Gut-Microbiome Signatures Predicting Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. Search Strategy and Study Selection
2.4. Data Extraction and Risk-of-Bias Assessment
2.5. Effect Measures
2.6. Data Preparation and Synthesis Methods
2.7. Reporting Bias and Certainty Assessment
2.8. Cross Platform Harmonisation
3. Results
4. Discussion
4.1. Summary of Evidence
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ding, R.; Li, J.; Wu, T.; Shen, Z.; Li, S.; Zhang, Y.; Dong, C.; Shang, Z.; Zhou, H.; et al. Efficacy and safety of the “watch-and-wait” approach for rectal cancer with clinical complete response after neoadjuvant chemoradiotherapy: A meta-analysis. Surg. Endosc. 2022, 36, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.I.; Jang, B.S.; Chang, J.H.; Kim, E.; Lee, T.H.; Park, J.H.; Chie, E.K. Relationships between the Microbiome and Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Cancer Res Treat. 2024; Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, M.; Shin, Y.; Lee, Y.; Kim, T.-J. Optimizing Cancer Treatment Through Gut Microbiome Modulation. Cancers 2025, 17, 1252. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, M.; Kwon, J.; Shin, H.J.; Moon, S.M.; Kim, S.B.; Shin, U.S.; Han, Y.H.; Kim, Y. Butyrate enhances the efficacy of radiotherapy via FOXO3A in colorectal cancer patient-derived organoids. Int. J. Oncol. 2020, 57, 1307–1318. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef]
- Dziubańska-Kusibab, P.J.; Berger, H.; Battistini, F.; Bouwman, B.A.M.; Iftekhar, A.; Katainen, R.; Cajuso, T.; Crosetto, N.; Orozco, M.; Aaltonen, L.A.; et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 2020, 26, 1063–1069. [Google Scholar] [CrossRef]
- Benej, M.; Hoyd, R.; Kreamer, M.; Wheeler, C.E.; Grencewicz, D.J.; Choueiry, F.; Chan, C.H.F.; Zakharia, Y.; Ma, Q.; Dodd, R.D.; et al. The Tumor Microbiome Reacts to Hypoxia and Can Influence Response to Radiation Treatment in Colorectal Cancer. Cancer Res. Commun. 2024, 4, 1690–1701. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, H.; Wang, L.; Lin, Z.; Jiang, C.; Chen, X.; Wang, K.; Liu, L.; Shao, L.; Pan, J.; Li, J.; et al. Methylglyoxal from gut microbes boosts radiosensitivity and radioimmunotherapy in rectal cancer by triggering endoplasmic reticulum stress and cGAS-STING activation. J. Immunother. Cancer 2023, 11, e007840. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, B.; Xiao, Y.; Liu, J.; Wang, Q.; Xiao, H.; Jin, Y.; Liu, Z.; Chen, Z.; Li, Y.; et al. Roseburia intestinalis sensitizes colorectal cancer to radiotherapy through the butyrate/OR51E1/RALB axis. Cell Rep. 2024, 43, 113846, Erratum in Cell Rep. 2024, 43, 114407. [Google Scholar] [CrossRef]
