Joint Metabolomics and Transcriptomics Reveal Rewired Glycerophospholipid and Arginine Metabolism as Components of BRCA1-Induced Metabolic Reprogramming in Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Intact Cell NMR Spectroscopy
2.3. Metabolite Extraction
2.4. Aqueous Phase NMR Spectroscopy
2.5. Aqueous Phase Liquid Chromatography–Mass Spectrometry
2.6. Organic-Phase High-Performance Thin-Layer Chromatography
2.7. RNA Extraction and Quantitative RT-PCR
2.8. Univariate Statistics
2.9. OPLS Data Processing of NMR Spectra
2.10. O2PLS of Joint Metabolites/Transcripts
2.11. Pathway Analysis
3. Results
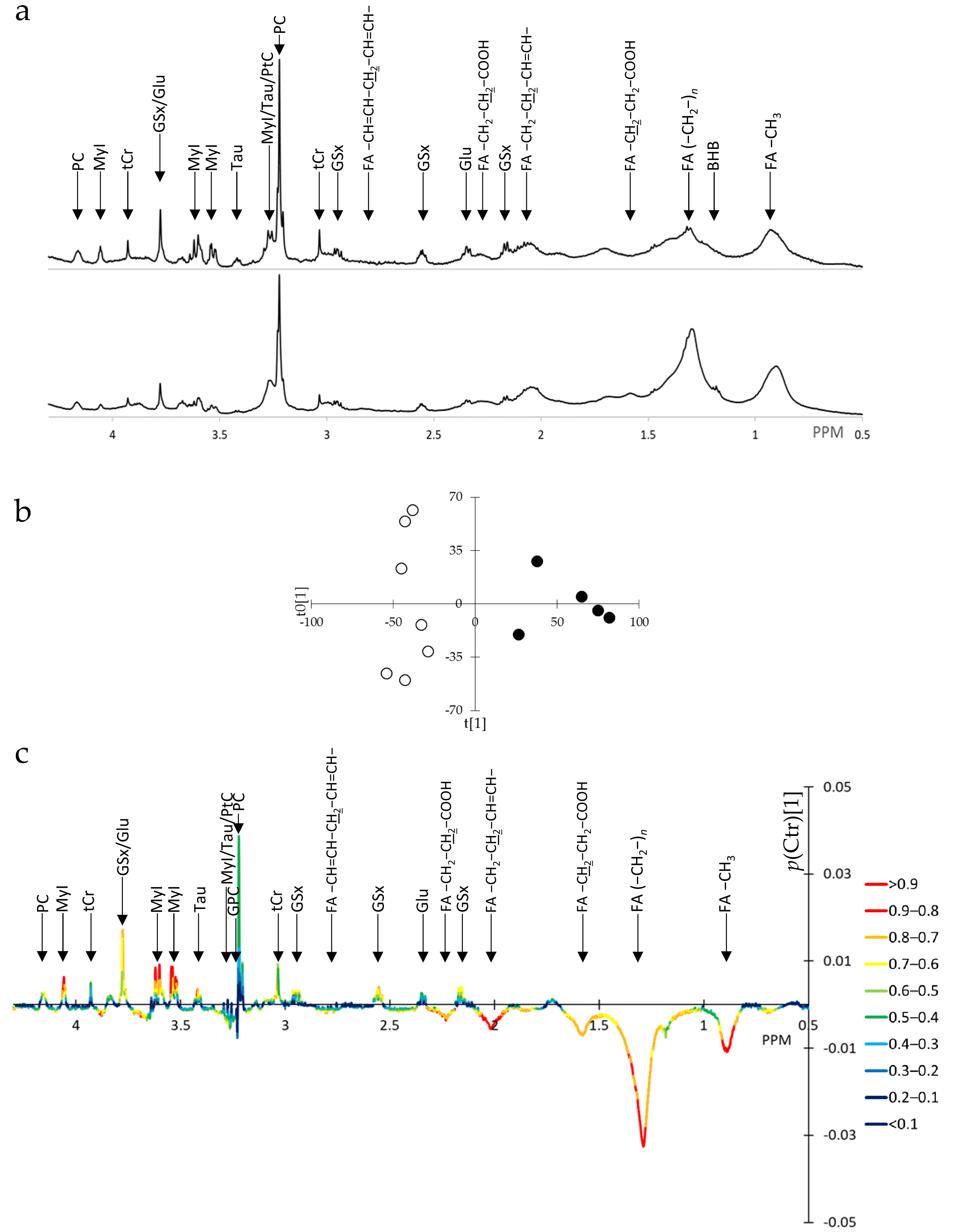
