A. Methicillin-Resistant Staphylococcus aureus in Diabetic Foot Infections: Protein Profiling, Virulence Determinants, and Antimicrobial Resistance. Appl. Sci. 2022, 12, 10803. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Moussa, I.; Anagreyyah, S.; AlGhamdi, A.; Alqarni, A.; Aljohani, A.; Hemeg, H.A.; Almuzaini, A.M.; Alzaben, F.; et al. Using Protein Fingerprinting for Identifying and Discriminating Methicillin Resistant Staphylococcus aureus Isolates from Inpatient and Outpatient Clinics. Diagnostics 2023, 13, 2825. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Z.; Cong, L.; Shan, M.; Zhu, Z.; Li, Y. Rapid identification of methicillin-resistant Staphylococcus aureus by MALDI-TOF MS: A meta-analysis. Biotechnol. Appl. Biochem. 2023, 70, 1217–1229. [Google Scholar] [CrossRef]
- Alksne, L.; Makarova, S.; Avsejenko, J.; Cibrovska, A.; Trofimova, J.; Valcina, O. Determination of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis by MALDI-TOF MS in clinical isolates from Latvia. Clin. Mass Spectrom. 2020, 16, 33–39. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, Y.; Lizou, Y.; Li, J.; Zhang, R. Evaluation of Staphylococcus aureus Subtyping Module for Methicillin-Resistant Staphylococcus aureus Detection Based on Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Front. Microbiol. 2019, 10, 2504. [Google Scholar] [CrossRef] [PubMed]
- Josten, M.; Dischinger, J.; Szekat, C.; Reif, M.; Al-Sabti, N.; Sahl, H.G.; Parcina, M.; Bekeredjian-Ding, I.; Bierbaum, G. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2014, 304, 1018–1023. [Google Scholar] [CrossRef]
- Paskova, V.; Chudejova, K.; Sramkova, A.; Kraftova, L.; Jakubu, V.; Petinaki, E.A.; Zemlickova, H.; Neradova, K.; Papagiannitsis, C.C.; Hrabak, J. Insufficient repeatability and reproducibility of MALDI-TOF MS-based identification of MRSA. Folia Microbiol. 2020, 65, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.D.; Wang, H.; Karichu, J.; Richter, S.S. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 2016, 86, 257–261. [Google Scholar] [CrossRef]
- Du, Z.; Yang, R.; Guo, Z.; Song, Y.; Wang, J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2002, 74, 5487–5491. [Google Scholar] [CrossRef]
- Edwards-Jones, V.; Claydon, M.A.; Evason, D.J.; Walker, J.; Fox, A.J.; Gordon, D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000, 49, 295–300. [Google Scholar] [CrossRef]
- Kim, J.M.; Kim, I.; Chung, S.H.; Chung, Y.; Han, M.; Kim, J.S. Rapid Discrimination of Methicillin-Resistant Staphylococcus aureus by MALDI-TOF MS. Pathogens 2019, 8, 214. [Google Scholar] [CrossRef]
- Sogawa, K.; Watanabe, M.; Ishige, T.; Segawa, S.; Miyabe, A.; Murata, S.; Saito, T.; Sanda, A.; Furuhata, K.; Nomura, F. Rapid Discrimination between Methicillin-Sensitive and Methicillin-Resistant Staphylococcus aureus Using MALDI-TOF Mass Spectrometry. Biocontrol Sci. 2017, 22, 163–169. [Google Scholar] [CrossRef]