- Shao, Z.; Xu, Y.; Zhang, X.; Zou, C.; Xie, R. Changes in serum uric acid, serum uric acid/serum creatinine ratio, and gamma-glutamyltransferase might predict the efficacy of neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer. Strahlenther. Onkol. 2024, 200, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Lv, Q.; Zou, N.; Zhang, Y.; Zhang, J.; Tang, Q.; Chou, S.H.; Lu, L.; He, J. Influence of neo-adjuvant radiotherapy on the intestinal microbiota of rectal cancer patients. J. Cancer Res. Clin. Oncol. 2023, 149, 6085–6096. [Google Scholar] [CrossRef]
- Cicalini, I.; Chiarelli, A.M.; Chiacchiaretta, P.; Perpetuini, D.; Rosa, C.; Mastrodicasa, D.; d’Annibale, M.; Trebeschi, S.; Serafini, F.L.; Cocco, G.; et al. Multi-Omics Staging of Locally Advanced Rectal Cancer Predicts Treatment Response: A Pilot Study. La Radiol. Med. 2024, 129, 712–726. [Google Scholar] [CrossRef]
- Wang, H.; Jia, H.; Gao, Y.; Zhang, H.; Fan, J.; Zhang, L.; Ren, F.; Yin, Y.; Cai, Y.; Zhu, J.; et al. Serum metabolic traits reveal therapeutic toxicities and responses of neoadjuvant chemoradiotherapy in patients with rectal cancer. Nat. Commun. 2022, 13, 7802. [Google Scholar] [CrossRef]
- Zhao, M.; Jiang, G.; Zhou, H.; Li, J.; Xiang, W.; Li, S.; Wang, H.; Zhou, J. Gut microbiota: A potential target for improved cancer therapy. J. Cancer Res. Clin. Oncol. 2023, 149, 541–552. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Bmj 2021, 10, 89. [Google Scholar]
- Shi, W.; Shen, L.; Zou, W.; Wang, J.; Yang, J.; Wang, Y.; Liu, B.; Xie, L.; Zhu, J.; Zhang, Z. The Gut Microbiome is Associated with Therapeutic Responses and Toxicities of Neoadjuvant Chemoradiotherapy in Rectal Cancer Patients-A Pilot Study. Front. Cell. Infect. Microbiol. 2020, 10, 562463. [Google Scholar] [CrossRef]