3.1. SUM1315-BRCA1 Intact Cells Exhibit Reduction in Intracellular Fatty Acid Levels
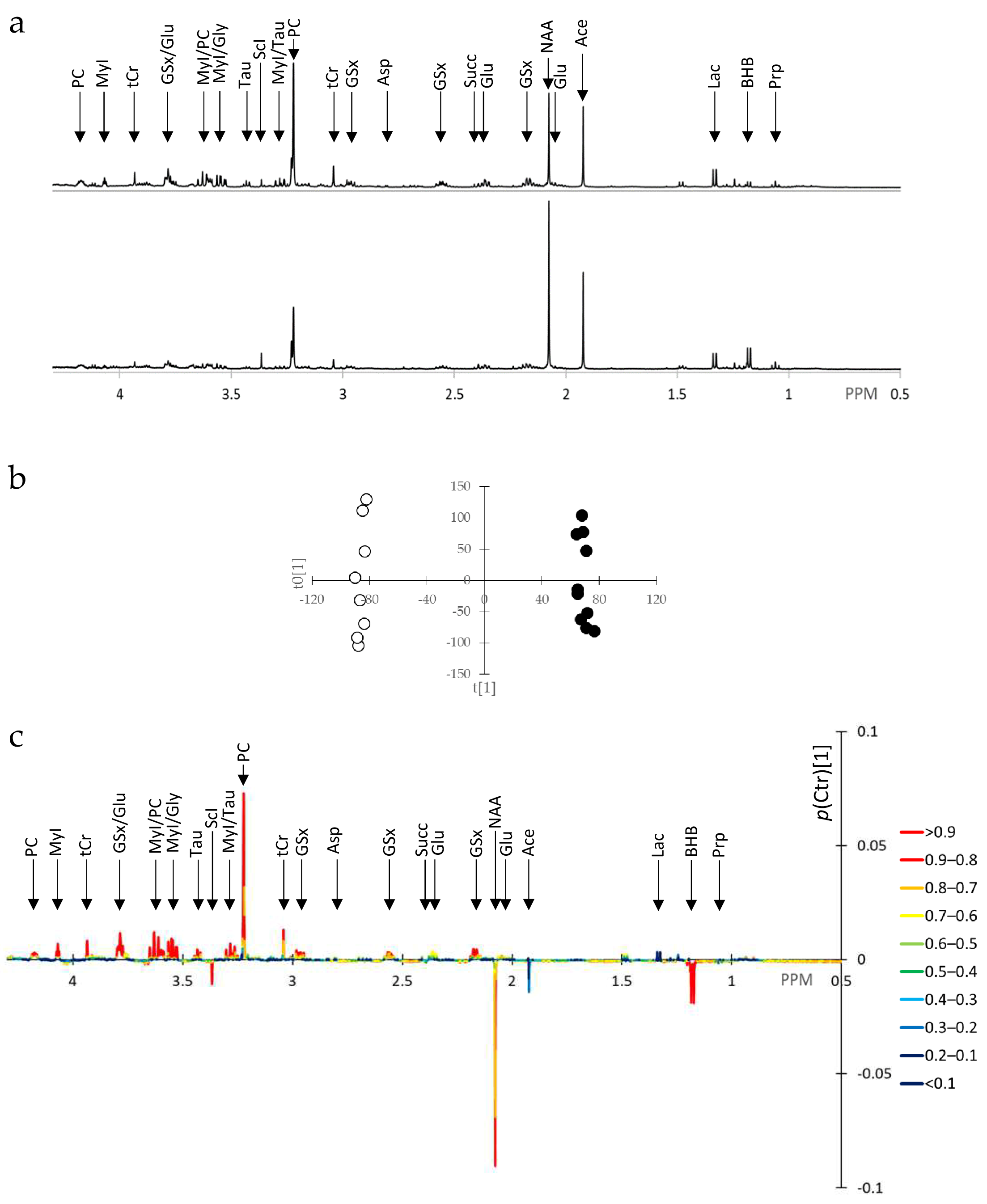
3.2. SUM1315-BRCA1 Water-Soluble Extracts Exhibit Numerous Metabolite Alterations
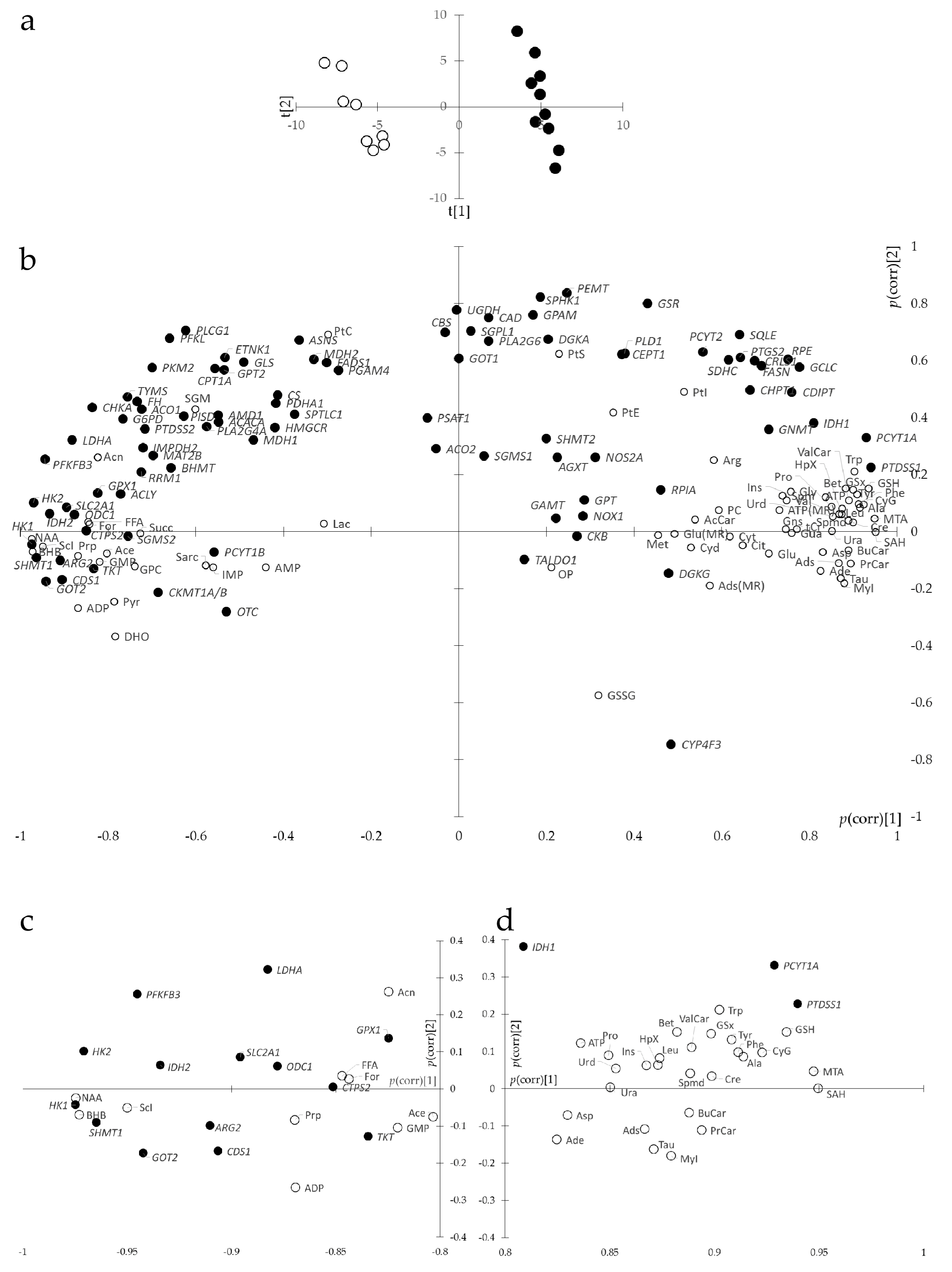
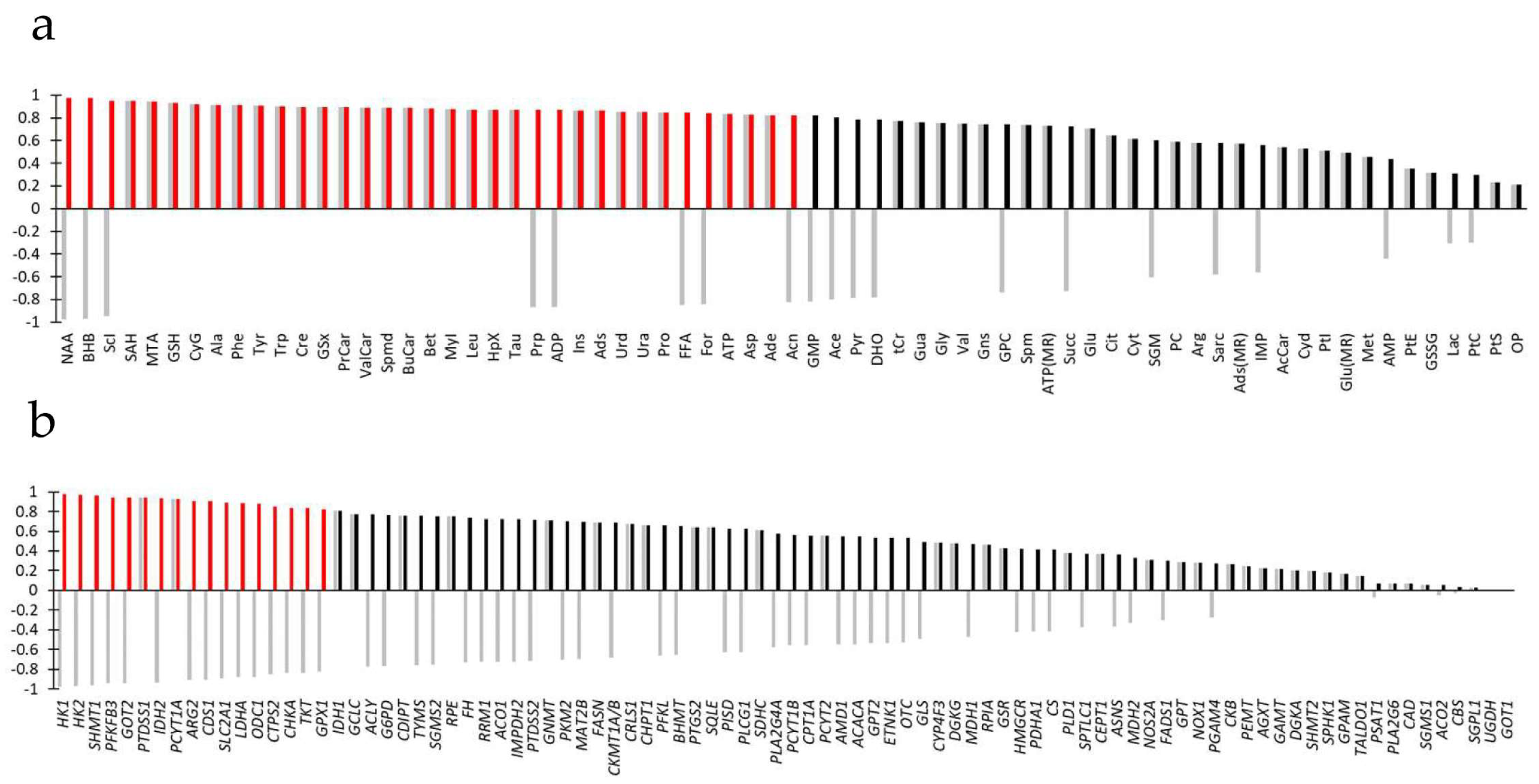
3.3. Joint Metabolite/Transcript Analysis Using O2PLS Identifies Most Important Metabolites and Genes of Metabolic Reprogramming
3.4. Joint Metabolite/Transcript Analysis Using ORA Identifies Most Important Pathways of Metabolic Reprogramming
3.5. Combining Results of Joint Data Processing
- (1)
