Metabolomic Profiling of Hepatitis B-Associated Liver Disease Progression: Chronic Hepatitis B, Cirrhosis, and Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Information and Data Collection
2.2. Serum Preparation for Nuclear Magnetic Resonance (NMR)
2.3. 1H-NMR Data Processing and Multivariate Statistical Analysis
3. Results
3.1. The Clinical Characteristics of the Study Population of Patients with the Progression of HBV-Associated Liver Disease
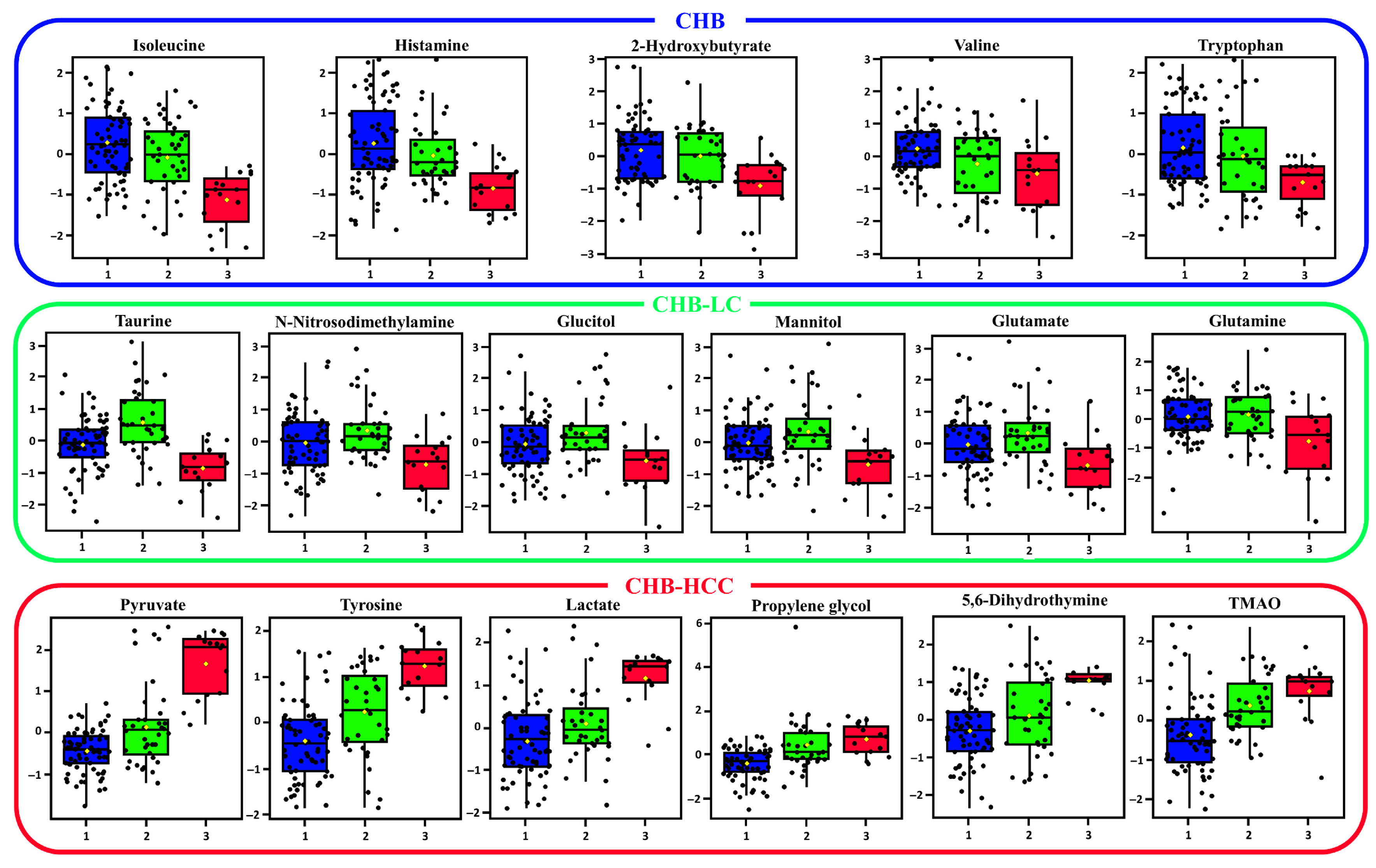
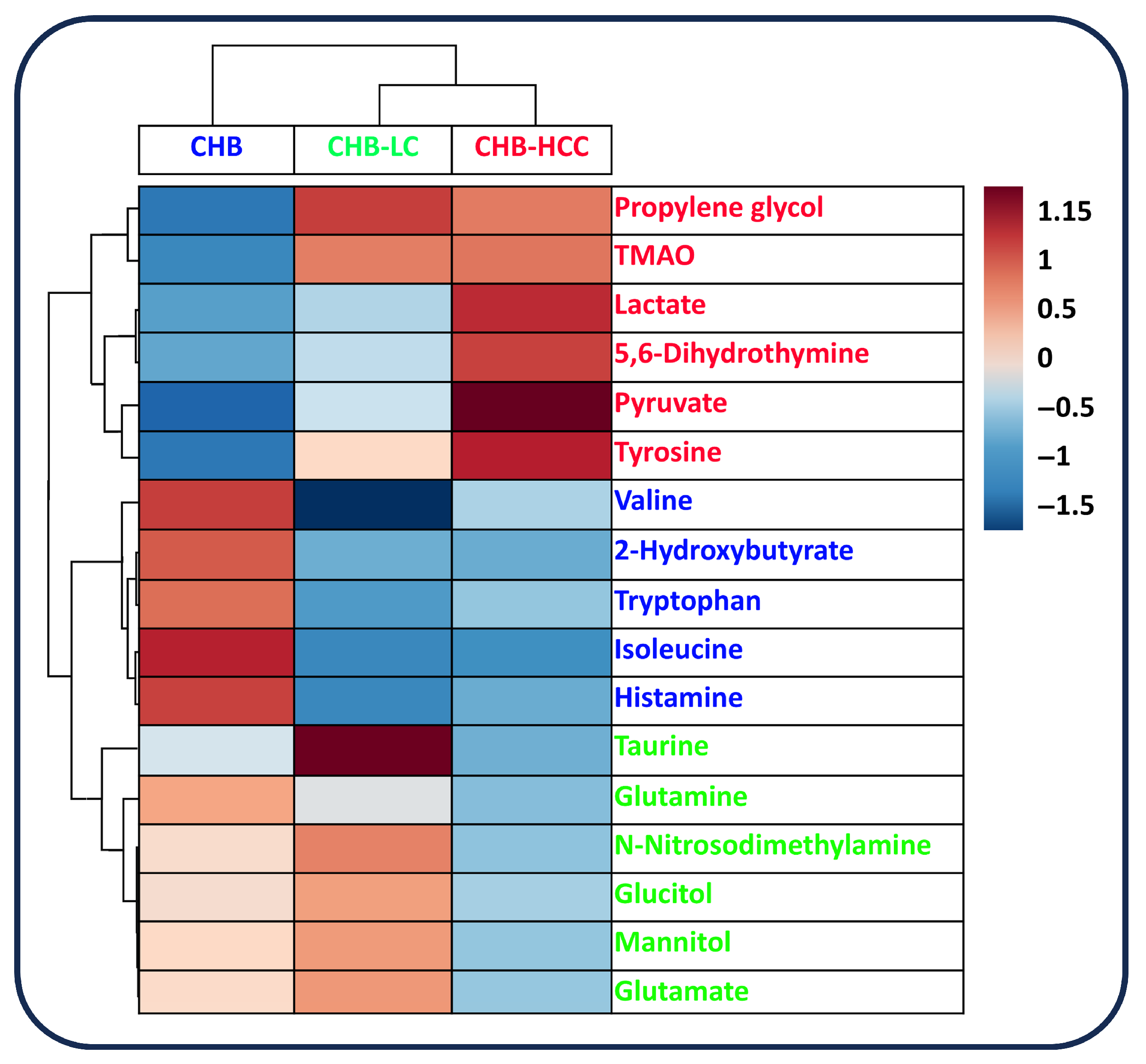
3.2. The Identification of Serum Metabolites in Patients with the Progression of HBV-Associated Liver Disease
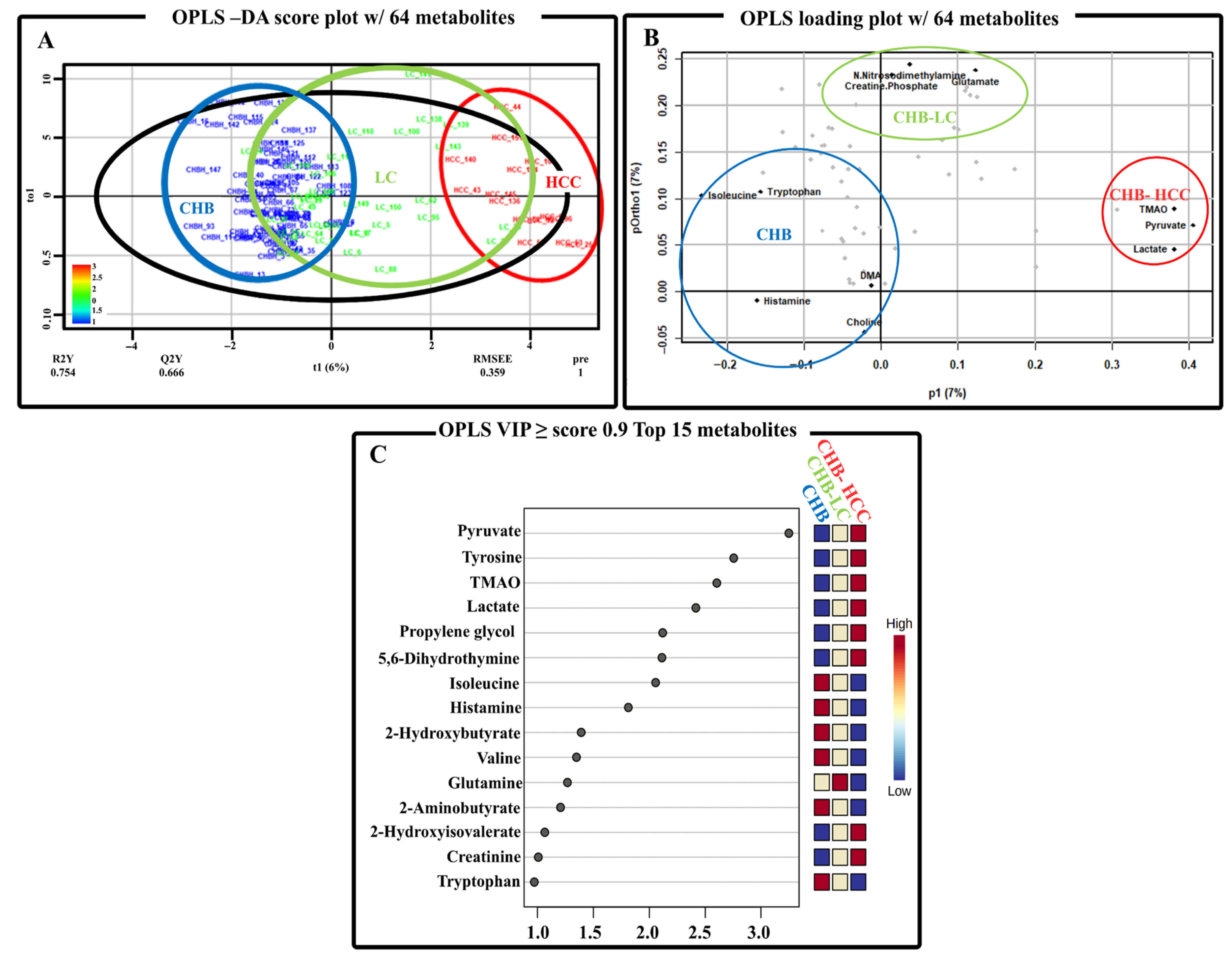
3.3. The Multivariate Analysis of Serum Metabolite Data from Patients with the Progression of HBV-Associated Liver Disease
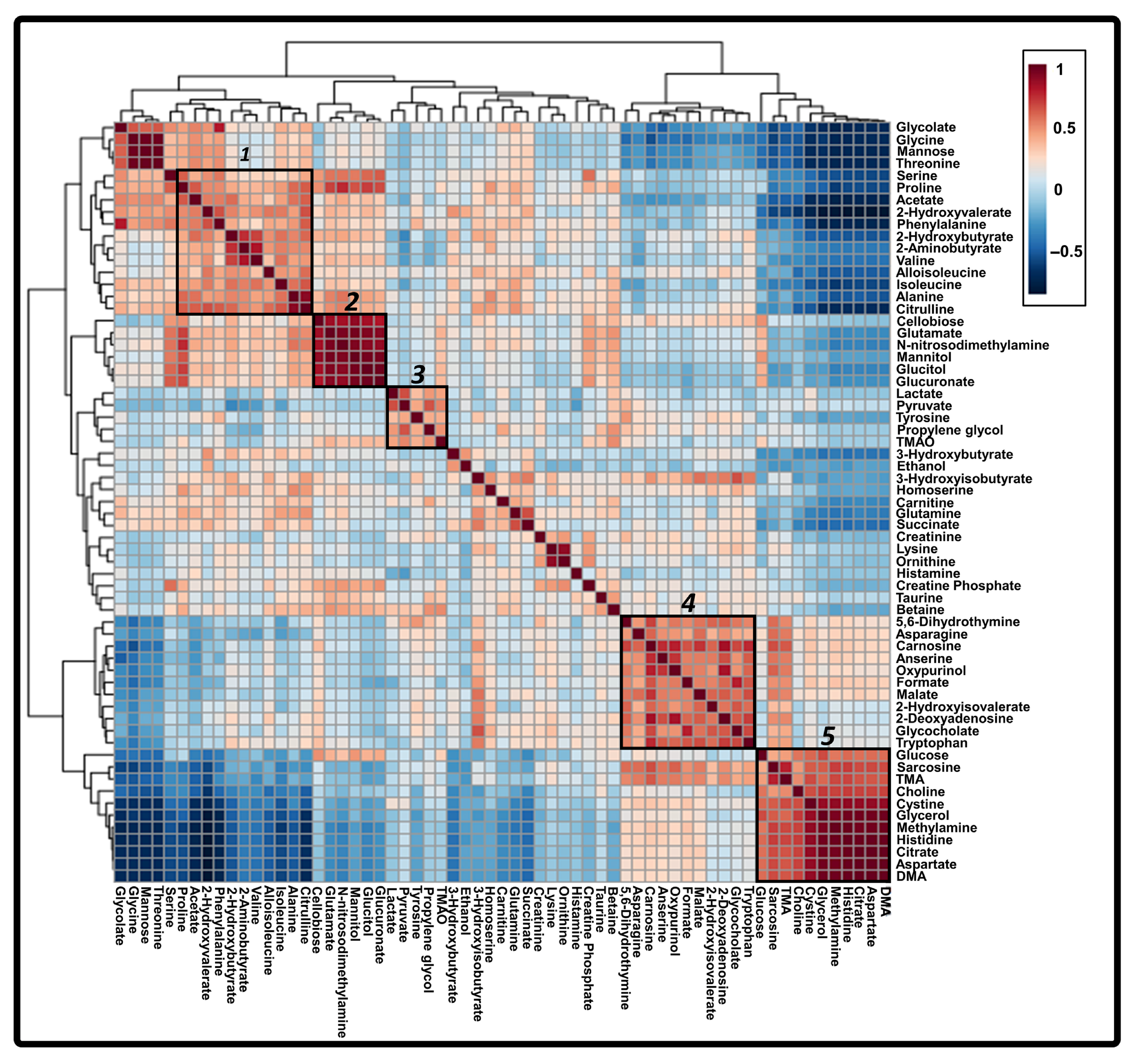
3.4. Correlated Metabolites of Six Core Discriminatory Metabolites in the Progression of HBV-Associated Liver Disease
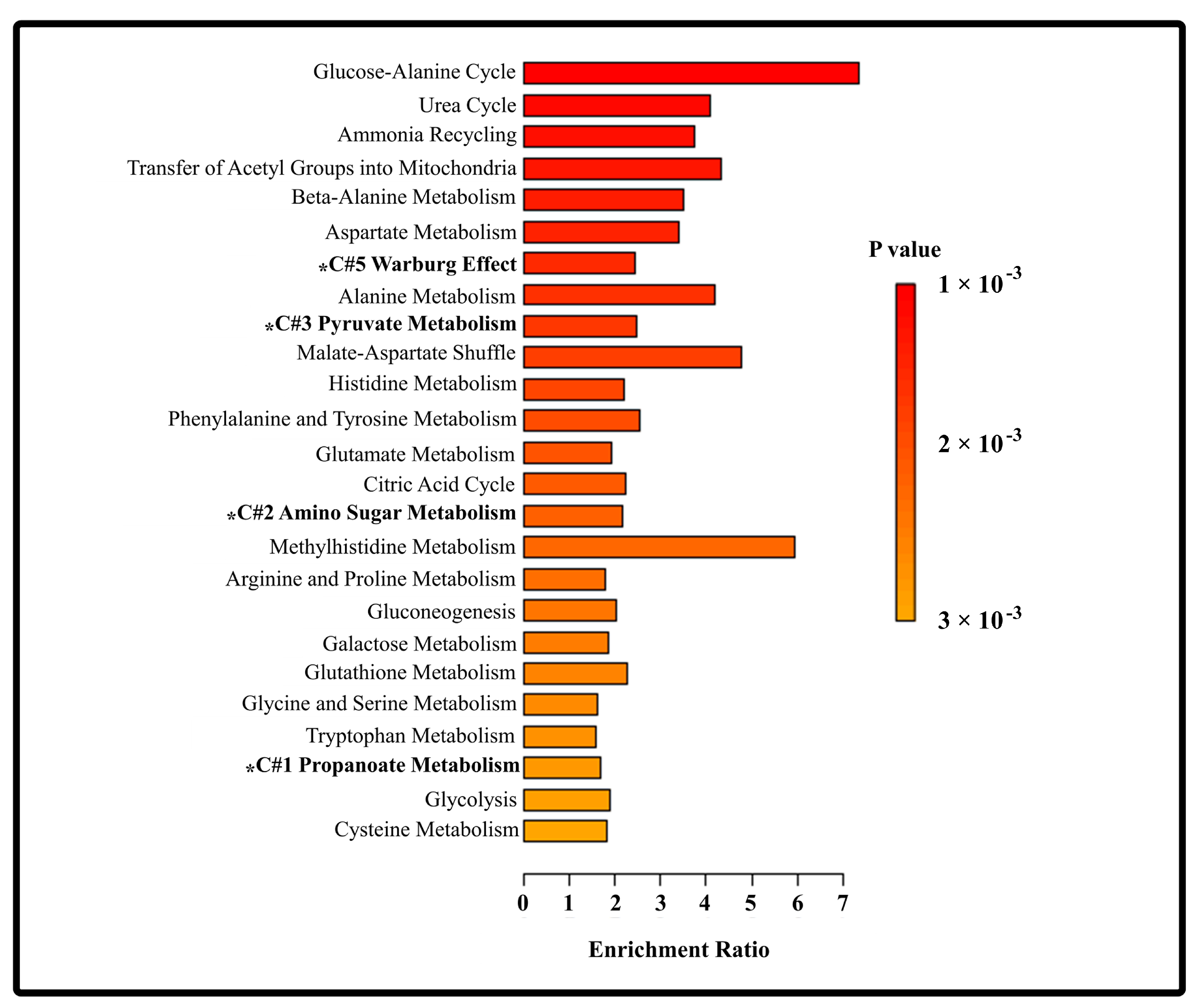
3.5. Metabolic Pathways Affected by the Progression of HBV-Associated Liver Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, H.; Chang, L.; Yan, Y.; Wang, L. Hepatitis B Virus Pre-S Region: Clinical Implications and Applications. Rev. Med. Virol. 2020, 31, e2201. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Cheng, J.; Fan, C.; Shi, X.; Cao, Y.; Sun, B.; Ding, H.; Hu, C.; Dong, F.; Yan, X. Serum Metabolomics to Identify the Liver Disease-Specific Biomarkers for the Progression of Hepatitis to Hepatocellular Carcinoma. Sci. Rep. 2015, 5, 18175. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond Biomarkers and towards Mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Gitto, S.; Schepis, F.; Andreone, P.; Villa, E. Study of the Serum Metabolomic Profile in Nonalcoholic Fatty Liver Disease: Research and Clinical Perspectives. Metabolites 2018, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, G.N.; Nagana Gowda, G.A.; Djukovic, D.; Raftery, D. Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach. Metabolites 2020, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Masarone, M.; Troisi, J.; Aglitti, A.; Torre, P.; Colucci, A.; Dallio, M.; Federico, A.; Balsano, C.; Persico, M. Untargeted Metabolomics as a Diagnostic Tool in NAFLD: Discrimination of Steatosis, Steatohepatitis and Cirrhosis. Metabolomics 2021, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Huang, C.; Gao, L.; Xu, Y.