- Yi, Y.; Shen, L.; Shi, W.; Xia, F.; Zhang, H.; Wang, Y.; Zhang, J.; Wang, Y.; Sun, X.; Zhang, Z.; et al. Gut Microbiome Components Predict Response to Neoadjuvant Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer: A Prospective, Longitudinal Study. Clin. Cancer Res. 2021, 27, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Shang, F.; Chen, C.; Zhou, H.; Fan, J.; Yang, M.; Nie, X.; Liu, L.; Cai, K.; Liu, H. Microbial Characteristics of Locally Advanced Rectal Cancer Patients After Neoadjuvant Chemoradiation Therapy According to Pathologic Response. Cancer Manag. Res. 2021, 13, 2655–2667. [Google Scholar] [CrossRef]
- Huang, X.; Chen, C.; Xie, W.; Zhou, C.; Tian, X.; Zhang, Z.; Wang, Q.; Chang, H.; Xiao, W.; Zhang, R.; et al. Metagenomic Analysis of Intratumoral Microbiome Linking to Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2023, 117, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.; Wang, Y.; Sui, X.; Fan, J.; Li, S.; Lei, X.; Shi, C.; Sun, W.; Song, M.; Wang, H.; et al. Gut microbiota-mediated nucleotide synthesis attenuates the response to neoadjuvant chemoradiotherapy in rectal cancer. Cancer Cell 2023, 41, 124–138.e6. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Jin, C.; Yue, K.; Sheng, D.; Zhang, T.; Dou, X.; Liu, J.; Jing, H.; Zhang, L.; et al. Prospective, longitudinal analysis of the gut microbiome in patients with locally advanced rectal cancer predicts response to neoadjuvant concurrent chemoradiotherapy. J. Transl. Med. 2023, 21, 221. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, M.; Batool, S.S.; Zhao, Y.; Yu, Z.; Zhou, J.; Liu, K.; Huang, J. Metagenomic analysis reveals effects of gut microbiome in response to neoadjuvant chemoradiotherapy in advanced rectal cancer. Genomics 2024, 116, 110951. [Google Scholar] [CrossRef]
- Boldrini, L.; Chiloiro, G.; Franco, S.D.; Romano, A.; Smiljanic, L.; Tran, E.H.; Bono, F.; Davies, D.C.; Lopetuso, L.; Bonis, M.D.; et al. MOREOVER: Multiomics MR-guided radiotherapy optimization in locally advanced rectal cancer. Radiat. Oncol. 2024, 19, 94. [Google Scholar] [CrossRef]
- Yang, Z.; Ma, J.; Han, J.; Li, A.; Liu, G.; Sun, Y.; Zheng, J.; Zhang, J.; Chen, G.; Xu, R.; et al. Gut microbiome model predicts response to neoadjuvant immunotherapy plus chemoradiotherapy in rectal cancer. Med 2024, 5, 1293–1306.e4. [Google Scholar] [CrossRef]
- Takenaka, I.K.T.M.; Bartelli, T.F.; Defelicibus, A.; Sendoya, J.M.; Golubicki, M.; Robbio, J.; Serpa, M.S.; Branco, G.P.; Santos, L.B.C.; Claro, L.C.L.; et al. Exome and Tissue-Associated Microbiota as Predictive Markers of Response to Neoadjuvant Treatment in Locally Advanced Rectal Cancer. Front. Oncol. 2022, 12, 809441. [Google Scholar] [CrossRef]
