Nutri-Epigenetic Regulation of Vitamin D—Impact on Metabolism and Biological Functions: Narrative Review
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
3. Development
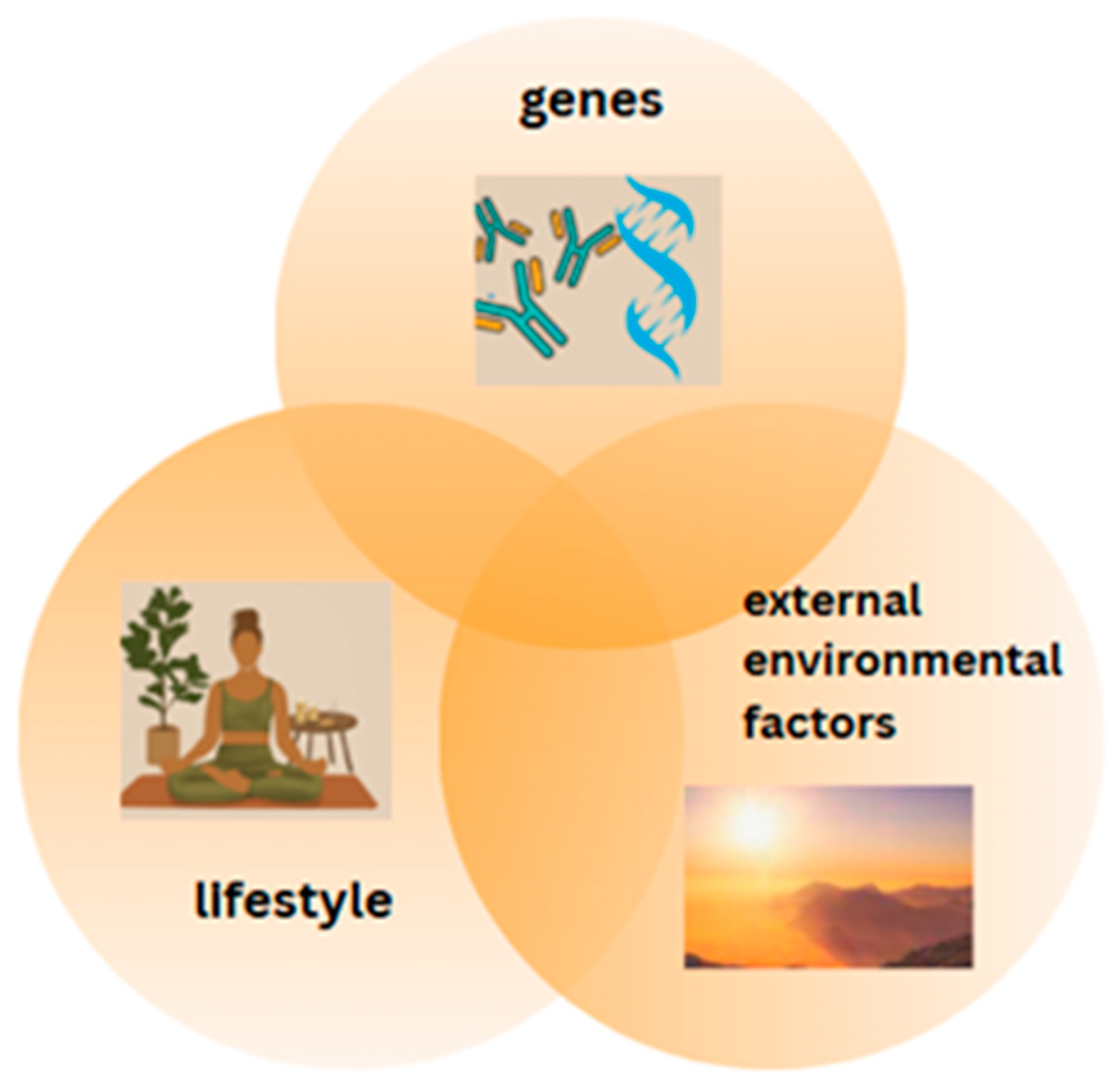
3.1. The Influence of Intracellular Factors and Environmental Stimuli on Epigenetic Modifications
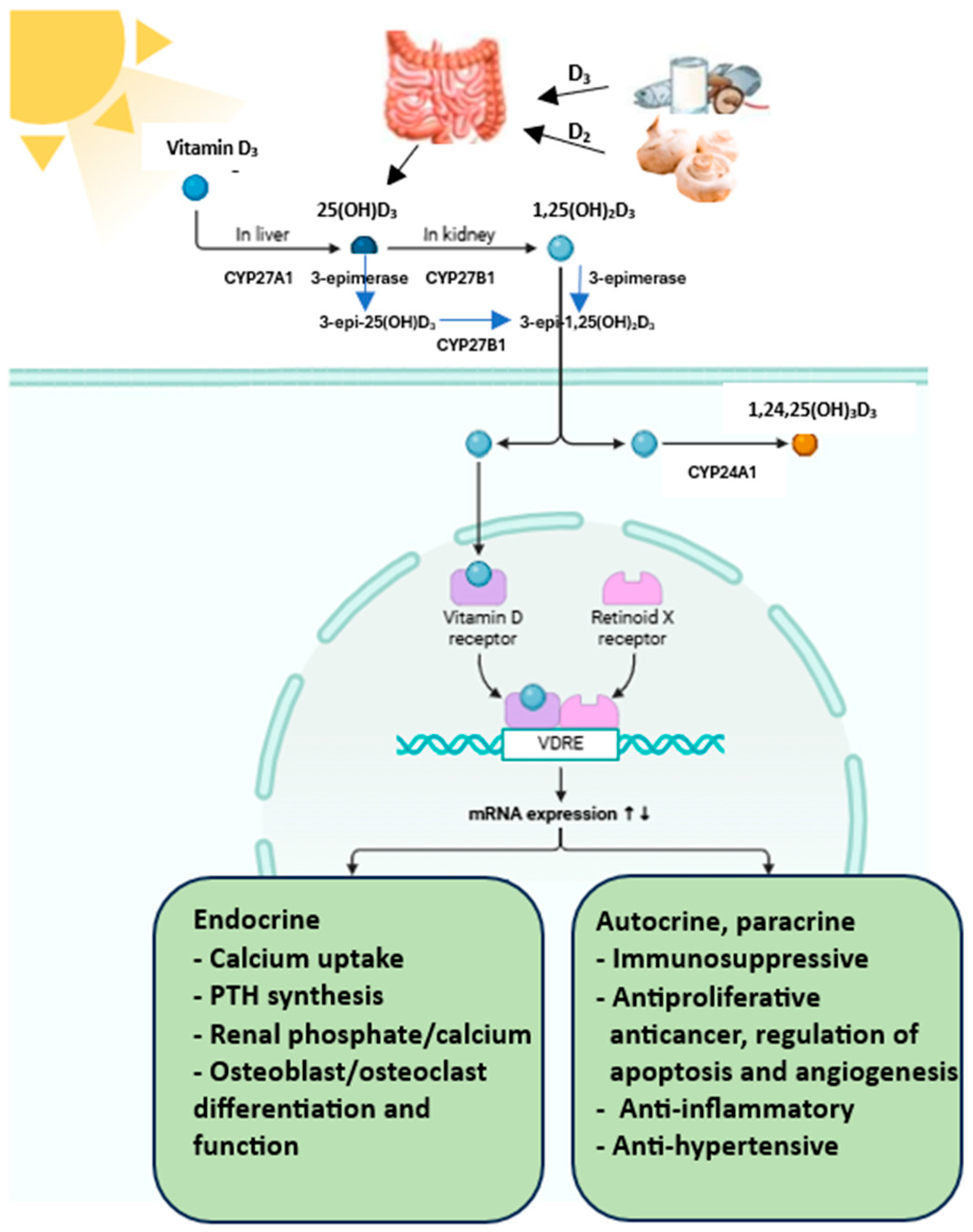
3.2. Metabolism of Vitamin D
3.3. Genetic Determinants of Vitamin D Metabolism/Key Genes Associated with Vitamin D Metabolism
3.3.1. VDR Receptor
3.3.2. NADSYN1/DHCR7
3.3.3. Vitamin D Hydroxylases
CYP2R1
CYP27A1
CYP27B1
CYP24A1
3.3.4. GC (Group-Specific Component)
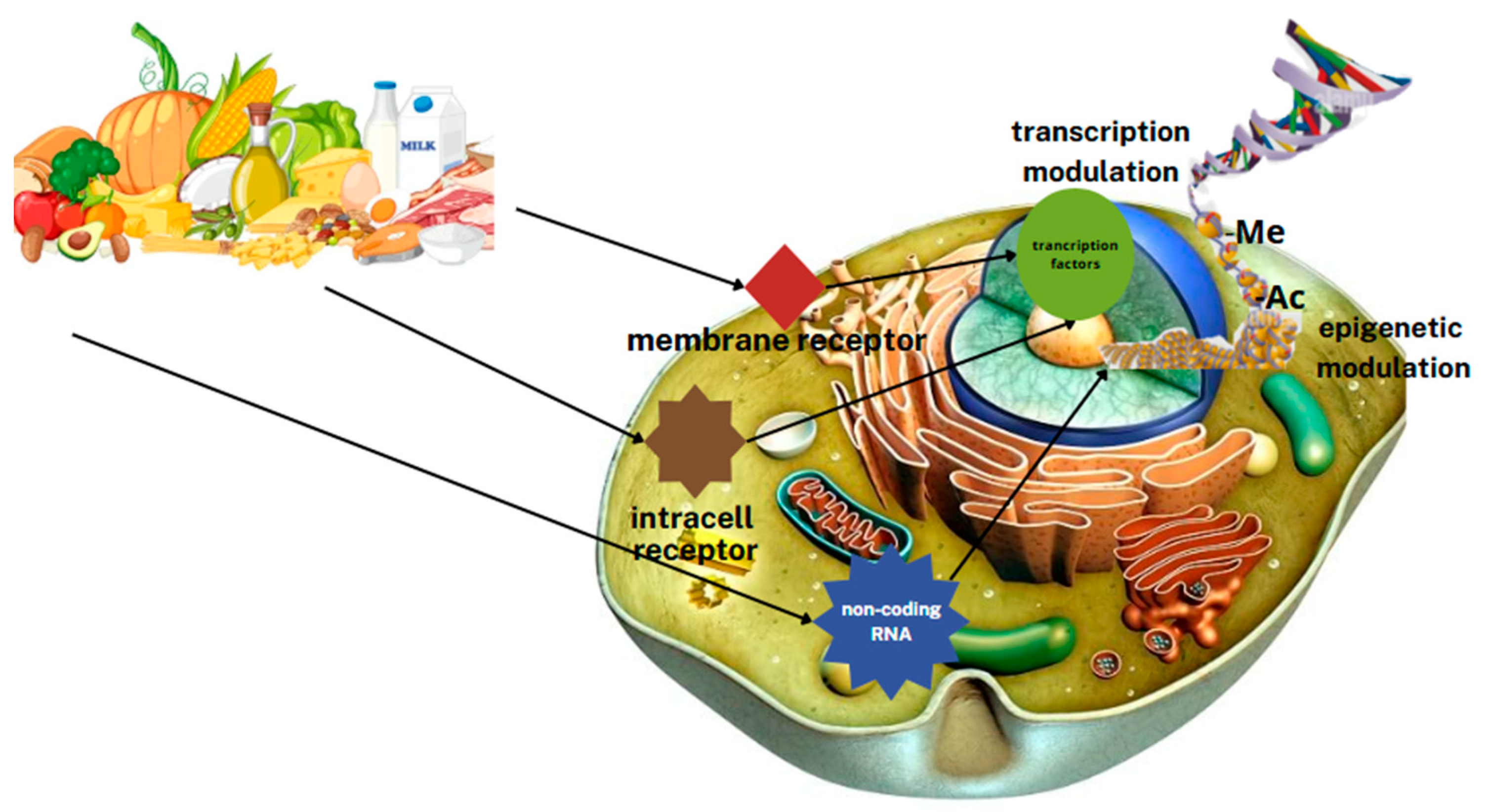
3.4. Nutritional Epigenomics
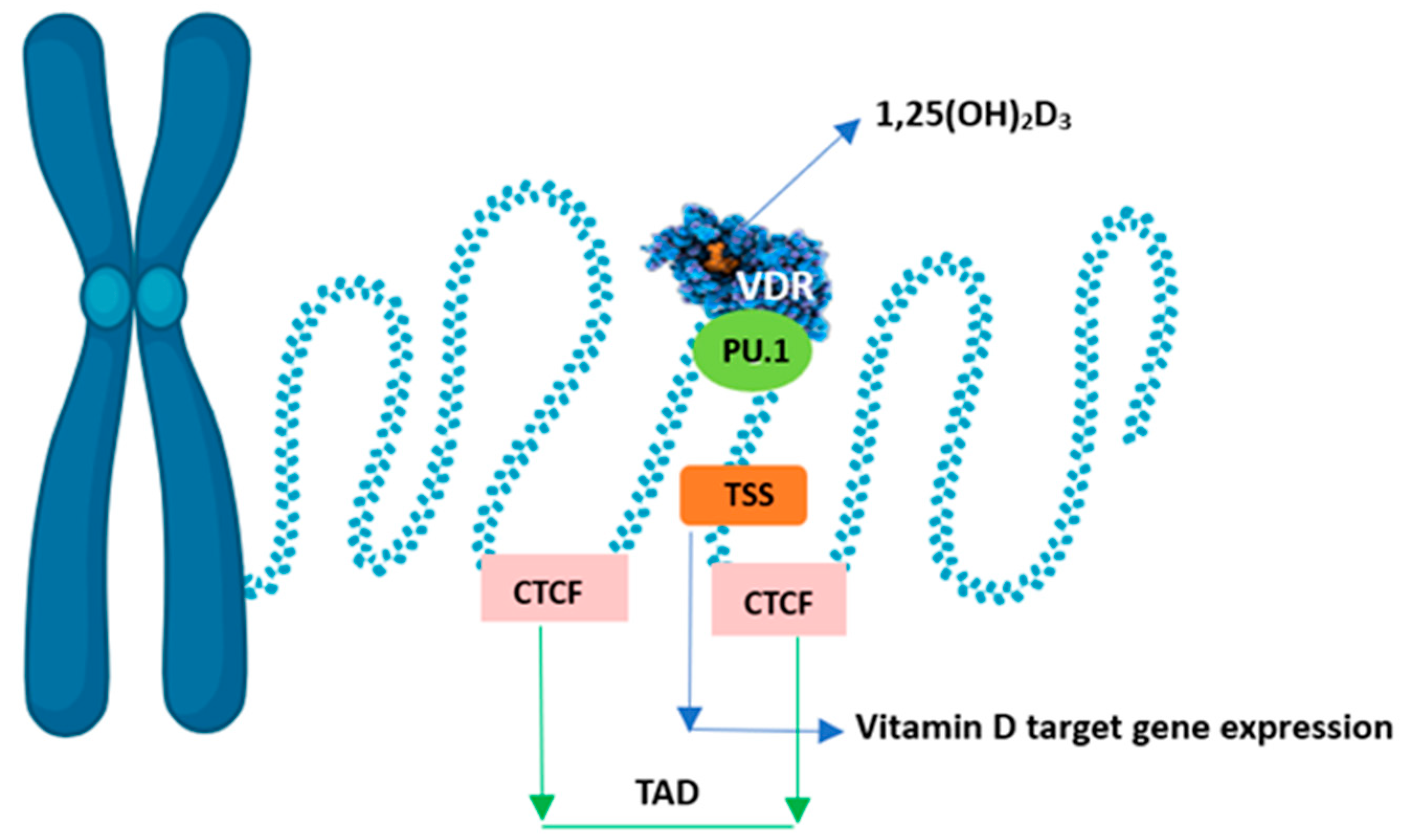
3.5. Epigenetics of Vitamin D
3.5.1. Effect of Vitamin D on DNA Methylation
3.5.2. Effect of Vitamin D on Chromatin
3.5.3. Histone Modifications
3.5.4. MicroRNA
3.6. Polymorphisms of Vitamin D-Related Genes
3.6.1. VDR Gene Polymorphism
3.6.2. NADSYN1/DHCR7 Gene Polymorphism
3.6.3. CYP2R1 Gene Polymorphism
3.6.4. CYP27B1 Gene Polymorphism
3.6.5. CYP24A1 Gene Polymorphism
3.6.6. GC Gene Polymorphism
3.7. Personalisation of Vitamin D Supplementation in Response to Genetic Differences–Future Directions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baethge, C.; Goldbeck-Wood, S.; Mertens, S. SANRA—A scale for the quality assessment of narrative review articles. Res. Integr. Peer Rev. 2019, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Snegarova, V.; Naydenova, D. Vitamin D: A Review of its Effects on Epigenetics and Gene Regulation. Folia Med. 2020, 62, 662–667. [Google Scholar] [CrossRef]
- Dauncey, M.J.; White, P.; Burton, K.A.; Katsumata, M. Nutrition-hormone receptor-gene interactions: Implications for development and disease. Proc. Nutr. Soc. 2001, 60, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, C.; Madeo, G.; Macchia, A.; Donato, K.; Cristoni, S.; Ceccarini, M.R.; Beccari, T.; Laconelli, A.; Aquilanti, B.; Matera, G.; et al. Nutrigenomics: SNPs correlated to vitamins’ deficiencies. Clin. Ter. 2023, 174, 173–182. [Google Scholar]
- Fenech, M.; Baghurst, P.; Luderer, W.; Turner, J.; Record, S.; Ceppi, M.; Bonassi, S. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability—Results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis 2005, 26, 991–999. [Google Scholar] [PubMed]
- Mierziak, J.; Kostyn, K.; Boba, A.; Czemplik, M.; Kulma, A.; Wojtasik, W. Influence of the Bioactive Diet Components on the Gene Expression Regulation. Nutrients 2021, 13, 3673. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, X.; Shi, H.; Su, S.; Harshfield, G.A.; Gutin, B.; Snieder, H.; Dong, Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J. Pediatr. 2013, 162, 1004–1009.e1. [Google Scholar] [CrossRef] [PubMed]
- Fetahu, I.S.; Höbaus, J.; Kállay, E. Vitamin D and the epigenome. Front. Physiol. 2014, 5, 164. [Google Scholar] [CrossRef]
- Pereira-Santos, M.; Costa, P.R.; Assis, A.M.; Santos, C.A.; Santos, D.B. Obesity and vitamin D deficiency: A systematic review and meta-analysis. Obes. Rev. 2015, 16, 341–349. [Google Scholar] [CrossRef]
- Sergeev, I.N. Vitamin D Status and Vitamin D-Dependent Apoptosis in Obesity. Nutrients 2020, 12, 1392. [Google Scholar] [CrossRef]