- Wang, Y.R.; Chen, Q.; Cui, S.H.; Li, F.Q. Characterization of Staphylococcus aureus isolated from clinical specimens by matrix assisted laser desorption/ionization time-of-flight mass spectrometry. Biomed. Environ. Sci. 2013, 26, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tien, N.; Liu, Y.C.; Cho, D.Y.; Chen, J.W.; Tsai, Y.T.; Huang, Y.C.; Chao, H.J.; Chen, C.J. Rapid Identification of Methicillin-Resistant Staphylococcus aureus Using MALDI-TOF MS and Machine Learning from over 20,000 Clinical Isolates. Microbiol. Spectr. 2022, 10, e0048322. [Google Scholar] [CrossRef]
- Fan, L.-P.; Tang, X.; Bai, X.; Cheng, H.; Zeng, C.; Huang, S.; Liao, W.; Huang, Q.-S.; Du, F.-L.; Dan Wei, D.; et al. Rapid identification of MRSA directly from sterile body fluids by co-magnetic bead enrichment and MALDI-TOF mass spectrometry. Microchem. J. 2024, 197, 109739. [Google Scholar] [CrossRef]
- Gao, W.; Li, B.; Ling, L.; Zhang, L.; Yu, S. MALDI-TOF MS method for differentiation of methicillin-sensitive and methicillin-resistant Staphylococcus aureus using (E)-Propyl alpha-cyano-4-Hydroxyl cinnamylate. Talanta 2022, 244, 123405. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Barker, T.H.; Stone, J.C.; Sears, K.; Klugar, M.; Leonardi-Bee, J.; Tufanaru, C.; Aromataris, E.; Munn, Z. Revising the JBI quantitative critical appraisal tools to improve their applicability: An overview of methods and the development process. JBI Evid. Synth. 2023, 21, 478–493. [Google Scholar] [CrossRef]
- McGuinness, L.A.; Higgins, J.P.T. Risk-of-bias VISualization (robvis): An R package and Shiny web app for visualizing risk-of-bias assessments. Res. Synth. Methods 2021, 12, 55–61. [Google Scholar] [CrossRef]
- Joanna Briggs Institute (JBI). Checklist for Analytical Cross Sectional Studies. Available online: https://jbi.global/critical-appraisal-tools (accessed on 12 December 2024).
- MedCalc Software Ltd.; Ostend, B. MedCalc Statistical Software Version: 23.0.8. Available online: https://www.medcalc.org (accessed on 20 May 2025).
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Mergenhagen, K.A.; Starr, K.E.; Wattengel, B.A.; Lesse, A.J.; Sumon, Z.; Sellick, J.A. Determining the Utility of Methicillin-Resistant Staphylococcus aureus Nares Screening in Antimicrobial Stewardship—PubMed. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 1142–1148. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Stewardship Programmes in Health-Care Facilities in Low- and Middle-Income Countries—A WHO Practical Toolkit; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Commission, E. EU Guidelines for the Prudent Use of Antimicrobials in Human Health; 2017/C 212/01; European Union: Brussels, Belgium, 2017; Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:52017XC0701(01) (accessed on 20 May 2024).
- Kingston, R.; Vella, V.; Pouwels, K.B.; Schmidt, J.E.; Abdelatif El-Abasiri, R.A.; Reyna-Villasmil, E.; Hassoun-Kheir, N.; Harbarth, S.; Rodriguez-Bano, J.; Tacconelli, E.; et al. Excess resource use and cost of drug-resistant infections for six key pathogens in Europe: A systematic review and Bayesian meta-analysis. Clin. Microbiol. Infect. 2024, 30 (Suppl. S1), S26–S36. [Google Scholar] [CrossRef] [PubMed]
- Alatoom, A.A.; Cunningham, S.A.; Ihde, S.M.; Mandrekar, J.; Patel, R. Comparison of direct colony method versus extraction method for identification of gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011, 49, 2868–2873. [Google Scholar] [CrossRef]