- Duan, T.; Ren, Z.; Jiang, H.; Ding, Y.; Wang, H.; Wang, F. Gut microbiome signature in response to neoadjuvant chemoradiotherapy in patients with rectal cancer. Front. Microbiol. 2025, 16, 1543507. [Google Scholar] [CrossRef]
- Sun, L.; Qu, J.; Ke, X.; Zhang, Y.; Xu, H.; Lv, N.; Leng, J.; Zhang, Y.; Guan, A.; Feng, Y.; et al. Interaction between intratumoral microbiota and tumor mediates the response of neoadjuvant therapy for rectal cancer. Front. Microbiol. 2023, 14, 1229888. [Google Scholar] [CrossRef]
- Abukhiran, I.M.; Masaadeh, A.H.; Byrne, J.D.; Bosch, D.E. Mucosal Microbiome Markers of Complete Pathologic Response to Neoadjuvant Therapy in Rectal Carcinoma. Cancer Res. Commun. 2025, 5, 756–766. [Google Scholar] [CrossRef]
- Then, C.K.; Paillas, S.; Moomin, A.; Misheva, M.D.; Moir, R.A.; Hay, S.M.; Bremner, D.; Roberts, K.S.; Smith, E.E.; Heidari, Z.; et al. Dietary fibre supplementation enhances radiotherapy tumour control and alleviates intestinal radiation toxicity. Microbiome 2024, 12, 89. [Google Scholar] [CrossRef]
- Poonacha, K.N.T.; Villa, T.G.; Notario, V. The Interplay among Radiation Therapy, Antibiotics and the Microbiota: Impact on Cancer Treatment Outcomes. Antibiotics 2022, 11, 331. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Cheng, X.; Meng, C.; Zhao, H.; Fu, W.; Gao, Y.; Ma, L.; Yang, Z.; Yao, H.; et al. Unraveling the role of gut microbiome in predicting adverse events in neoadjuvant therapy for rectal cancer. Hum. Vaccines Immunother. 2024, 20, 2430087. [Google Scholar] [CrossRef]
- Yang, R.; Liu, W.; Cai, S.; Feng, X.; Chen, Y.; Cheng, X.; Ma, J.; Ma, W.; Tian, Z.; Yang, W. Evaluation of the efficacy of probiotics in the chemoradiotherapy of colorectal cancer: A meta-analysis of Randomized Controlled Trials. BMC Gastroenterol. 2025, 25, 312. [Google Scholar] [CrossRef]
- Scott, A.J.; Kennedy, E.B.; Berlin, J.; Brown, G.; Chalabi, M.; Cho, M.T.; Cusnir, M.; Dorth, J.; George, M.; Kachnic, L.A.; et al. Management of Locally Advanced Rectal Cancer: ASCO Guideline. JCO Oncol. Pract. 2024, 42, 3355–3375. [Google Scholar] [CrossRef]