- Wimalawansa, S.J. Vitamin D Deficiency: Effects on Oxidative Stress, Epigenetics, Gene Regulation, and Aging. Biology 2019, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Karlic, H.; Varga, F. Impact of vitamin D metabolism on clinical epigenetics. Clin. Epigenet. 2011, 2, 55–61. [Google Scholar] [CrossRef]
- Wang, H.; Chen, W.; Li, D.; Yin, X.; Zhang, X.; Olsen, N.; Zheng, S.G. Vitamin D and Chronic Diseases. Aging Dis. 2017, 8, 346–353. [Google Scholar] [CrossRef]
- Bell, D.S. Protean manifestations of vitamin D deficiency, part 2: Deficiency and its association with autoimmune disease, cancer, infection, asthma, dermopathies, insulin resistance, and type 2 diabetes. South. Med. J. 2011, 104, 335–339. [Google Scholar] [CrossRef]
- Aiello, G.; Lombardo, M.; Baldelli, S. Exploring Vitamin D Synthesis and Function in Cardiovascular Health: A Narrative Review. Appl. Sci. 2024, 14, 4339. [Google Scholar] [CrossRef]
- Jiang, X.; Kiel, D.P.; Kraft, P. The genetics of vitamin D. Bone 2019, 126, 59–77. [Google Scholar] [CrossRef]
- Webb, A.R.; Holick, M.F. The role of sunlight in the cutaneous production of vitamin D3. Annu. Rev. Nutr. 1988, 8, 375–399. [Google Scholar] [CrossRef]
- Kift, R.; Rhodes, L.E.; Farrar, M.D.; Webb, A.R. Is Sunlight Exposure Enough to Avoid Wintertime Vitamin D Deficiency in United Kingdom Population Groups? Int. J. Environ. Res. Public Health 2018, 15, 1624. [Google Scholar] [CrossRef]
- Rice, S.A.; Carpenter, M.; Fityan, A.; Vearncombe, L.M.; Ardern-Jones, M.; Jackson, A.A.; Cooper, C.; Baird, J.; Healy, E. Limited exposure to ambient ultraviolet radiation and 25-hydroxyvitamin D levels: A systematic review. Br. J. Dermatol. 2015, 172, 652–661. [Google Scholar] [CrossRef]
- Saponaro, F.; Saba, A.; Zucchi, R. An Update on Vitamin D Metabolism. Int. J. Mol. Sci. 2020, 21, 6573. [Google Scholar] [CrossRef]
- Prabhu, A.V.; Luu, W.; Li, D.; Sharpe, L.J.; Brown, A.J. DHCR7: A vital enzyme switch between cholesterol and vitamin D production. Prog. Lipid Res. 2016, 64, 138–151. [Google Scholar] [CrossRef]
- Delrue, C.; Speeckaert, M.M. Vitamin D and Vitamin D-Binding Protein in Health and Disease. Int. J. Mol. Sci. 2023, 24, 4642. [Google Scholar] [CrossRef]
- Mulligan, G.B.; Licata, A. Taking vitamin D with the largest meal improves absorption and results in higher serum levels of 25-hydroxyvitamin D. J. Bone Miner. Res. 2010, 25, 928–930. [Google Scholar] [CrossRef]
- Jones, G.; Prosser, D.E.; Kaufmann, M. Cytochrome P450-mediated metabolism of vitamin D. J. Lipid Res. 2014, 55, 13–31. [Google Scholar] [CrossRef]
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, Molecular Mechanism of Action, and Pleiotropic Effects. Physiol. Rev. 2016, 96, 365–408. [Google Scholar] [CrossRef]
- Portales-Castillo, I.; Simic, P. PTH, FGF-23, Klotho and Vitamin D as regulators of calcium and phosphorus: Genetics, epigenetics and beyond. Front. Endocrinol. 2022, 13, 992666. [Google Scholar] [CrossRef]
- Young, K.; Beggs, M.R.; Grimbly, C.; Alexander, R.T. Regulation of 1 and 24 hydroxylation of vitamin D metabolites in the proximal tubule. Exp. Biol. Med. 2022, 247, 1103–1111. [Google Scholar] [CrossRef]