-J.; Chia, S.E.; Chen, S.; Li, N.; Yu, K.; Ling, Q.; Cheng, Q.; et al. Identification of serum biomarkers associated with hepatitis B virus-related hepatocellular carcinoma and liver cirrhosis using mass-spectrometry-based metabolomics. Metabolomics 2015, 11, 1526–1538. [Google Scholar] [CrossRef]
- Cai, F.-F.; Song, Y.-N.; Lu, Y.-Y.; Zhang, Y.; Hu, Y.-Y.; Su, S.-B. Analysis of plasma metabolic profile, characteristics and enzymes in the progression from chronic hepatitis B to hepatocellular carcinoma. Aging 2020, 12, 14949–14965. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.Y.; Wu, Q.Q.; Yin, Q.Q.; Dai, Y.N.; Huang, Y.C.; Zheng, W.; Hui, T.C.; Chen, M.J.; Wang, M.S.; Zhang, J.J.; et al. LC/MS-based global metabolomic identification of serum biomarkers differentiating hepatocellular carcinoma from chronic hepatitis B and liver cirrhosis. ACS Omega 2021, 6, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Lu, Q.; Liu, X.; Cong, H.; Zhao, L.; Wang, H.; Lin, D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009, 100, 782–785. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hong, Z.; Tan, G.; Dong, X.; Yang, G.; Zhao, L.; Chen, X.; Zhu, Z.; Lou, Z.; Qian, B.; et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int. J. Cancer 2014, 135, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xie, G.; Wang, X.; Fan, J.; Qiu, Y.; Zheng, X.; Qi, X.; Cao, Y.; Su, M.; Wang, X.; et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell. Proteom. 2011, 10, M110.004945. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.X.; Huang, C.J.; Yang, Z.G. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J. Gastroenterol. 2016, 22, 8161–8167. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhu, Y.; Cong, Q.; Wu, C. Metabonomics Research Progress on Liver Diseases. Can. J. Gastroenterol. Hepatol. 2017, 2017, e8467192. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.-L.; Yang, S.-S. Metabolic Signature of Hepatic Fibrosis: From Individual Pathways to Systems Biology. Cells 2019, 8, 1423. [Google Scholar] [CrossRef] [PubMed]