- Cuenod, A.; Foucault, F.; Pfluger, V.; Egli, A. Factors Associated With MALDI-TOF Mass Spectral Quality of Species Identification in Clinical Routine Diagnostics. Front. Cell. Infect. Microbiol. 2021, 11, 646648. [Google Scholar] [CrossRef] [PubMed]
- Boucherabine, S.; Giddey, A.; Nassar, R.; Al-Hroub, H.M.; Mohamed, L.; Harb, M.; Soares, N.C.; Senok, A. Proteomic and metabolomic profiling of methicillin resistant versus methicillin sensitive Staphylococcus aureus using a simultaneous extraction protocol. Front. Microbiol. 2024, 15, 1402796. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, J.; Vougas, K.; Shah, A.; Shah, H.; Misra, R.; Mkrtchyan, H.V. Comparative Proteomic Profiling of Methicillin-Susceptible and Resistant Staphylococcus aureus. Proteomics 2020, 20, e1900221. [Google Scholar] [CrossRef]
- Deurenberg, R.H.; Vink, C.; Kalenic, S.; Friedrich, A.W.; Bruggeman, C.A.; Stobberingh, E.E. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2007, 13, 222–235. [Google Scholar] [CrossRef]
- Blackmon, S.; Avendano, E.E.; Balaji, S.; Argaw, S.A.; Morin, R.A.; Nirmala, N.; Doron, S.; Nadimpalli, M.L. Neighborhood-level income and MRSA infection risk in the USA: Systematic review and meta-analysis. BMC Public Health 2025, 25, 1074. [Google Scholar] [CrossRef] [PubMed]
- Azzam, A.; Khaled, H.; Fayed, H.M.; Mansour, Y.; Eldalil, M.; Elshennawy, E.; Salem, H.; Elkatan, H.A. Prevalence, antibiogram, and risk factors of methicillin-resistant Staphylococcus aureus (MRSA) asymptomatic carriage in Africa: A systematic review and meta-analysis. BMC Infect. Dis. 2025, 25, 505. [Google Scholar] [CrossRef] [PubMed]
- Nikmanesh, Y.; Foolady Azarnaminy, A.; Avishan, P.; Taheri, M.; Sabeghi, P.; Najibzadeh, E.; Khaledi, A. A Middle East systematic review and meta-analysis of prevalence and antibiotic susceptibility pattern in MRSA Staphylococcus aureus isolated from patients with cystic fibrosis. J. Health Popul. Nutr. 2022, 41, 26. [Google Scholar] [CrossRef]
- Patil, S.S.; Suresh, K.P.; Shinduja, R.; Amachawadi, R.G.; Chandrashekar, S.; Pradeep, S.; Kollur, S.P.; Syed, A.; Sood, R.; Roy, P.; et al. Prevalence of Methicillin-resistant Staphylococcus aureus in India: A Systematic Review and Meta-analysis. Oman Med. J. 2022, 37, e440. [Google Scholar] [CrossRef]
- Rodenbeck, M.; Ayobami, O.; Eckmanns, T.; Pletz, M.W.; Bleidorn, J.; Markwart, R. Clinical epidemiology and case fatality due to antimicrobial resistance in Germany: A systematic review and meta-analysis, 1 January 2010 to 31 December 2021. Eurosurveillance 2023, 28, 2200672. [Google Scholar] [CrossRef]
- Chatterjee, S.S.; Chen, L.; Joo, H.S.; Cheung, G.Y.; Kreiswirth, B.N.; Otto, M. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e28781. [Google Scholar] [CrossRef]
- Qin, L.; McCausland, J.W.; Cheung, G.Y.; Otto, M. PSM-Mec-A Virulence Determinant that Connects Transcriptional Regulation, Virulence, and Antibiotic Resistance in Staphylococci. Front. Microbiol. 2016, 7, 1293. [Google Scholar] [CrossRef]
- Gagnaire, J.; Dauwalder, O.; Boisset, S.; Khau, D.; Freydiere, A.M.; Ader, F.; Bes, M.; Lina, G.; Tristan, A.; Reverdy, M.E.; et al. Detection of Staphylococcus aureus delta-toxin production by whole-cell MALDI-TOF mass spectrometry. PLoS ONE 2012, 7, e40660. [Google Scholar] [CrossRef] [PubMed]