- Quarles, L.D. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nat. Rev. Endocrinol. 2012, 8, 276–286. [Google Scholar] [CrossRef]
- Shimada, T.; Kakitani, M.; Yamazaki, Y.; Hasegawa, H.; Takeuchi, Y.; Fujita, T.; Fukumoto, S.; Tomizuka, K.; Yamashita, T. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Investig. 2004, 113, 561–568. [Google Scholar] [CrossRef]
- Bikle, D.D. Vitamin D metabolism, mechanism of action, and clinical applications. Chem. Biol. 2014, 21, 319–329. [Google Scholar] [CrossRef]
- Bailey, D.; Veljkovic, K.; Yazdanpanah, M.; Adeli, K. Analytical measurement and clinical relevance of vitamin D(3) C3-epimer. Clin. Biochem. 2013, 46, 190–196. [Google Scholar] [CrossRef]
- Abouzid, M.; Karaźniewicz-Łada, M.; Pawlak, K.; Burchardt, P.; Kruszyna, Ł.; Główka, F. Measurement of plasma 25-hydroxyvitamin D2, 25-hydroxyvitamin D3 and 3-epi-25-hydroxyvitamin D3 in population of patients with cardiovascular disease by UPLC-MS/MS method. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1159, 122350. [Google Scholar] [CrossRef]
- Meza-Meza, M.R.; Ruiz-Ballesteros, A.I.; de la Cruz-Mosso, U. Functional effects of vitamin D: From nutrient to immunomodulator. Crit. Rev. Food Sci. Nutr. 2022, 62, 3042–3062. [Google Scholar] [CrossRef]
- Carlberg, C. Vitamin D Genomics: From In Vitro to In Vivo. Front. Endocrinol. 2018, 9, 250. [Google Scholar] [CrossRef]
- Maestro, M.A.; Molnár, F.; Mouriño, A.; Carlberg, C. Vitamin D receptor 2016: Novel ligands and structural insights. Expert Opin. Ther. Pat. 2016, 26, 1291–1306. [Google Scholar] [CrossRef]
- Carlberg, C. Vitamin D and Its Target Genes. Nutrients 2022, 14, 1354. [Google Scholar] [CrossRef]
- Pike, J.W.; Meyer, M.B.; Lee, S.M.; Onal, M.; Benkusky, N.A. The vitamin D receptor: Contemporary genomic approaches reveal new basic and translational insights. J. Clin. Investig. 2017, 127, 1146–1154. [Google Scholar] [CrossRef]
- Haussler, M.R.; Jurutka, P.W.; Mizwicki, M.; Norman, A.W. Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2vitamin D3: Genomic and non-genomic mechanisms. Best. Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 543–559. [Google Scholar] [CrossRef]
- Shaffer, P.L.; Gewirth, D.T. Structural analysis of RXR-VDR interactions on DR3 DNA. J. Steroid Biochem. Mol. Biol. 2004, 89–90, 215–219. [Google Scholar] [CrossRef]
- Trochoutsou, A.I.; Kloukina, V.; Samitas, K.; Xanthou, G. Vitamin-D in the Immune System: Genomic and Non-Genomic Actions. Mini Rev. Med. Chem. 2015, 15, 953–963. [Google Scholar] [CrossRef]
- Seraphin, G.; Rieger, S.; Hewison, M.; Capobianco, E.; Lisse, T.S. The impact of vitamin D on cancer: A mini review. J. Steroid Biochem. Mol. Biol. 2023, 231, 106308. [Google Scholar] [CrossRef] [PubMed]
- Tuoresmäki, P.; Väisänen, S.; Neme, A.; Heikkinen, S.; Carlberg, C. Patterns of genome-wide VDR locations. PLoS ONE 2014, 9, e96105. [Google Scholar] [CrossRef]
- Hii, C.S.; Ferrante, A. The Non-Genomic Actions of Vitamin D. Nutrients 2016, 8, 135. [Google Scholar] [CrossRef] [PubMed]
- Khanal, R.C.; Nemere, I. The ERp57/GRp58/1,25D3-MARRS receptor: Multiple functional roles in diverse cell systems. Curr. Med. Chem. 2007, 14, 1087–1093. [Google Scholar] [CrossRef]