- Alexandrescu, L.; Nicoara, A.D.; Tofolean, D.E.; Herlo, A.; Nelson Twakor, A.; Tocia, C.; Trandafir, A.; Dumitru, A.; Dumitru, E.; Aftenie, C.F.; et al. Healing from Within: How Gut Microbiota Predicts IBD Treatment Success—A Systematic Review. Int. J. Mol. Sci. 2024, 25, 8451. [Google Scholar] [CrossRef]
- Dumitru, A.; Matei, E.; Cozaru, G.C.; Chisoi, A.; Alexandrescu, L.; Popescu, R.C.; Butcaru, M.P.; Dumitru, E.; Rugină, S.; Tocia, C. Endotoxin Inflammatory Action on Cells by Dysregulated-Immunological-Barrier-Linked ROS-Apoptosis Mechanisms in Gut-Liver Axis. Int. J. Mol. Sci. 2024, 25, 2472. [Google Scholar] [CrossRef] [PubMed]
- Alexandrescu, L.; Suceveanu, A.P.; Stanigut, A.M.; Tofolean, D.E.; Axelerad, A.D.; Iordache, I.E.; Herlo, A.; Nelson Twakor, A.; Nicoara, A.D.; Tocia, C.; et al. Intestinal Insights: The Gut Microbiome’s Role in Atherosclerotic Disease: A Narrative Review. Microorganisms 2024, 12, 2341. [Google Scholar] [CrossRef] [PubMed]
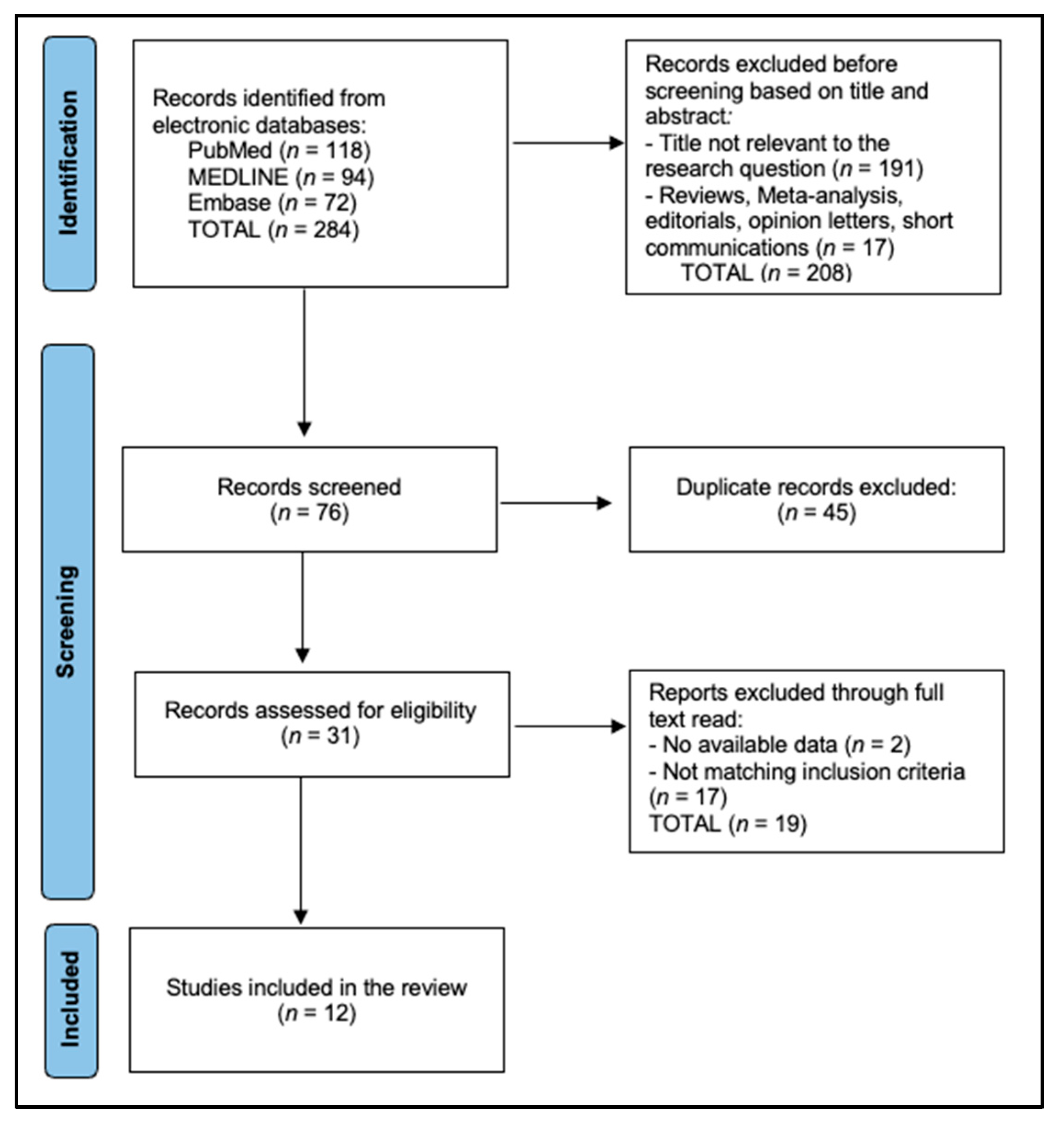
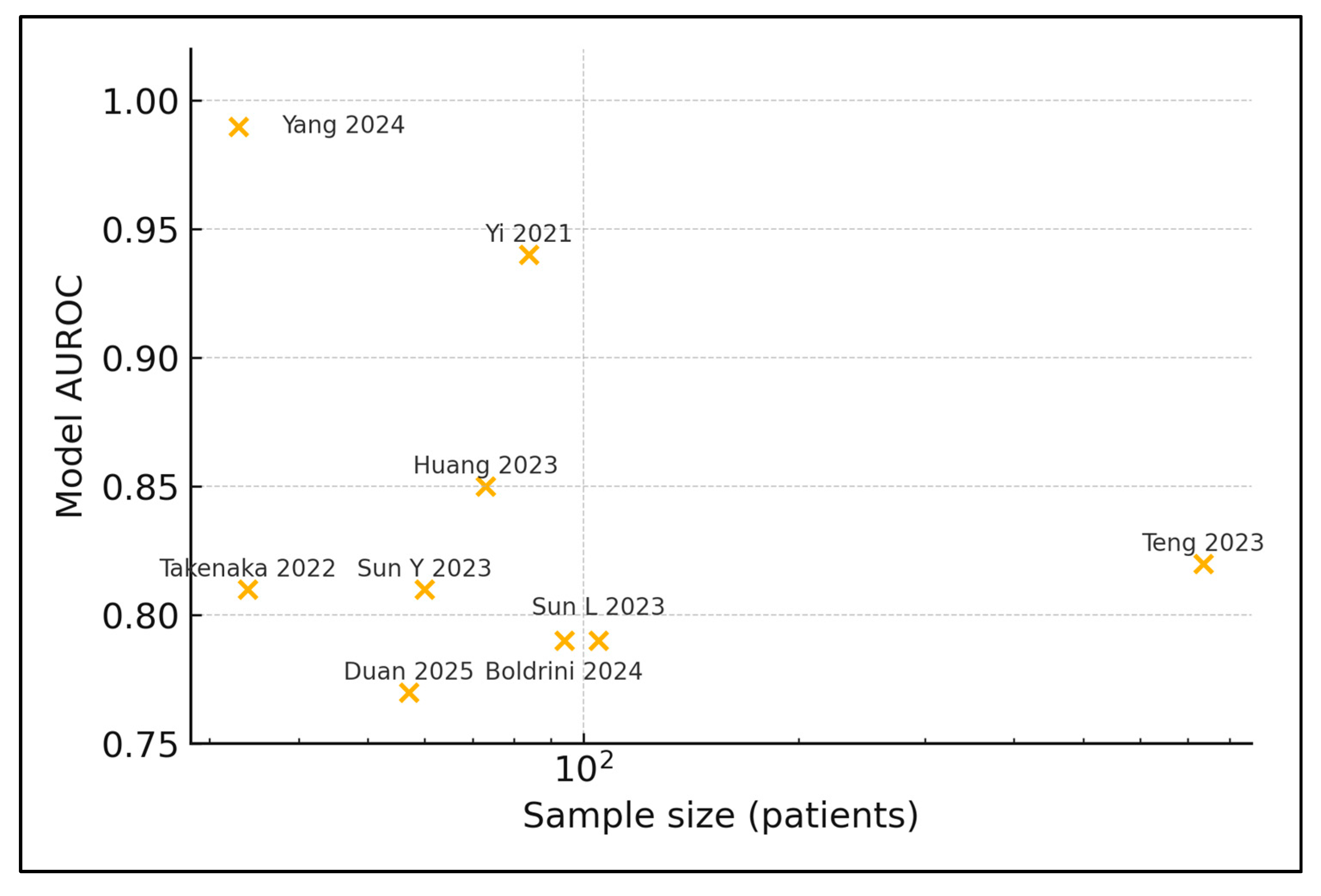
| # | First Author, Year | Country | n Patients | Sample Source | Sequencing | Response Metric | Predictive Model | AUROC/Accuracy |
|---|---|---|---|---|---|---|---|---|
| 1 | Shi 2020 [21] | China | 22 | Stool | 16S | TRG 0–1 vs. 2–3 | LEfSe + LDA | NR |
| 2 | Yi 2021 [22] | China | 84 | Stool | 16S | pCR vs. non | Random Forest | 0.94/0.74 |
| 3 | Fan 2021 [23] | China | 57 | Stool | 16S | pCR vs. non | Logistic regression | NR |
| 4 | Huang 2023 [24] | China | 73 | Tumour biopsy | WGS | GR vs. PR | Random Forest | 0.85/0.88 |
| 5 | Teng 2023 [25] | China | 735 | Stool | 16S | GR vs. PR | Gradient Boosting | 0.82 (internal) |
| 6 | Sun Y 2023 [26] | China | 60 | Stool | 16S + cytokines | GR vs. PR | SVM | 0.81/0.78 |
| 7 | Chen 2024 [27] | China | NR | Stool | Shotgun | pCR vs. non | Random Forest | NR |
| 8 | Boldrini 2024 [28] | Italy | 94 | Stool + plasma | 16S + metabolome | pCR vs. non | XGBoost | 0.79 (bootstrap) |
| 9 | Yang 2024 [29] | China | 33 | Stool | Shotgun | pCR vs. non | CNN deep-learn | 0.99/0.78 |
| 10 | Takenaka 2022 [30] | Brazil/Argentina | 34 | Tumour FFPE | WGS | TRG 0–1 vs ≥2 | Elastic Net | 0.81 (CV) |
| 11 | Duan 2025 [31] | China | 57 | Stool | 16S | GR vs. PR | Random Forest | 0.77 (5-fold CV) |
| 12 | Sun L 2023 [32] | China | 105 | Intratumour | 16S | TRG 0–1 vs. ≥2 | LASSO | 0.79 |
| # | Study | Enriched in Responders | Enriched in Non-Responders (Most Significant) | Statistic (p ≤ 0.05 Unless NR) |