- Cortes, M.; Garcia-Cañaveras, J.C.; Pareja, E.; Lahoz, A. Liver Transplantation Biomarkers in the Metabolomics Era. In Biomarkers in Liver Disease; Preedy, V., Patel, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 99–128, ISBN-10: 9789400776746. [Google Scholar]
- Pu, W.; Wang, X.; Zhong, X.; Zhao, D.; Zeng, Z.; Cai, W.; Zhong, Y.; Huang, J.; Tang, D.; Dai, Y. Dysregulation of lipid metabolism in the pseudolobule promotes region-specific autophagy in hepatitis B liver cirrhosis. Hepatol. Commun. 2023, 7, e0187. [Google Scholar] [CrossRef] [PubMed]
- He, M.J.; Pu, W.; Wang, X.; Zhong, X.; Zhao, D.; Zeng, Z.; Cai, W.; Liu, J.; Huang, J.; Tang, D.; et al. Spatial metabolomics on liver cirrhosis to hepatocellular carcinoma progression. Cancer Cell Int. 2022, 22, 366. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Unger, K.; Alunni-Fabbroni, M.; Hirner-Eppeneder, H.; Öcal, E.; Zitzelsberger, H.; Mayerle, J.; Malfertheiner, P.; Ricke, J. Metabolomic Analysis of Human Cirrhosis and Hepatocellular Carcinoma: A Pilot Study. Dig. Dis. Sci. 2024, 69, 2488–2501. [Google Scholar] [CrossRef] [PubMed]
- Fitian, A.I.; Nelson, D.R.; Liu, C.; Xu, Y.; Ararat, M.; Cabrera, R. Integrated metabolomic profiling of hepatocellular carcinoma in hepatitis C cirrhosis through GC/MS and UPLC/MS-MS. Liver Int. 2014, 34, 1428–1444. [Google Scholar] [CrossRef] [PubMed]
- Manchel, A.; Mahadevan, R.; Bataller, R.; Hoek, J.B.; Vadigepalli, R. Genome-Scale Metabolic Modeling Reveals Sequential Dysregulation of Glutathione Metabolism in Livers from Patients with Alcoholic Hepatitis. Metabolites 2022, 12, 1157. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Izumi, N.; Charlton, M.R.; Sata, M. Branched-Chain Amino Acids as Pharmacological Nutrients in Chronic Liver Disease. Hepatology 2011, 54, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Meng, H.; Liu, S.; Wang, Y.; Yang, X.; Lu, F.; Wang, H. Identification of Key Metabolic Changes during Liver Fibrosis Progression in Rats Using a Urine and Serum Metabolomics Approach. Sci. Rep. 2017, 7, 11433. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Nishimura, T.; Aizawa, N.; Takashima, T.; Ikeda, N.; Yuri, Y.; Fujiwara, A.; Yoshihara, K.; Yoshioka, R.; Kawata, S.; et al. Branched chain amino acid-to-tyrosine ratio: Not only an indicator of the amino acid imbalance. Dig. Med. Res. 2022, 6, 1. [Google Scholar] [CrossRef]