- Mavar, M.; Sorić, T.; Bagarić, E.; Sarić, A.; Matek Sarić, M. The Power of Vitamin D: Is the Future in Precision Nutrition through Personalized Supplementation Plans? Nutrients 2024, 16, 1176. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Riley, E.M.; Meyer, M.B.; Benkusky, N.A.; Plum, L.A.; DeLuca, H.F.; Pike, J.W. 1,25-Dihydroxyvitamin D3 Controls a Cohort of Vitamin D Receptor Target Genes in the Proximal Intestine That Is Enriched for Calcium-regulating Components. J. Biol. Chem. 2015, 290, 18199–18215. [Google Scholar] [CrossRef]
- Carlberg, C.; Raczyk, M.; Zawrotna, N. Vitamin D: A master example of nutrigenomics. Redox Biol. 2023, 62, 102695. [Google Scholar] [CrossRef]
- Demay, M.B.; Pittas, A.G.; Bikle, D.D.; Diab, D.L.; Kiely, M.E.; Lazaretti-Castro, M.; Lips, P.; Mitchell, D.M.; Murad, M.H.; Powers, S.; et al. Vitamin D for the Prevention of Disease: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2024, 109, 1907–1947, Erratum in J. Clin. Endocrinol. Metab. 2025, 110, e916. [Google Scholar] [CrossRef]
- Grant, W.B.; Wimalawansa, S.J.; Pludowski, P.; Cheng, R.Z. Vitamin D: Evidence-Based Health Benefits and Recommendations for Population Guidelines. Nutrients 2025, 17, 277. [Google Scholar] [CrossRef]
- Płudowski, P.; Kos-Kudła, B.; Walczak, M.; Fal, A.; Zozulińska-Ziółkiewicz, D.; Sieroszewski, P.; Peregud-Pogorzelski, J.; Lauterbach, R.; Targowski, T.; Lewiński, A.; et al. Guidelines for Preventing and Treating Vitamin D Deficiency: A 2023 Update in Poland. Nutrients 2023, 15, 695. [Google Scholar] [CrossRef]
- King, R.J.; Singh, P.K.; Mehla, K. The cholesterol pathway: Impact on immunity and cancer. Trends Immunol. 2022, 43, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Bikle, D.D.; Christakos, S. New aspects of vitamin D metabolism and action—Addressing the skin as source and target. Nat. Rev. Endocrinol. 2020, 16, 234–252. [Google Scholar] [CrossRef]
- Prabhu, A.V.; Luu, W.; Sharpe, L.J.; Brown, A.J. Cholesterol-mediated Degradation of 7-Dehydrocholesterol Reductase Switches the Balance from Cholesterol to Vitamin D Synthesis. J. Biol. Chem. 2016, 291, 8363–8373. [Google Scholar] [CrossRef]
- Kuan, V.; Martineau, A.R.; Griffiths, C.J.; Hyppönen, E.; Walton, R. DHCR7 mutations linked to higher vitamin D status allowed early human migration to Northern latitudes. BMC Evol. Biol. 2013, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.N.; Tan, B.H.; Pan, Y.; Ong, C.E. The CYP2R1 Enzyme: Structure, Function, Enzymatic Properties and Genetic Polymorphism. J. Pharm. Pharm. Sci. 2021, 4, 94–112. [Google Scholar] [CrossRef]
- Voltan, G.; Cannito, M.; Ferrarese, M.; Ceccato, F.; Camozzi, V. Vitamin D: An Overview of Gene Regulation, Ranging from Metabolism to Genomic Effects. Genes 2023, 14, 1691. [Google Scholar] [CrossRef]
- Nurminen, V.; Neme, A.; Seuter, S.; Carlberg, C. The impact of the vitamin D-modulated epigenome on VDR target gene regulation. Biochim. Biophys. Acta 2018, 1861, 697–705. [Google Scholar] [CrossRef]
- Bikle, D.D.; Patzek, S.; Wang, Y. Physiologic and pathophysiologic roles of extra renal CYP27b1: Case report and review. Bone Rep. 2018, 8, 255–267, Erratum in Bone Rep. 2021, 14, 101084. [Google Scholar] [CrossRef]