|---|---|---|---|---|
| 1 | Shi 2020 [21] | Shuttleworthia, Howardella | Leptotrichia, Peptostreptococcus | LDA > 2 |
| 2 | Yi 2021 [22] | Blautia wexlerae, Roseburia hominis | Fusobacterium nucleatum | ΔRA +4.6% |
| 3 | Fan 2021 [23] | Thermus | Proteobacteria | OR 2.1 |
| 4 | Huang 2023 [24] | Coprococcus comes | Pseudomonas azotoformans | AUC contribution 9% |
| 5 | Teng 2023 [25] | Intestinimonas butyriciproducens | Bacteroides vulgatus | HR 1.74 |
| 6 | Sun Y 2023 [26] | Lachnospiraceae NK4A136 | Prevotella copri | LDA > 2 |
| 7 | Chen 2024 [27] | Bifidobacterium longum | Enterococcus faecalis | ΔRA +3.2% |
| 8 | Boldrini 2024 [28] | Akkermansia muciniphila | Escherichia coli (toxigenic) | OR 2.5 |
| 9 | Yang 2024 [29] | Eubacterium limosum | Streptococcus equinus | AUROC boost +0.04 |
| 10 | Takenaka 2022 [30] | Bacteroides uniformis | Prevotella spp | q = 0.04 |
| 11 | Duan 2025 [31] | Subdoligranulum variabile | Klebsiella pneumoniae | LDA > 3 |
| 12 | Sun L 2023 [32] | NR | Alistipes | AUC 0.702 |
| # | Study | Dominant Pathway Response | Sample Storage | Bio-Informatics Pipeline | External Validation |
|---|---|---|---|---|---|
| 1 | Shi 2020 [21] | Fatty-acid metabolism ↑ responders | −80 °C | QIIME 2/PICRUSt | No |
| 2 | Yi 2021 [22] | Butyrate synthesis ↑ responders | −20 °C | mothur/RandomForest | Yes |
| 3 | Fan 2021 [23] | Arginine/proline catabolism ↑ non-resp. | −80 °C | QIIME 1/STAMP | No |
| 4 | Huang 2023 [24] | Histidine catabolism ↑ non-resp. | Liquid N2 | Kraken2/HUMAnN 3 | Yes |
| 5 | Teng 2023 [25] | Nucleotide-biosynthesis ↑ non-resp. | −80 °C | Deblur/LEfSe | No |
| 6 | Sun Y 2023 [26] | Glyoxylate cycle ↑ responders | −80 °C | QIIME 2/MaAsLin 2 | Yes |
| 7 | Chen 2024 [27] | Sulphur-assimilation ↑ responders | −80 °C | MetaPhlAn 4/HUMAnN 3 | No |
| 8 | Boldrini 2024 [28] | Tryptophan–kynurenine ↑ toxicity | −80 °C | DADA2/MixOmics | No |
| 9 | Yang 2024 [29] | Taurine/hypotaurine ↑ responders | −80 °C | MetaPhlAn 3/CNN | Yes |
| 10 | Takenaka 2022 [30] | DNA-repair modules ↑ non-resp. | FFPE | HUMAnN 3 | Yes |
| 11 | Duan 2025 [31] | SCFA biosynthesis ↑ responders | −80 °C | QIIME 2/RandomForest | No |
| 12 | Sun L 2023 [32] | Nitrogen fixation ↑ non-resp. | −80 °C | QIIME 2/LEfSe | No |
| Taxon (Genus/Species) | Direction | Median Δ-Abundance (%) | Supporting Studies (n) |
|---|---|---|---|