- Lo, E.K.K.; Felicianna; Xu, J.H.; Zhan, Q.; Zeng, Z.; El-Nezami, H. The Emerging Role of Branched-Chain Amino Acids in Liver Diseases. Biomedicines 2022, 10, 1444. [Google Scholar] [CrossRef] [PubMed]
- Andrisic, L.; Dudzik, D.; Barbas, C.; Milkovic, L.; Grune, T.; Zarkovic, N. Short overview on metabolomics approach to study pathophysiology of oxidative stress in cancer. Redox Biol. 2018, 14, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, G.; Wang, X.; Zhou, M.; Yu, H.; Lin, Y.; Du, G.; Luo, G.; Liu, P. Urinary metabolite profiling offers potential for differentiation of liver-kidney yin deficiency and dampness-heat internal smoldering syndromes in posthepatitis B cirrhosis patients. Evid. Based Complement. Alternat. Med. 2015, 2015, 464969. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Liu, X.; Liu, Y.; Zhu, H.; Zhang, Y.; Zhang, Z.; Guo, H.; Yuan, W.; Qing, Z.; Zhang, N.; et al. Metabolic profiling: Shedding new light on the pathophysiological process and prognosis of HBV-related acute-on-chronic liver failure. Int. J. Clin. Exp. Med. 2017, 10, 7597–7610. [Google Scholar]
- Alonso-Peña, M.; Sanchez-Martin, A.; Sanchon-Sanchez, P.; Soto-Muñiz, M.; Espinosa-Escudero, R.; Marin, J.J.G. Pharmacogenetics of hepatocellular carcinoma and cholangiocarcinoma. Cancer Drug Resist. 2019, 2, 680–709. [Google Scholar] [CrossRef] [PubMed]
- Fages, A.; Duarte-Salles, T.; Stepien, M.; Ferrari, P.; Fedirko, V.; Pontoizeau, C.; Trichopoulou, A.; Aleksandrova, K.; Tjønneland, A.; Olsen, A.; et al. Metabolomic profiles of hepatocellular carcinoma in a European prospective cohort. BMC Med. 2015, 13, 242. [Google Scholar] [CrossRef] [PubMed]
