Development of Sucrose-Utilizing Escherichia coli Nissle 1917 for Efficient Heparosan Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmid Construction
2.2. Chromosomal Gene Knockout in EcN
2.3. Shake Flask Cultivation for Heparonsan Production
2.4. Analytical Methods
3. Results
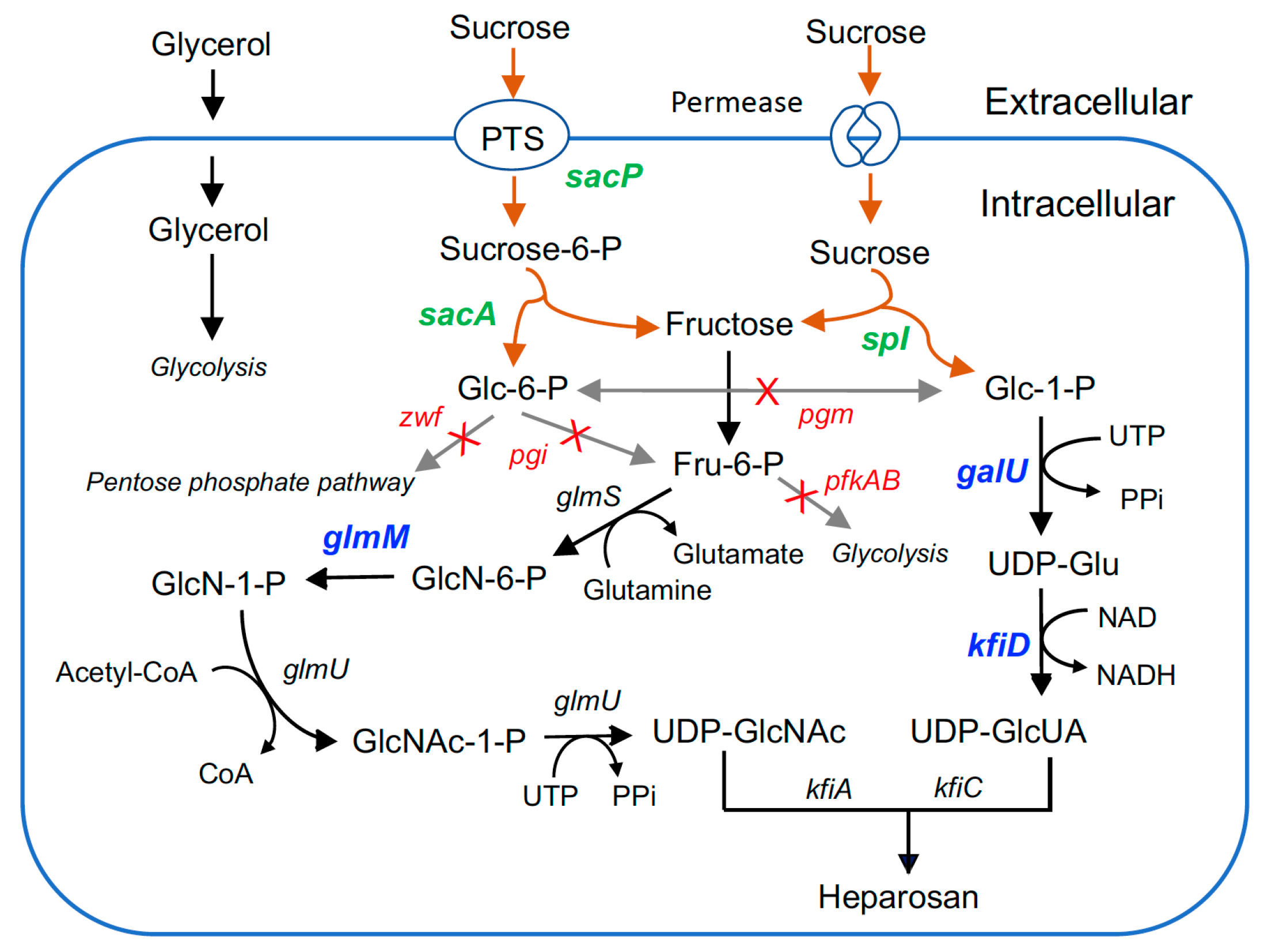
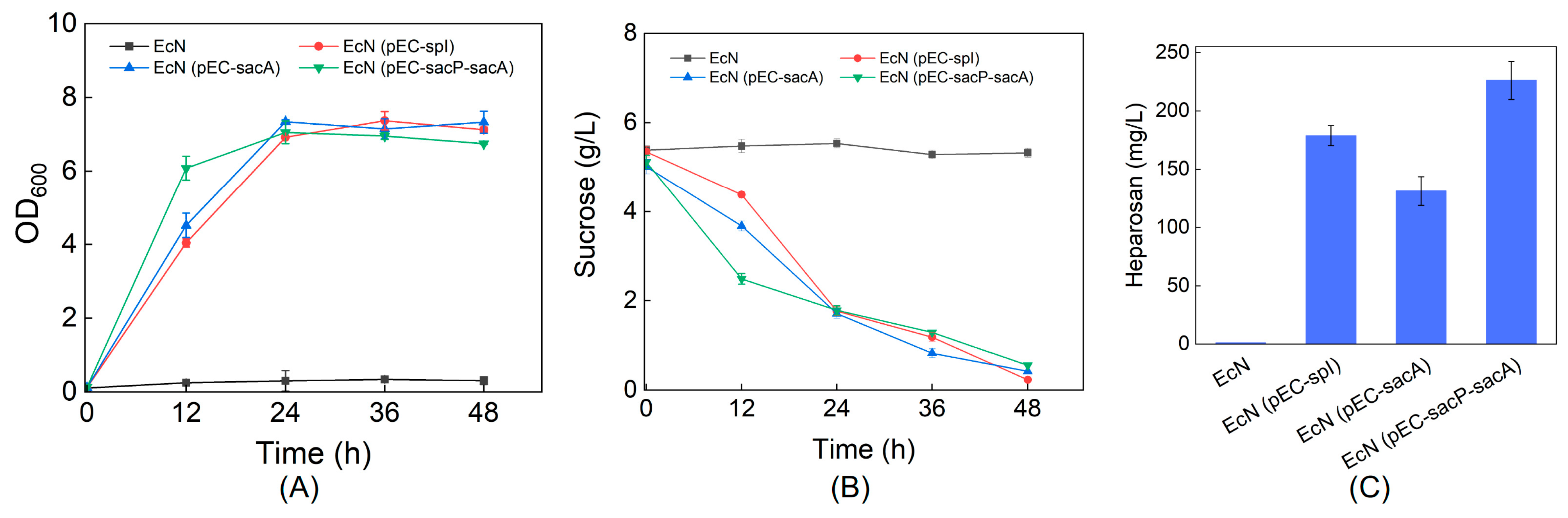
3.1. Establishment of Sucrose Utilization Pathways in EcN
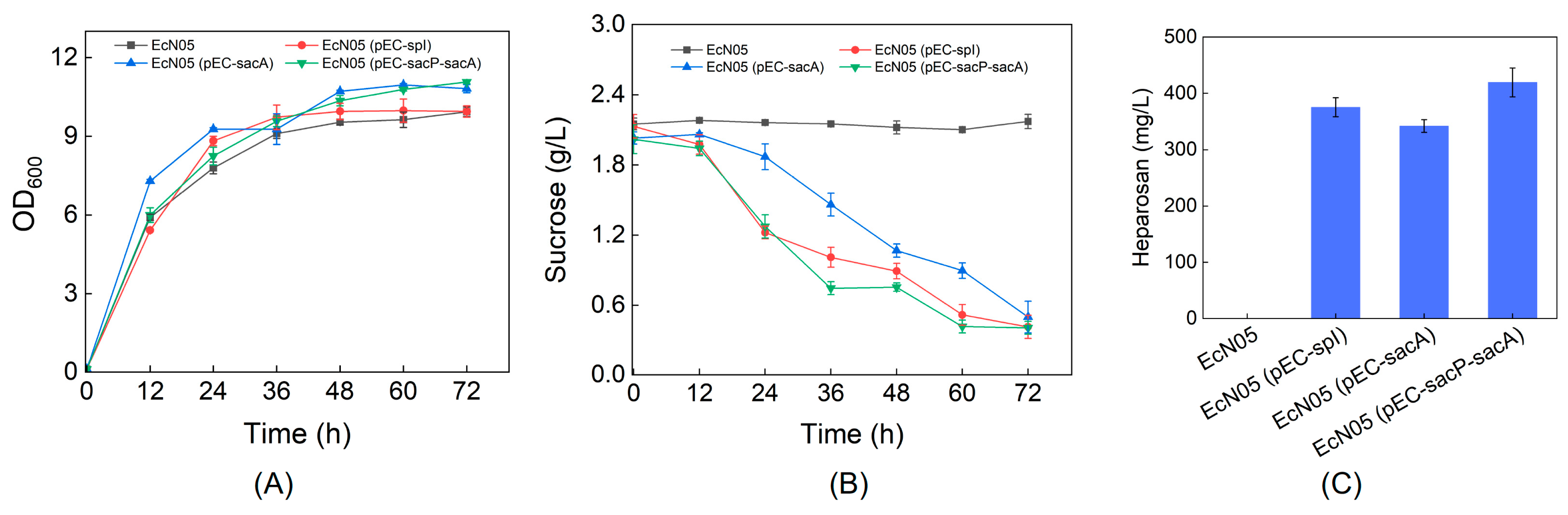
3.2. Heparosan Production Using Sucrose as the Carbon Source
3.3. Improved Heparosan Production Using Mixed Carbon Sources
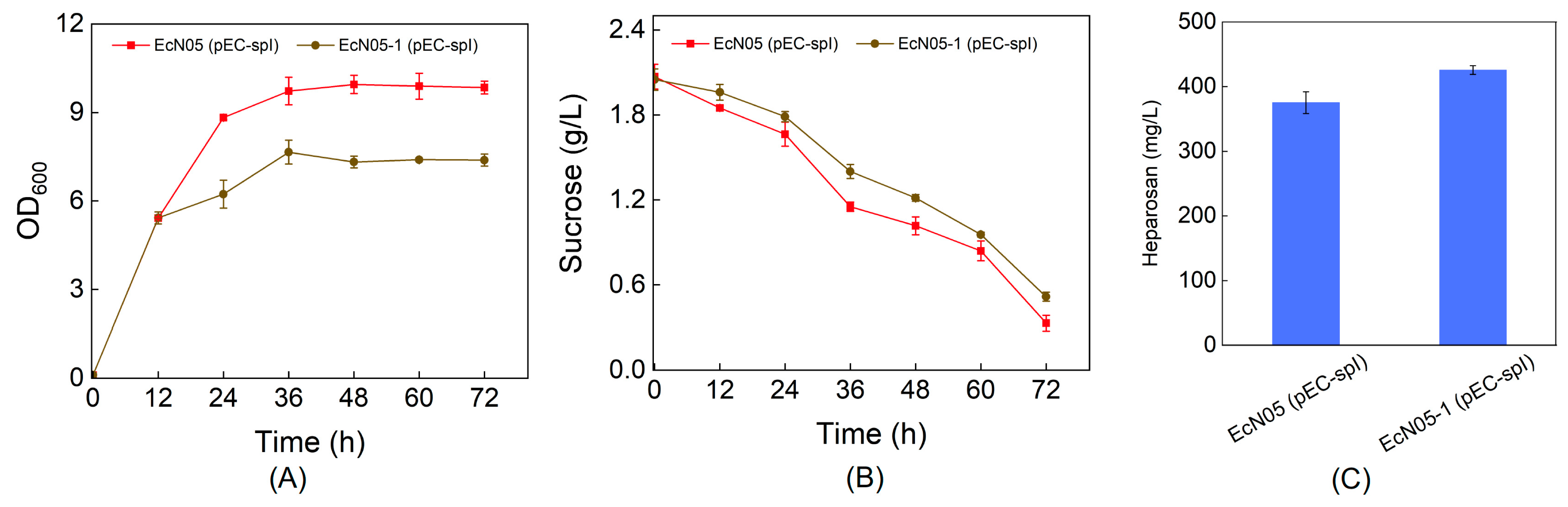
3.4. Deletion of Phosphoglucomutase to Improve Heparosan Production
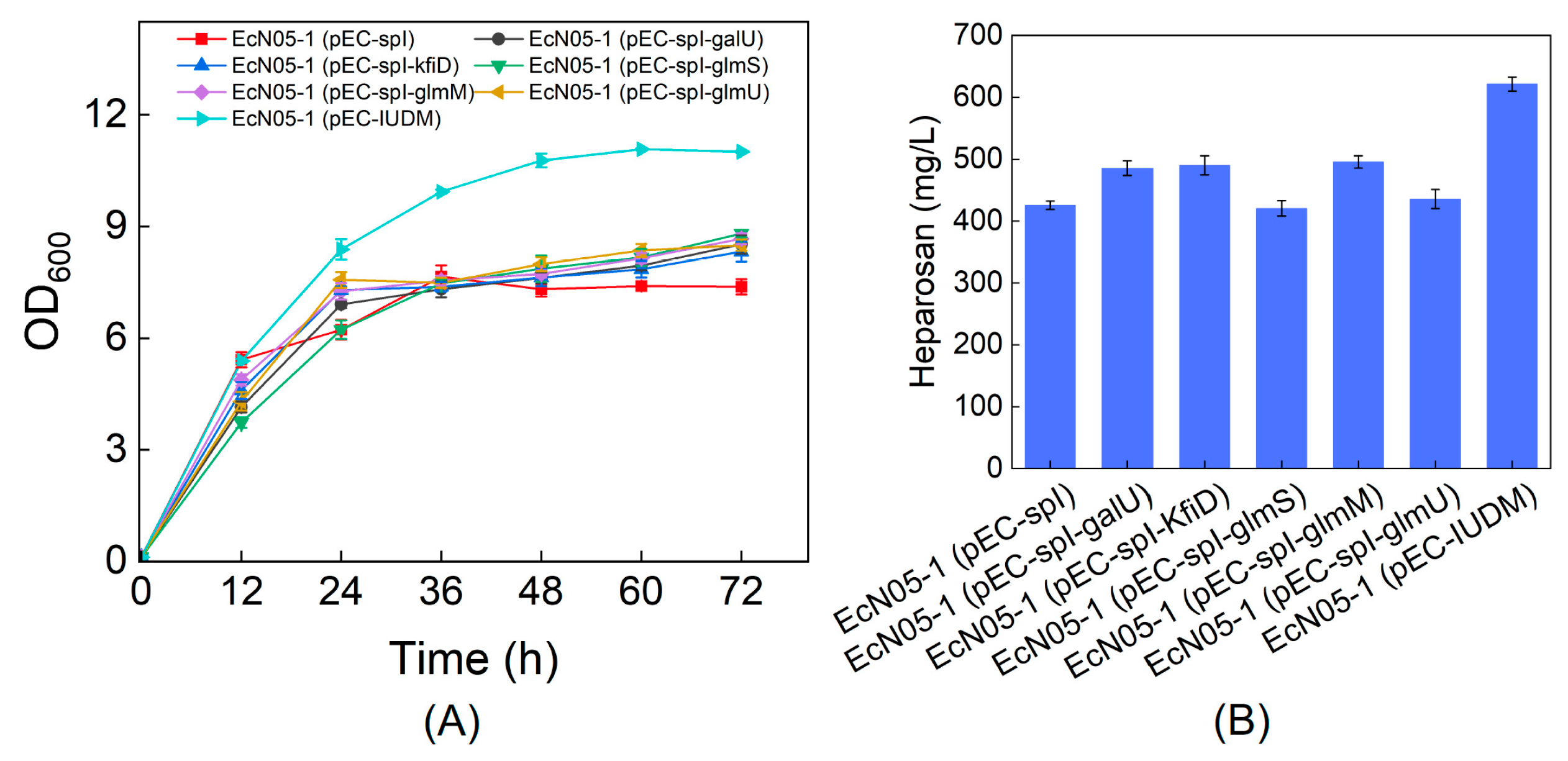
3.5. Overexpression of Key Enzymes to Improve Heparosan Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sonnenborn, U.; Schulze, J. The non-pathogenic Escherichia coli strain Nissle 1917—Features of a versatile probiotic. Microb. Ecol. Healt. Dis. 2009, 21, 122–158. [Google Scholar] [CrossRef]