- A first metabolism superset was centered around bioenergetic metabolism and contained 7 pathways (‘Central carbon metabolism in cancer’, ‘Citrate cycle (TCA cycle)’, ‘Pyruvate metabolism’, ‘HIF-1 signaling pathway’, ’Pentose phosphate pathway’, ‘Glycolysis or Gluconeogenesis’, and ‘Butanoate metabolism’), 7 downregulated genes (SLC2A1, HK1, HK2, PFKFB3, LDHA, IDH2, and TKT), 8 increased metabolites (Ala, Asp, Leu, Phe, Pro, Tyr, and ATP) and 3 decreased metabolites (For, Acn, and BHB).
- (2)
- A second metabolism superset was centered around glycerophospholipid metabolism and contained 3 pathways (‘Glycerophospholipid metabolism’, ‘Choline metabolism in cancer’, and ‘Inositol phosphate metabolism’), 2 upregulated genes (PTDSS1 and PCYT1A), 2 downregulated genes (CDS1 and CHKA), 1 increased metabolite (MyI), and 2 decreased metabolites (Sci and FFA).
- (3)
- A third metabolism superset was centered around glutathione metabolism and contained 3 pathways (‘Glutathione metabolism’, ‘Cysteine and methionine metabolism’, and ‘Taurine and hypotaurine metabolism’), 5 downregulated genes (GOT2, LDHA, IDH2, ODC1, and GPX1), 9 increased metabolites (GSH, GSx, CyG, Spmd, Asp, MTA, SAH, Ala, and Tau) and 1 decreased metabolite (Ace).
- (4)
- A fourth metabolism superset was centered around nucleic acid metabolism and contained 4 pathways (‘One-carbon pool by folate’, ‘Purine metabolism’, ‘Pantothenate and CoA biosynthesis’ and ‘Pyrimidine metabolism’), 2 downregulated genes (SHMT1 and CTPS2), 10 increased metabolites (Asp, Ade, HpX, Ins, Ads, Ura, Urd, ATP, Bet, and SAH) and 1 decreased metabolite (ADP).
- (5)
- A fifth metabolism superset was centered around Arg metabolism and contained 5 pathways (‘Arginine and proline metabolism’, ‘Alanine, aspartate, and glutamate metabolism’, ‘Glycine, serine, and threonine metabolism’, Arginine biosynthesis’, and ‘Glyoxylate and dicarboxylate metabolism’), 4 downregulated genes (ARG2, ODC1, GOT2, and SHMT1), 7 increased metabolites (Pro, Spmd, Cre, Bet, Trp, Asp, and Ala), and 2 decreased metabolites (NAA and For).