- Prosser, D.E.; Jones, G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem. Sci. 2004, 29, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Streeten, E.A.; Zarbalian, K.; Damcott, C.M. CYP24A1 mutations in idiopathic infantile hypercalcemia. N. Engl. J. Med. 2011, 365, 1741–1742. [Google Scholar]
- Jones, G. Pharmacokinetics of vitamin D toxicity. Am. J. Clin. Nutr. 2008, 88, 582S–586S. [Google Scholar] [CrossRef] [PubMed]
- White, P.; Cooke, N. The multifunctional properties and characteristics of vitamin D-binding proteins. Trends Endocrinol. Metab. 2000, 11, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C. Nutrigenomics of Vitamin D. Nutrients 2019, 11, 676. [Google Scholar] [CrossRef]
- Carlberg, C. Molecular endocrinology of vitamin D on the epigenome level. Mol. Cell. Endocrinol. 2017, 453, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Riveram, C.M.; Ren, B. Mapping human epigenomes. Cell 2013, 155, 39–55. [Google Scholar] [CrossRef]
- Meyer, C.A.; Liu, X.S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat. Rev. Genet. 2014, 15, 709–721. [Google Scholar] [CrossRef]
- Forouhari, A.; Heidari-Beni, M.; Veisi, S.; Poursafa, P.; Kelishadi, R. Effect of epigenetics on vitamin D levels: A systematic review until December 2020. Arch. Public Health 2023, 81, 106. [Google Scholar] [CrossRef]
- Beckett, E.L.; Duesing, K.; Martin, C.; Jones, P.; Furst, J.; King, K.; Niblett, S.; Yates, Z.; Veysey, M.; Lucock, M. Relationship between methylation status of vitamin D-related genes, vitamin D levels, and methyl-donor biochemistry. J. Nutr. Intermed. Metab. 2016, 6, 8–15. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Carlberg, C.; Campbell, M.J. Vitamin D receptor signaling mechanisms: Integrated actions of a well-defined transcription factor. Steroids 2013, 78, 127–136. [Google Scholar] [CrossRef]
- Chen, H.; Lin, R.J.; Xie, W.; Wilpitz, D.; Evans, R.M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 1999, 98, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Hanel, A.; Malmberg, H.R.; Carlberg, C. Genome-wide effects of chromatin on vitamin D signaling. J. Mol. Endocrinol. 2020, 64, R45–R56. [Google Scholar] [CrossRef] [PubMed]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 952–961. [Google Scholar] [CrossRef]
- Apprato, G.; Fiz, C.; Fusano, I.; Bergandi, L.; Silvagno, F. Natural Epigenetic Modulators of Vitamin D Receptor. Appl. Sci. 2020, 10, 4096. [Google Scholar] [CrossRef]
- DeLucas, M.; Sánchez, J.; Palou, A.; Serra, F. The Impact of Diet on miRNA Regulation and Its Implications for Health: A Systematic Review. Nutrients 2024, 16, 770. [Google Scholar] [CrossRef]
- Zeljic, K.; Supic, G.; Magic, Z. New insights into vitamin D anticancer properties: Focus on miRNA modulation. Mol. Genet. Genom. 2017, 292, 511–524. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Mohri, T.; Nakajima, M.; Takagi, S.; Komagata, S.; Yokoi, T. MicroRNA regulates human vitamin D receptor. Int. J. Cancer 2009, 125, 1328–1333. [Google Scholar] [CrossRef]
- Giangreco, A.A.; Nonn, L. The sum of many small changes: microRNAs are specifically and potentially globally altered by vitamin D3 metabolites. J. Steroid Biochem. Mol. Biol. 2013, 136, 86–93. [Google Scholar] [CrossRef]