- Teilhet, C.; Morvan, D.; Joubert-Zakeyh, J.; Biesse, A.-S.; Pereira, B.; Massoulier, S.; Dechelotte, P.; Pezet, D.; Buc, E.; Lamblin, G.; et al. Specificities of Human Hepatocellular Carcinoma Developed on Non-Alcoholic Fatty Liver Disease in Absence of Cirrhosis Revealed by Tissue Extracts 1H-NMR Spectroscopy. Metabolites 2017, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, S.E.; Abdel-Samiee, M.; El-Said, H.; Youssef, M.I.; ElZohry, H.A.; Abdelsameea, E.; Moaz, I.; Abdelwahab, S.F.; Elaskary, S.A.; Zaher, E.M.; et al. Evaluation of Amino Acids Profile as Non-Invasive Biomarkers of Hepatocellular Carcinoma in Egyptians. Trop. Med. Infect. Dis. 2022, 7, 437. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Cho, E.J.; Yu, K.-S.; Jang, I.-J.; Yoon, J.-H.; Park, T.; Cho, J.-Y. Comprehensive Metabolomic Search for Biomarkers to Differentiate Early Stage Hepatocellular Carcinoma from Cirrhosis. Cancers 2019, 11, 1497. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.Y.-L.; U, M.R.A.; Cox, I.J.; Taylor-Robinson, S.D. The Role of Mass Spectrometry in Hepatocellular Carcinoma Biomarker Discovery. Metabolites 2023, 13, 1059. [Google Scholar] [CrossRef] [PubMed]
- U, M.R.A.; Shen, E.Y.; Cartlidge, C.; Alkhatib, A.; Thursz, M.R.; Waked, I.; Gomaa, A.I.; Holmes, E.; Sharma, R.; Taylor-Robinson, S.D. Optimized Systematic Review Tool: Application to Candidate Biomarkers for the Diagnosis of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2022, 31, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, Z.; Luo, L.; Shu, D.; Wang, K. Metabolomics in hepatocellular carcinoma: From biomarker discovery to precision medicine. Front. Med. Technol. 2023, 4, 1065506. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The Human Serum Metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Zar, T.; Graeber, C.; Perazella, M.A. Reviews: Recognition, Treatment, and Prevention of Propylene Glycol Toxicity. Semin. Dial. 2007, 20, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.C.W.; Carella, M.A.; Papa, S.; Bubici, C. High Expression of Glycolytic Genes in Cirrhosis Correlates With the Risk of Developing Liver Cancer. Front. Cell. Dev. Biol. 2018, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Liu, C.; Qin, M.; Zhang, X.; Xi, T.; Yuan, S.; Hao, H.; Xiong, J. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm. Sin. B. 2022, 12, 558–580. [Google Scholar] [CrossRef] [PubMed]