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BRCA1/BRCA1 | Breast cancer susceptibility gene 1 (gene/protein) |
| FA | Fatty acid |
| ACC1 | Acetyl-CoA carboxylase 1 |
| ROS | Reactive oxygen species |
| HIF-1 | Hypoxia-inducible-factor 1 |
| GOT2 | Glutamic-oxaloacetic transaminase 2 |
| NNMT | Nicotinamide N-methyltransferase |
| Ade | Adenine |
| PtS | Phosphatidylserine |
| PtC | Phosphatidylcholine |
| GSH/GSSG | Reduced/oxidized glutathione |
| Arg | Arginine |
| TNBC | Triple-negative breast cancer |
| Cre | Creatine |
| OPLS | Orthogonal partial least squares |
| CV-ANOVA | Cross-validated ANOVA |
| O2PLS | Two-way orthogonal partial least squares |
| ORA | Overrepresentation analysis |
| GSx | Total glutathione |
| tCr | Total creatine |
| MyI | Myoinositol |
| Tau | Taurine |
| Glu | Glutamic acid |
| BHB | 3-Hydroxybutyric acid |
| NAA | N-acetyl-L-aspartic acid |
| PC | Phosphorylcholine |
| ScI | Scylloinositol |
| TCA | Tricarboxylic acid cycle |
| ACoA | Acetyl-CoA |
| AKG | Alphaketoglutarate |
| NADH | Reduced nicotinamide adenine dinucleotide |
| NADPH | Reduced nicotinamide adenine dinucleotide phosphate |
| AMPK | AMP-activated protein kinase |
References
- Rosen, E.M.; Fan, S.; Ma, Y. BRCA1 Regulation of Transcription. Cancer Lett. 2006, 236, 175–185. [Google Scholar] [CrossRef]
- Moreau, K.; Dizin, E.; Ray, H.; Luquain, C.; Lefai, E.; Foufelle, F.; Billaud, M.; Lenoir, G.M.; Venezia, N.D. BRCA1 Affects Lipid Synthesis through Its Interaction with Acetyl-CoA Carboxylase. J. Biol. Chem. 2006, 281, 3172–3181. [Google Scholar] [CrossRef]
- Bae, I.; Fan, S.; Meng, Q.; Rih, J.K.; Kim, H.J.; Kang, H.J.; Xu, J.; Goldberg, I.D.; Jaiswal, A.K.; Rosen, E.M. BRCA1 Induces Antioxidant Gene Expression and Resistance to Oxidative Stress. Cancer Res. 2004, 64, 7893–7909. [Google Scholar] [CrossRef]
- Saha, T.; Rih, J.K.; Rosen, E.M. BRCA1 Down-regulates Cellular Levels of Reactive Oxygen Species. FEBS Lett. 2009, 583, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Gorrini, C.; Baniasadi, P.S.; Harris, I.S.; Silvester, J.; Inoue, S.; Snow, B.; Joshi, P.A.; Wakeham, A.; Molyneux, S.D.; Martin, B.; et al. BRCA1 Interacts with Nrf2 to Regulate Antioxidant Signaling and Cell Survival. J. Exp. Med. 2013, 210, 1529–1544. [Google Scholar] [CrossRef]
- Privat, M.; Radosevic-Robin, N.; Aubel, C.; Cayre, A.; Penault-Llorca, F.; Marceau, G.; Sapin, V.; Bignon, Y.-J.; Morvan, D. BRCA1 Induces Major Energetic Metabolism Reprogramming in Breast Cancer Cells. PLoS ONE 2014, 9, e102438. [Google Scholar] [CrossRef]
- Chiyoda, T.; Hart, P.C.; Eckert, M.A.; McGregor, S.M.; Lastra, R.R.; Hamamoto, R.; Nakamura, Y.; Yamada, S.D.; Olopade, O.I.; Lengyel, E.; et al. Loss of BRCA1 in the Cells of Origin of Ovarian Cancer Induces Glycolysis: A Window of Opportunity for Ovarian Cancer Chemoprevention. Cancer Prev. Res. 2017, 10, 255–266. [Google Scholar] [CrossRef]
- Kanakkanthara, A.; Kurmi, K.; Ekstrom, T.L.; Hou, X.; Purfeerst, E.R.; Heinzen, E.P.; Correia, C.; Huntoon, C.J.; O’Brien, D.; Wahner Hendrickson, A.E.; et al. BRCA1 Deficiency Upregulates NNMT, Which Reprograms Metabolism and Sensitizes Ovarian Cancer Cells to Mitochondrial Metabolic Targeting Agents. Cancer Res. 2019, 79, 5920–5929. [Google Scholar] [CrossRef] [PubMed]