- Lisse, T.S.; Chun, R.F.; Rieger, S.; Adams, J.S.; Hewison, M. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J. Bone Miner. Res. 2013, 28, 1478–1488. [Google Scholar] [CrossRef]
- Pan, Y.Z.; Gao, W.; Yu, A.M. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab. Dispos. 2009, 37, 2112–2117. [Google Scholar] [CrossRef]
- Li, F.; Zhang, A.; Shi, Y.; Ma, Y.; Du, Y. 1alpha,25-Dihydroxyvitamin D3 prevents the differentiation of human lung fibroblasts via microRNA-27b targeting the vitamin D receptor. Int. J. Mol. Med. 2015, 36, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Komagata, S.; Nakajima, M.; Takagi, S.; Mohri, T.; Taniya, T.; Yokoi, T. Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol. Pharmacol. 2009, 76, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Kasiappan, R.; Shen, Z.; Tse, A.K.; Jinwal, U.; Tang, J.; Lungchukiet, P.; Sun, Y.; Kruk, P.; Nicosia, S.V.; Zhang, X.; et al. 1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J. Biol. Chem. 2012, 287, 41297–41309. [Google Scholar] [CrossRef]
- Ting, H.J.; Messing, J.; Yasmin-Karim, S.; Lee, Y.F. Identification of microRNA-98 as a therapeutic target inhibiting prostate cancer growth and a biomarker induced by vitamin D. J. Biol. Chem. 2013, 288, 1–9. [Google Scholar] [CrossRef]
- Beckett, E.L.; Martin, C.; Duesing, K.; Jones, P.; Furst, J.; Yates, Z.; Veysey, M.; Lucock, M. Vitamin D receptor genotype modulates the correlation between vitamin D and circulating levels of let-7a/b and vitamin D intake in an elderly cohort. J. Nutrigenet Nutrigenom. 2015, 7, 264–273. [Google Scholar] [CrossRef]
- McGrath, J.J.; Saha, S.; Burne, T.H.; Eyles, D.W. A systematic review of the association between common single nucleotide polymorphisms and 25-hydroxyvitamin D concentrations. J. Steroid Biochem. Mol. Biol. 2010, 121, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Sadeghnia, H.R.; Tabatabaeizadeh, S.A.; Bahrami-Taghanaki, H.; Behboodi, N.; Esmaeili, H.; Ferns, G.A.; Mobarhan, M.G.; Avan, A. Genetic and epigenetic factors influencing vitamin D status. J. Cell. Physiol. 2018, 233, 4033–4043. [Google Scholar]
- Pineda-Lancheros, L.E.; Gálvez-Navas, J.M.; Rojo-Tolosa, S.; Membrive-Jiménez, C.; Valverde-Merino, M.I.; Martínez-Martínez, F.; Sánchez-Martín, A.; Ramírez-Tortosa, M.; Pérez-Ramírez, C.; Jiménez-Morales, A. Polymorphisms in VDR, CYP27B1, CYP2R1, GC and CYP24A1 Genes as Biomarkers of Survival in Non-Small Cell Lung Cancer: A Systematic Review. Nutrients 2023, 15, 1525. [Google Scholar] [CrossRef]
- Schuch, N.J.; Garcia, V.C.; Vívolo, S.R.; Martini, L.A. Relationship between Vitamin D Receptor gene polymorphisms and the components of metabolic syndrome. Nutr. J. 2013, 12, 96. [Google Scholar] [CrossRef]
- Dastani, Z.; Li, R.; Richards, B. Genetic Regulation of Vitamin D Levels. Calcif. Tissue Int. 2013, 92, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Usategui-Martín, R.; De Luis-Román, D.A.; Fernández-Gómez, J.M.; Ruiz-Mambrilla, M.; Pérez-Castrillón, J.L. Vitamin D Receptor (VDR) Gene Polymorphisms Modify the Response to Vitamin D Supplementation: A Systematic Review and Meta-Analysis. Nutrients 2022, 14, 360. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Bornman, L. Cdx-2 polymorphism in the vitamin D receptor gene (VDR) marks VDR expression in monocyte/macrophages through VDR promoter methylation. Immunogenetics 2018, 70, 523–532. [Google Scholar] [CrossRef]