- Thelen, P.; Graeber, S.; Schmidt, E.; Hamprecht, A. A side-by-side comparison of the new VITEK MS PRIME and the MALDI Biotyper sirius in the clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1355–1363. [Google Scholar] [CrossRef]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Rafailidis, P.I.; Ioannidou, E.N.; Falagas, M.E.; Rafailidis, P.I.; Ioannidou, E.N.; Falagas, M.E. Ampicillin/Sulbactam. Drugs 2007, 67, 13. [Google Scholar] [CrossRef] [PubMed]


| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Articles published in English language. | Articles published in other languages. |
| Articles published until 27 July 2024. Free full-text access. | Abstract of congress, reports, reviews/state-of-the-art articles. |
| Articles reporting findings from any country. | Studies involving veterinary infections. |
| Original scientific articles on the topic: | Other infectious agents. |
| MRSA and MSSA human infections. | Identification of isolates by MALDI-TOF MS. |
| Identification of biomarkers of S. aureus. |
| Author | Sample | Study Location | Methodology | Results | ||||
|---|---|---|---|---|---|---|---|---|
| Reference Method | MALDI-TOF Analyser | Sample Preparation | MRSA Biomarkers (Da) | MSSA Biomarkers (Da) | AI Models | |||
| Abalkhail & Elbehiry, 2022 [21] | 22 MRSA and 26 MSSA | Saudi Arabia | mecA gene PCR | MALDI Biotyper (Bruker Daltonics) | I-PEM *1 | 5530, 6580, 6710, and 6820 | 2771, 2996, 3720, 4480, 4540, and 6310 | PCA |
| Alksne et al., 2020 [24] | 26 MRSA and 28 MSSA | Latvia | mecA gene PCR | Autoflex Speed MS (Bruker Daltonics) | I-PEM *1 O-PEM *1 ICM *1 | ICM and O-PEM: PSM-mec (2414 ± 2) ICM, O-PEM, and I-PEM: delta toxin (3006 ± 2) | ND | NA |
| Du et al., 2002 [29] | 35 MRSA and 41 MSSA | China | mecA gene PCR | linear MALDI-TOF MS (Micromass UK Ltd.) | O-PEM *2 | Main peaks: 2413.01 and 2453.54 | Main peaks: 2547.91, 2585.28, 2686.45, and 2723.17 | NA |
| Edwards-Jones et al., 2000 [30] | 7 MRSA and 7 MSSA | UK | PFGE and phage typing | Kompact MALDI 2 linear TOF MS (Kratos Analytical) | O-PEM *2 | 891, 1140, 1165, 1229, 2127, 2454, and 3045 | 2548 and 2647 | NA |
| Elbehiry et al., 2023 [22] | 197 MRSA and 129 MSSA | Saudi Arabia | Kirby–Bauer test | MALDI Biotyper (Bruker Daltonik) | I-PEM *1 | 3990, 4120, and 5850 | Lack of resistance peaks | PCA |
| Fan et al., 2024 [35] | 20 MRSA and 30 MSSA | China | Automated AST | VITEK MS (bioMérieux) | Amp-MB protocol O-PEM *3,5 | 4304.6889 (larger a.u. in MRSA) 3874.4304 and 6889 | 3041.2293 (larger a.u in MSSA) | NA |
| Gao et al., 2002 [36] | 21 MRSA and 41 MSSA | China | mecA gene PCR and Kirby–Bauer test | Autoflex max TOF/TOF MS (Bruker Daltonics) | I-PEM *4 | 4821 and 9645 | 2306 and 2322 (larger a.u in MSSA) | PCA |
| Hu et al., 2019 [25] | 241 MRSA and 106 MSSA | China | Kirby–Bauer test | MALDI-Biotyper (Bruker Daltonics) | O-PEM *1 | PSM-mec (2413 ± 2) | ND | Clinpro Tools |
| Josten et al., 2014 [26] | 356 S. aureus *7 | Germany | mecA gene PCR and Kirby–Bauer test | MALDI-TOF MS Biflex II (Bruker Daltonic) | O-PEM *1 | PSM-mec (CC5): 2415 ± 4 delta toxin: 3007 (most CC) and 3037 (CC1) | ND | NA |