- Van Der Groep, P.; Bouter, A.; Menko, F.H.; Van Der Wall, E.; Van Diest, P.J. High Frequency of HIF-1α Overexpression in BRCA1 Related Breast Cancer. Breast Cancer Res. Treat. 2008, 111, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Kim, H.J.; Rih, J.-K.; Mattson, T.L.; Kim, K.W.; Cho, C.-H.; Isaacs, J.S.; Bae, I. BRCA1 Plays a Role in the Hypoxic Response by Regulating HIF-1α Stability and by Modulating Vascular Endothelial Growth Factor Expression. J. Biol. Chem. 2006, 281, 13047–13056. [Google Scholar] [CrossRef]
- Hong, R.; Zhang, W.; Xia, X.; Zhang, K.; Wang, Y.; Wu, M.; Fan, J.; Li, J.; Xia, W.; Xu, F.; et al. Preventing BRCA 1/ZBRK 1 Repressor Complex Binding to the GOT2 Promoter Results in Accelerated Aspartate Biosynthesis and Promotion of Cell Proliferation. Mol. Oncol. 2019, 13, 959–977. [Google Scholar] [CrossRef]
- Zaidi, N.; Swinnen, J.V.; Smans, K. ATP-Citrate Lyase: A Key Player in Cancer Metabolism. Cancer Res. 2012, 72, 3709–3714. [Google Scholar] [CrossRef] [PubMed]
- Svensson, R.U.; Parker, S.J.; Eichner, L.J.; Kolar, M.J.; Wallace, M.; Brun, S.N.; Lombardo, P.S.; Van Nostrand, J.L.; Hutchins, A.; Vera, L.; et al. Inhibition of Acetyl-CoA Carboxylase Suppresses Fatty Acid Synthesis and Tumor Growth of Non-Small-Cell Lung Cancer in Preclinical Models. Nat. Med. 2016, 22, 1108–1119. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty Acid Synthase and the Lipogenic Phenotype in Cancer Pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Roig, B.; Rodríguez-Balada, M.; Samino, S.; Lam, E.W.-F.; Guaita-Esteruelas, S.; Gomes, A.R.; Correig, X.; Borràs, J.; Yanes, O.; Gumà, J. Metabolomics Reveals Novel Blood Plasma Biomarkers Associated to the BRCA1-Mutated Phenotype of Human Breast Cancer. Sci. Rep. 2017, 7, 17831. [Google Scholar] [CrossRef] [PubMed]
- Privat, M.; Aubel, C.; Arnould, S.; Communal, Y.; Ferrara, M.; Bignon, Y.-J. Breast Cancer Cell Response to Genistein Is Conditioned by BRCA1 Mutations. Biochem. Biophys. Res. Commun. 2009, 379, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Morvan, D.; Demidem, A. Metabolomics by Proton Nuclear Magnetic Resonance Spectroscopy of the Response to Chloroethylnitrosourea Reveals Drug Efficacy and Tumor Adaptive Metabolic Pathways. Cancer Res. 2007, 67, 2150–2159. [Google Scholar] [CrossRef]
- Bylesjö, M.; Eriksson, D.; Kusano, M.; Moritz, T.; Trygg, J. Data Integration in Plant Biology: The O2PLS Method for Combined Modeling of Transcript and Metabolite Data. Plant J. 2007, 52, 1181–1191. [Google Scholar] [CrossRef]
- Rios Garcia, M.; Steinbauer, B.; Srivastava, K.; Singhal, M.; Mattijssen, F.; Maida, A.; Christian, S.; Hess-Stumpp, H.; Augustin, H.G.; Müller-Decker, K.; et al. Acetyl-CoA Carboxylase 1-Dependent Protein Acetylation Controls Breast Cancer Metastasis and Recurrence. Cell Metab. 2017, 26, 842–855.e5. [Google Scholar] [CrossRef]
- Keenan, M.M.; Liu, B.; Tang, X.; Wu, J.; Cyr, D.; Stevens, R.D.; Ilkayeva, O.; Huang, Z.; Tollini, L.A.; Murphy, S.K.; et al. ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-Ketoglutarate. PLoS Genet. 2015, 11, e1005599. [Google Scholar] [CrossRef]