- Lu, L.; Sheng, H.; Li, H.; Gan, W.; Liu, C.; Zhu, J.; Loos, R.J.; Lin, X. Associations between common variants in GC and DHCR7/NADSYN1 and vitamin D concentration in Chinese Hans. Hum. Genet. 2012, 131, 505–512. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Liu, Y.; Qu, H.; Qu, S.; Wang, W.; Ren, L. The GC, CYP2R1 and DHCR7 genes are associated with vitamin D levels in northeastern Han Chinese children. Swiss Med. Wkly. 2012, 142, w13636. [Google Scholar] [CrossRef]
- Duan, L.; Xue, Z.; Ji, H.; Zhang, D.; Wang, Y. Effects of CYP2R1 gene variants on vitamin D levels and status: A systematic review and meta-analysis. Gene 2018, 678, 361–369. [Google Scholar] [CrossRef]
- Ramos-Lopez, E.; Brück, P.; Jansen, T.; Herwig, J.; Badenhoop, K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab. Res. Rev. 2007, 23, 631–636. [Google Scholar] [CrossRef]
- Khayyatzadeh, S.S.; Mehramiz, M.; Esmaeily, H.; Mirmousavi, S.J.; Khajavi, L.; Salehkhani, F.N.; Hanachi, P.; Bahrami-Taghanaki, H.; Eslami, S.; Vatanparast, H.; et al. A variant in CYP2R1 predicts circulating vitamin D levels after supplementation with high-dose of vitamin D in healthy adolescent girls. J. Cell. Physiol. 2019, 234, 13977–13983. [Google Scholar] [CrossRef] [PubMed]
- Nissen, J.; Rasmussen, L.B.; Ravn-Haren, G.; Andersen, E.W.; Hansen, B.; Andersen, R.; Mejborn, H.; Madsen, K.H.; Vogel, U. Common variants in CYP2R1 and GC genes predict vitamin D concentrations in healthy Danish children and adults. PLoS ONE 2014, 9, e89907. [Google Scholar] [CrossRef]
- Ahn, J.; Yu, K.; Stolzenberg-Solomon, R.; Simon, K.C.; McCullough, M.L.; Gallicchio, L.; Jacobs, E.J.; Ascherio, A.; Helzlsouer, K.; Jacobs, K.B.; et al. Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 2010, 19, 2739–2745. [Google Scholar] [CrossRef]
- Signorello, L.B.; Shi, J.; Cai, Q.; Zheng, W.; Williams, S.M.; Long, J.; Cohen, S.S.; Li, G.; Hollis, B.W.; Smith, J.R.; et al. Common variation in vitamin D pathway genes predicts circulating 25-hydroxyvitamin D Levels among African Americans. PLoS ONE 2011, 6, e28623. [Google Scholar] [CrossRef]
- Hu, Z.; Tao, S.; Liu, H.; Pan, G.; Li, B.; Zhang, Z. The Association between Polymorphisms of Vitamin D Metabolic-Related Genes and Vitamin D3 Supplementation in Type 2 Diabetic Patients. J. Diabetes Res. 2019, 2019, 8289741. [Google Scholar] [CrossRef] [PubMed]
- Usama; Khan, A.; Raza, M.K..; Abbas, K.; Mansour, L.; Ali, A.; Imran, M. Genetic insights into vitamin D deficiency: A case-control study of GC and CYP24A1 gene polymorphisms. Steroids 2025, 218, 109607. [Google Scholar] [CrossRef]
- Tebben, P.J.; Milliner, D.S.; Horst, R.L.; Harris, P.C.; Singh, R.J.; Wu, Y.; Foreman, J.W.; Chelminski, P.R.; Kumar, R. Hypercalcemia, hypercalciuria, and elevated calcitriol concentrations with autosomal dominant transmission due to CYP24A1 mutations: Effects of ketoconazole therapy. J. Clin. Endocrinol. Metab. 2012, 97, E423–E427. [Google Scholar] [CrossRef]