- Reissbrodt, R.; Hammes, W.P.; dal Bello, F.; Prager, R.; Fruth, A.; Hantke, K.; Rakin, A.; Starcic-Erjavec, M.; Williams, P.H. Inhibition of growth of Shiga toxin-producing Escherichia coli by nonpathogenic Escherichia coli. FEMS Microbiol. Lett. 2009, 290, 62–69. [Google Scholar] [CrossRef]
- Chen, H.; Lei, P.; Ji, H.; Yang, Q.; Peng, B.; Ma, J.; Fang, Y.; Qu, L.; Li, H.; Wu, W.; et al. Advances in Escherichia coli Nissle 1917 as a customizable drug delivery system for disease treatment and diagnosis strategies. Mater. Today Bio 2023, 18, 100543. [Google Scholar] [CrossRef]
- Redenti, A.; Im, J.; Redenti, B.; Li, F.; Rouanne, M.; Sheng, Z.; Sun, W.; Gurbatri, C.R.; Huang, S.; Komaranchath, M.; et al. Probiotic neoantigen delivery vectors for precision cancer immunotherapy. Nature 2024, 635, 453–461. [Google Scholar] [CrossRef]
- Yan, X.; Liu, X.Y.; Zhang, D.; Zhang, Y.D.; Li, Z.H.; Liu, X.; Wu, F.; Chen, G.Q. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell Mol. Immunol. 2021, 18, 2344–2357. [Google Scholar] [CrossRef]