- Galdieri, L.; Vancura, A. Acetyl-CoA Carboxylase Regulates Global Histone Acetylation. J. Biol. Chem. 2012, 287, 23865–23876. [Google Scholar] [CrossRef]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.-H.; Sullivan, L.B.; Cheng, T.; Yang, Y.; Linehan, W.M.; Chandel, N.S.; DeBerardinis, R.J. Reductive Carboxylation Supports Growth in Tumour Cells with Defective Mitochondria. Nature 2011, 481, 385–388. [Google Scholar] [CrossRef]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive Glutamine Metabolism by IDH1 Mediates Lipogenesis under Hypoxia. Nature 2012, 481, 380–384. [Google Scholar] [CrossRef]
- Li, J.; Yu, T.; Zeng, P.; Tian, J.; Liu, P.; Qiao, S.; Wen, S.; Hu, Y.; Liu, Q.; Lu, W.; et al. Wild-Type IDH2 Is a Therapeutic Target for Triple-Negative Breast Cancer. Nat. Commun. 2024, 15, 3445. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, G.; Longo, V.D. Ketone Bodies in Cell Physiology and Cancer. Am. J. Physiol. Cell Physiol. 2024, 326, C948–C963. [Google Scholar] [CrossRef]
- Nagarajan, S.R.; Butler, L.M.; Hoy, A.J. The Diversity and Breadth of Cancer Cell Fatty Acid Metabolism. Cancer Metab. 2021, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Zhang, X.; Wang, G.; Zhang, W.; Cai, Z.; Pan, B.-S.; Gu, H.; Xu, C.; Jin, G.; Xu, X.; et al. Inositol Serves as a Natural Inhibitor of Mitochondrial Fission by Directly Targeting AMPK. Mol. Cell 2021, 81, 3803–3819.e7. [Google Scholar] [CrossRef] [PubMed]
- Paone, A.; Marani, M.; Fiascarelli, A.; Rinaldo, S.; Giardina, G.; Contestabile, R.; Paiardini, A.; Cutruzzolà, F. SHMT1 Knockdown Induces Apoptosis in Lung Cancer Cells by Causing Uracil Misincorporation. Cell Death Dis. 2014, 5, e1525. [Google Scholar] [CrossRef]
- Prokesch, A.; Pelzmann, H.J.; Pessentheiner, A.R.; Huber, K.; Madreiter-Sokolowski, C.T.; Drougard, A.; Schittmayer, M.; Kolb, D.; Magnes, C.; Trausinger, G.; et al. N-Acetylaspartate Catabolism Determines Cytosolic Acetyl-CoA Levels and Histone Acetylation in Brown Adipocytes. Sci. Rep. 2016, 6, 23723. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucaora, T.; Morvan, D. Joint Metabolomics and Transcriptomics Reveal Rewired Glycerophospholipid and Arginine Metabolism as Components of BRCA1-Induced Metabolic Reprogramming in Breast Cancer Cells. Metabolites 2025, 15, 534. https://doi.org/10.3390/metabo15080534
Lucaora T, Morvan D. Joint Metabolomics and Transcriptomics Reveal Rewired Glycerophospholipid and Arginine Metabolism as Components of BRCA1-Induced Metabolic Reprogramming in Breast Cancer Cells. Metabolites. 2025; 15(8):534. https://doi.org/10.3390/metabo15080534
Chicago/Turabian StyleLucaora, Thomas, and Daniel Morvan. 2025. "Joint Metabolomics and Transcriptomics Reveal Rewired Glycerophospholipid and Arginine Metabolism as Components of BRCA1-Induced Metabolic Reprogramming in Breast Cancer Cells" Metabolites 15, no. 8: 534. https://doi.org/10.3390/metabo15080534
APA StyleLucaora, T., & Morvan, D. (2025). Joint Metabolomics and Transcriptomics Reveal Rewired Glycerophospholipid and Arginine Metabolism as Components of BRCA1-Induced Metabolic Reprogramming in Breast Cancer Cells. Metabolites, 15(8), 534. https://doi.org/10.3390/metabo15080534