- Spring, S.; Premathilake, H.; Bradway, C.; Shili, C.; DeSilva, U.; Carter, S.; Pezeshki, A. Effect of Very Low-Protein Diets Supplemented with Branched-Chain Amino Acids on Energy Balance, Plasma Metabolomics and Fecal Microbiome of Pigs. Sci. Rep. 2020, 10, 15859. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The Gut-Liver Axis in Liver Disease: Pathophysiological Basis for Therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Kawada, N. Role of the Gut–Liver Axis in Liver Inflammation, Fibrosis, and Cancer: A Special Focus on the Gut Microbiota Relationship. Hepatol. Commun. 2019, 3, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Gatarek, P.; Kaluzna-Czaplinska, J. Trimethylamine N-Oxide (TMAO) in Human Health. EXCLI J. 2021, 20, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.R.; Nguyen, M.H.; Lim, J.K. Hepatocellular carcinoma in patients with non-alcoholic fatty liver disease. World J. Gastroenterol. 2016, 22, 8294–8303. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Duan, Y.; Yang, L.; Schnabl, B. Small metabolites, possible big changes: A microbiota-centered view of non-alcoholic fatty liver disease. Gut 2019, 68, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-M.; Liu, Y.; Zhou, R.-F.; Chen, X.-L.; Wang, C.; Tan, X.-Y.; Wang, L.-J.; Zheng, R.-D.; Zhang, H.-W.; Ling, W.-H.; et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci. Rep. 2016, 6, 19076. [Google Scholar] [CrossRef] [PubMed]
- León-Mimila, P.; Villamil-Ramírez, H.; Li, X.; Shih, D.; Hui, S.; Ocampo-Medina, E.; López-Contreras, B.; Morán-Ramos, S.; Olivares-Arevalo, M.; Grandini-Rosales, P.; et al. Trimethylamine N-oxide levels are associated with NASH in obese subjects with type 2 diabetes. Diabetes Metab. 2021, 47, 101183. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, X.; Xu, J.; Xue, C.; Xue, Y.; Wang, Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 2014, 118, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Barrea, L.; Annunziata, G.; Muscogiuri, G.; Di Somma, C.; Laudisio, D.; Maisto, M.; de Alteriis, G.; Tenore, G.C.; Colao, A.; Savastano, S. Trimethylamine-N-oxide (TMAO) as Novel Potential Biomarker of Early Predictors of Metabolic Syndrome. Nutrients 2018, 10, 1971. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives. Biomolecules 2022, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Shariff, M.I.; Gomaa, A.I.; Cox, I.J.; Patel, M.; Williams, H.R.; Crossey, M.M.; Thillainayagam, A.V.; Thomas, H.C.; Waked, I.; Khan, S.A.; et al. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: A validation study. J. Proteome Res. 2011, 10, 1828–1836. [Google Scholar] [CrossRef] [PubMed]