- Braun, A.; Bichlmaier, R.; Cleve, H. Molecular analysis of the gene for the human vitamin-D-binding protein (group-specific component): Allelic differences of the common genetic GC types. Hum. Genet. 1992, 89, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Lauridsen, A.L.; Vestergaard, P.; Nexo, E. Mean serum concentration of vitamin D-binding protein (Gc globulin) is related to the Gc phenotype in women. Clin. Chem. 2001, 47, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Yun, F.; Oczak, M.; Wong, B.Y.; Vieth, R.; Cole, D.E. Common genetic variants of the vitamin D binding protein (DBP) predict differences in response of serum 25-hydroxyvitamin D [25(OH)D] to vitamin D supplementation. Clin. Biochem. 2009, 42, 1174–1177. [Google Scholar] [CrossRef]
- Rozmus, D.; Płomiński, J.; Augustyn, K.; Cieślińska, A. rs7041 and rs4588 Polymorphisms in Vitamin D Binding Protein Gene (VDBP) and the Risk of Diseases. Int. J. Mol. Sci. 2022, 23, 933. [Google Scholar] [CrossRef]
- Hyppönen, E.; Vimaleswaran, K.S.; Zhou, A. Genetic Determinants of 25-Hydroxyvitamin D Concentrations and Their Relevance to Public Health. Nutrients 2022, 14, 4408. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, L.J.; Xu, X.; Ye, A.; Travers-Gustafson, D.; Zhou, B.; Wang, H.W.; Zhang, W.; Lee Hamm, L.; Deng, H.W.; et al. DNA methylation levels of CYP2R1 and CYP24A1 predict vitamin D response variation. J. Steroid Biochem. Mol. Biol. 2014, 144, 207–214. [Google Scholar] [CrossRef]
- Carlberg, C.; Haq, A. The concept of the personal vitamin D response index. J. Steroid Biochem. Mol. Biol. 2018, 175, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Saksa, N.; Neme, A.; Ryynänen, J.; Uusitupa, M.; de Mello, V.D.; Voutilainen, S.; Nurmi, T.; Virtanen, J.K.; Tuomainen, T.P.; Carlberg, C. Dissecting high from low responders in a vitamin D3 intervention study. J. Steroid Biochem. Mol. Biol. 2015, 148, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Gospodarska, E.; Ghosh Dastidar, R.; Carlberg, C. Intervention Approaches in Studying the Response to Vitamin D3 Supplementation. Nutrients 2023, 29, 3382. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalówka, M.; Górna, I.; Karaźniewicz-Łada, M.; Kusyk, D.; Przysławski, J.; Drzymała-Czyż, S. Nutri-Epigenetic Regulation of Vitamin D—Impact on Metabolism and Biological Functions: Narrative Review. Metabolites 2025, 15, 436. https://doi.org/10.3390/metabo15070436
Kowalówka M, Górna I, Karaźniewicz-Łada M, Kusyk D, Przysławski J, Drzymała-Czyż S. Nutri-Epigenetic Regulation of Vitamin D—Impact on Metabolism and Biological Functions: Narrative Review. Metabolites. 2025; 15(7):436. https://doi.org/10.3390/metabo15070436
Chicago/Turabian StyleKowalówka, Magdalena, Ilona Górna, Marta Karaźniewicz-Łada, Dominika Kusyk, Juliusz Przysławski, and Sławomira Drzymała-Czyż. 2025. "Nutri-Epigenetic Regulation of Vitamin D—Impact on Metabolism and Biological Functions: Narrative Review" Metabolites 15, no. 7: 436. https://doi.org/10.3390/metabo15070436
APA StyleKowalówka, M., Górna, I., Karaźniewicz-Łada, M., Kusyk, D., Przysławski, J., & Drzymała-Czyż, S. (2025). Nutri-Epigenetic Regulation of Vitamin D—Impact on Metabolism and Biological Functions: Narrative Review. Metabolites, 15(7), 436. https://doi.org/10.3390/metabo15070436