| Blautia wexlerae | ↑ Responder | 4.6 | [22,29] |
| Lachnospiraceae bacterium A4 | ↑ Responder | 3.8 | [26,29] |
| Functional Pathway (KEGG) | Associated Taxa | Direction | HR/AUROC for Endpoint | Study |
|---|---|---|---|---|
| Nucleotide-biosynthesis (purine/pyrimidine) | Bacteroides vulgatus | ↑ Resistance | HR for non-pCR = 1.74 (95% CI 1.2–2.6) | Teng 2023 [25] |
| Short-chain fatty-acid (butyrate) synthesis | Blautia, Roseburia | ↑ Sensitivity | AUROC 0.85 for pCR | Yi 2021 [22] |
| Histidine catabolism | Pseudomonas azotoformans | ↑ Resistance | AUROC 0.71 | Huang 2023 [24] |
| Taurine and hypotaurine metabolism | Eubacterium limosum | ↑ Sensitivity | OR for good response = 2.3 (p = 0.04) | Yang 2024 [29] |
| Region (Studies) | n Patients | Mean Fibre Intake (g day−1) | Recent Antibiotic Use (%) | Median BMI (kg m−2) | Public Screening Coverage (%) |
|---|---|---|---|---|---|
| East Asia (9) | 1048 | 26.1 | 32 | 24.7 | 49 |
| Europe (1) | 94 | 19.3 | 14 | 25.6 | 71 |
| S. America (1) | 113 | 22.7 | 21 | 28.4 | 43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domilescu, I.; Miutescu, B.; Horhat, F.G.; Popescu, A.; Nica, C.; Ghiuchici, A.M.; Gadour, E.; Sîrbu, I.; Hutanu, D. Gut-Microbiome Signatures Predicting Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer: A Systematic Review. Metabolites 2025, 15, 412. https://doi.org/10.3390/metabo15060412
Domilescu I, Miutescu B, Horhat FG, Popescu A, Nica C, Ghiuchici AM, Gadour E, Sîrbu I, Hutanu D. Gut-Microbiome Signatures Predicting Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer: A Systematic Review. Metabolites. 2025; 15(6):412. https://doi.org/10.3390/metabo15060412
Chicago/Turabian StyleDomilescu, Ielmina, Bogdan Miutescu, Florin George Horhat, Alina Popescu, Camelia Nica, Ana Maria Ghiuchici, Eyad Gadour, Ioan Sîrbu, and Delia Hutanu. 2025. "Gut-Microbiome Signatures Predicting Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer: A Systematic Review" Metabolites 15, no. 6: 412. https://doi.org/10.3390/metabo15060412
APA StyleDomilescu, I., Miutescu, B., Horhat, F. G., Popescu, A., Nica, C., Ghiuchici, A. M., Gadour, E., Sîrbu, I., & Hutanu, D. (2025). Gut-Microbiome Signatures Predicting Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer: A Systematic Review. Metabolites, 15(6), 412. https://doi.org/10.3390/metabo15060412