- Cox, I.J.; Aliev, A.E.; Crossey, M.M.; Dawood, M.; Al-Mahtab, M.; Akbar, S.M.; Rahman, S.; Riva, A.; Williams, R.; Taylor-Robinson, S.D. Urinary nuclear magnetic resonance spectroscopy of a Bangladeshi cohort with hepatitis-B hepatocellular carcinoma: A biomarker corroboration study. World J. Gastroenterol. 2016, 22, 4191–4200. [Google Scholar] [CrossRef] [PubMed]
| Criteria | CHB (n = 64) | CHB-LC (n = 35) | CHB-HCC (n = 15) | p-Value |
|---|---|---|---|---|
| Age (years) | 49 (23–75) | 51 (32–74) | 51 (33–83) | 0.6551 |
| Gender (Male) | 48 (75.0) | 24 (68.6) | 14 (93.3) | 0.2661 |
| Hypertension | 9 (14.1) | 8 (22.9) | 3 (20.0) | 0.1889 |
| Diabetes Mellitus | 10 (15.6) | 4 (11.4) | 2 (13.3) | 0.3528 |
| Hyperlipidemia | 5 (7.8) | 2(5.7) | 1 (6.6) | 0.5002 |
| Body Mass Index (kg/m2) | 24.8 (18.1–44.4) | 24.0 (18.1–29.0) | 23.1 (16.5–28.4) | 0.1778 |
| Stiffness (kPa) | 5.85 (3.3–9.9) | 12.4 (14.7–46.4) | 13.0 (6.0–33.8) | <0.0001 |
| Total Bilirubin (mg/dL) | 0.75 (0.2–12.7) | 0.95 (0.2–4.9) | 1.3 (0.4–19.9) | 0.02144 |
| Albumin (mg/dL) | 4.2 (2.9–4.8) | 4.0 (2.2–5.2) | 2.9 (2.1–4.3) | 0.002864 |
| AST (IU/dL) | 54 (12–2290) | 52.5 (20–1430) | 90 (51–369) | 0.01426 |
| ALT (IU/dL) | 84 (4–2140) | 42.5 (15–1402) | 52 (11–759) | 0.2416 |
| GGT (mg/dL) | 37 (11–767) | 53.5 (9–889) | 163 (13–535) | 0.02091 |
| Platelet (103/μL) | 206 (106–465) | 167 (22–300) | 159 (64–360) | 0.007749 |
| INR | 1.06 (0.93–1.37) | 1.16 (0.93–1.96) | 1.24 (0.96–2.27) | 0.005469 |
| Creatinine (mg/dL) | 0.75 (0.46–4.29) | 0.72 (0.50–3.52) | 0.90 (0.36–5.11) | 0.3903 |
| HBeAg (+) | 31 (48.4) | 15 (42.9) | 5 (33.3) | 0.5657 |
| HBV DNA (IU/L) | 2 × 106 (0–1 × 109) | 7 × 105 (0–2 × 108) | 1 × 106 (4.0 × 101–1 × 107) | 0.2458 |
| Metabolites | f. Value | p Value | FDR | Fisher’s LSD |
|---|---|---|---|---|
| Pyruvate | 52.265 | 9.27 × 10−17 | 5.84 × 10−15 | 2-1; 3-1; 3-2 |