| VITEK MS (bioMérieux) | ICM *5 | |||||||
| Kim et al., 2019 [31] | 320 S. aureus (database) 181 S. aureus (test sample) | Korea | mecA gene PCR | Microflex LT MALDI-TOF MS (Bruker Daltonics) | O-PEM *1 | SCCmec IV: 5541 (+) and 5053 (−) PSM-mec (SCCmec III specific): 2410 and 4607 At least one: 1975, 2410, 3890, 4607, and 6594 | 2194, 2339, and 2631 | BioNumerics (decision tree model) |
| Paskova et al., 2020 [27] | 35 MRSA | Multicentred | ND | microFlex MS (Bruker Daltonics) | O-PEM *1 | PSM-mec (2413 ± 3.00) and delta toxin (3006 ± 3.00) | ND | NA |
| Rhoads et al., 2016 [28] | 137 MRSA and 146 MSSA 12 MRSA USA 100-USA1200 | USA | mecA gene PCR | VITEK MS (bioMérieux) Bruker MicroFlex (Bruker Daltonics): USA isolates | ICM *5 | PSM-mec (2415 ± 2.00) | ND | NA |
| Sogawa et al., 2017 [32] | 50 MRSA and 50 MSSA (algorithm) 34 MRSA and 31 MSSA (test sample) | Japan | mecA gene PCR | Autoflex II TOF (Bruker Daltonics) | O-PEM *1 | 1888.1 (430.3 a.u.), 1935.9 (880.8 a.u.), 2867.9 (1490.9 a.u.), 3044.2 (20,061.4 a.u.) *6, and 4641.3 (260.0 a.u.) | 1935.9 (662.2 a.u.), and 2760.3 (1230.1 a.u.) | NL-SVM |
| Wang et al., 2013 [33] | 48 MRSA and 52 MSSA | China | mecA gene PCR | MALDI-Microflex (Bruker Daltonics) | I-PEM *1 | 3784 and 5700 (larger a.u. in MRSA) | 3784 and 5700 (smaller a.u. in MSSA) | Clinpro Tools (Genetic algorithm) |
| Yu et al., 2022 [34] | 4309 MRSA and 3949 MSSA (algorithm) 12,101 MS (external validation) | Taiwan | Automated AST | MicroflexLT MALDI-TOF MS (Bruker Daltonics) | O-PEM *1 | 6593.2 | 6550.0 | LightGBM |
| Performance Test | Alksne [24] | Hu [25] | Josten [26] | Rhoads [28] | Paskova [27] |
|---|---|---|---|---|---|
| Sensitivity (%) | 61 *1 | 60.2 | 94.7 *3 | 37 | 50/90 *4 |
| Specificity (%) | 100 *1 | 100 | 100 *3 | 100 | 100 |
| Reproducibility (%) | 87 *1 | ― | ― | ― | 33–100 *5 |
| Repeatability (%) | ― | 1.7/18.4 *2 | ― | ― | ― |
| Performance Test | Abalkhail [21] | Du [29] | Edwards-Jones [30] | Elbehiry [22] | Fan [35] | Gao [36] | Kim [31] | Sogawa [32] | Wang [33] | Yu [34] |
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (%) | 100 | 90.79 *1 | 85.71 *1 | 97.87 *1 | 96.0 *1 | 96.8 *1 | 87.6 | 89.0–100 | ND | ― |
| AUC | ― | ― | ― | ― | ― | ― | ― | ― | 0.78–0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, P.; Alho, I.; Ribeiro, E. MALDI-TOF MS Biomarkers for Methicillin-Resistant Staphylococcus aureus Detection: A Systematic Review. Metabolites 2025, 15, 540. https://doi.org/10.3390/metabo15080540
Santos P, Alho I, Ribeiro E. MALDI-TOF MS Biomarkers for Methicillin-Resistant Staphylococcus aureus Detection: A Systematic Review. Metabolites. 2025; 15(8):540. https://doi.org/10.3390/metabo15080540
Chicago/Turabian StyleSantos, Pedro, Irina Alho, and Edna Ribeiro. 2025. "MALDI-TOF MS Biomarkers for Methicillin-Resistant Staphylococcus aureus Detection: A Systematic Review" Metabolites 15, no. 8: 540. https://doi.org/10.3390/metabo15080540
APA StyleSantos, P., Alho, I., & Ribeiro, E. (2025). MALDI-TOF MS Biomarkers for Methicillin-Resistant Staphylococcus aureus Detection: A Systematic Review. Metabolites, 15(8), 540. https://doi.org/10.3390/metabo15080540