| Tyrosine | 24.954 | 1.09 × 10−9 | 3.43 × 10−18 | 2-1; 3-1; 3-2 |
| Lactate | 17.216 | 3.02 × 10−7 | 6.35 × 10−6 | 2-1; 3-1; 3-2 |
| Propylene glycol | 15.883 | 8.46 × 10−7 | 1.33 × 10−5 | 2-1; 3-1 |
| Isoleucine | 15.075 | 1.59 × 10−6 | 2.01 × 10−5 | 1-3; 2-3 |
| Taurine | 14.629 | 2.27 × 10−6 | 2.38 × 10−5 | 2-1; 1-3; 2-3 |
| TMAO | 13.295 | 6.60 × 10−6 | 5.19 × 10−5 | 2-1; 3-1 |
| Histamine | 8.857 | 0.00026851 | 0.0018796 | 1-3; 2-3 |
| 2-Hydroxybutyrate | 8.0749 | 0.00052966 | 0.0033368 | 1-3; 2-3 |
| N-Nitrosodimethylamine | 6.6211 | 0.0019149 | 0.010967 | 1-3; 2-3 |
| Mannitol | 5.8654 | 0.0037796 | 0.019843 | 1-3; 2-3 |
| Glutamate | 5.7421 | 0.0042263 | 0.020481 | 1-3; 2-3 |
| Valine | 5.3763 | 0.0058952 | 0.025792 | 1-2; 1-3 |
| Glutamine | 5.3315 | 0.006141 | 0.025792 | 1-3; 2-3 |
| Tryptophan | 4.7257 | 0.010707 | 0.042159 | 1-3; 2-3 |
| Glucitol | 4.6325 | 0.011668 | 0.043241 | 2-3 |
| 5,6-Dihydrothymine | 13.607 | 5.13 × 10−6 | 4.62 × 10−5 | 2-1; 3-1; 3-2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.; Baritugo, K.-A.G.; Kim, J.; Park, G.; Han, K.J.; Lee, S.; Sung, G.-H. Metabolomic Profiling of Hepatitis B-Associated Liver Disease Progression: Chronic Hepatitis B, Cirrhosis, and Hepatocellular Carcinoma. Metabolites 2025, 15, 504. https://doi.org/10.3390/metabo15080504
Oh J, Baritugo K-AG, Kim J, Park G, Han KJ, Lee S, Sung G-H. Metabolomic Profiling of Hepatitis B-Associated Liver Disease Progression: Chronic Hepatitis B, Cirrhosis, and Hepatocellular Carcinoma. Metabolites. 2025; 15(8):504. https://doi.org/10.3390/metabo15080504
Chicago/Turabian StyleOh, Junsang, Kei-Anne Garcia Baritugo, Jayoung Kim, Gyubin Park, Ki Jun Han, Sangheun Lee, and Gi-Ho Sung. 2025. "Metabolomic Profiling of Hepatitis B-Associated Liver Disease Progression: Chronic Hepatitis B, Cirrhosis, and Hepatocellular Carcinoma" Metabolites 15, no. 8: 504. https://doi.org/10.3390/metabo15080504
APA StyleOh, J., Baritugo, K.-A. G., Kim, J., Park, G., Han, K. J., Lee, S., & Sung, G.-H. (2025). Metabolomic Profiling of Hepatitis B-Associated Liver Disease Progression: Chronic Hepatitis B, Cirrhosis, and Hepatocellular Carcinoma. Metabolites, 15(8), 504. https://doi.org/10.3390/metabo15080504