- Yu, M.; Kim, J.; Ahn, J.H.; Moon, Y. Nononcogenic restoration of the intestinal barrier by E. coli-delivered human EGF. JCI Insight 2019, 4, e125166. [Google Scholar] [CrossRef]
- Datta, P.; Fu, L.; Brodfuerer, P.; Dordick, J.S.; Linhardt, R.J. High density fermentation of probiotic E. coli Nissle 1917 towards heparosan production, characterization, and modification. Appl. Microbiol. Biotechnol. 2021, 105, 1051–1062. [Google Scholar] [CrossRef]
- Yu, Y.; Gong, B.; Wang, H.; Yang, G.; Zhou, X. Chromosome evolution of Escherichia coli Nissle 1917 for high-level production of heparosan. Biotechnol. Bioeng. 2023, 120, 1081–1096. [Google Scholar] [CrossRef]
- Hu, S.; Zhao, L.; Hu, L.; Xi, X.; Zhang, Y.; Wang, Y.; Chen, J.; Chen, J.; Kang, Z. Engineering the probiotic bacterium Escherichia coli Nissle 1917 as an efficient cell factory for heparosan biosynthesis. Enzyme. Microb. Technol. 2022, 158, 110038. [Google Scholar] [CrossRef]
- Sheng, L.L.; Cai, Y.M.; Li, Y.; Huang, S.L.; Sheng, J.Z. Advancements in heparosan production through metabolic engineering and improved fermentation. Carbohydr. Polym. 2024, 331, 121881. [Google Scholar] [CrossRef]
- Sultana, R.; Kamihira, M. Bioengineered heparin: Advances in production technology. Biotechnol. Adv. 2024, 77, 108456. [Google Scholar] [CrossRef]
- Kim, J.R.; Kim, S.H.; Lee, S.Y.; Lee, P.C. Construction of homologous and heterologous synthetic sucrose utilizing modules and their application for carotenoid production in recombinant Escherichia coli. Bioresour. Technol. 2013, 130, 288–295. [Google Scholar] [CrossRef]
- Sabri, S.; Nielsen, L.K.; Vickers, C.E. Molecular control of sucrose utilization in Escherichia coli W, an efficient sucrose-utilizing strain. Appl. Environ. Microbiol. 2013, 79, 478–487. [Google Scholar] [CrossRef]
- Qu, Y.N.; Yan, H.J.; Guo, Q.; Li, J.L.; Ruan, Y.C.; Yue, X.Z.; Zheng, W.X.; Tan, T.W.; Fan, L.H. Biosynthesis of D-glucaric acid from sucrose with routed carbon distribution in metabolically engineered Escherichia coli. Metab. Eng. 2018, 47, 393–400. [Google Scholar] [CrossRef]
- He, X.; Chen, K.; Li, Y.; Wang, Z.; Zhang, H.; Qian, J.; Ouyang, P. Enhanced L-lysine production from pretreated beet molasses by engineered Escherichia coli in fed-batch fermentation. Bioprocess Biosyst. Eng. 2015, 38, 1615–1622. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, M.; Li, T.; Fu, L.; Cao, W.; Liu, H. Construction of efficient Streptococcus zooepidemicus strains for hyaluoronic acid production based on identification of key genes involved in sucrose metabolism. AMB Express 2016, 6, 121. [Google Scholar] [CrossRef]
- Stephens, C.; Martinez, M.; Leonardi, V.; Jaing, J.; Miller, A. The Scr and Csc pathways for sucrose utilization co-exist in E. coli, but only the Scr pathway is widespread in other Enterobacteriaceae. Front. Microbiol. 2024, 15, 1409295. [Google Scholar] [CrossRef]
- Lu, M.; Kleckner, N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 1994, 176, 5847–5851. [Google Scholar] [CrossRef]
- Bar-Peled, M.; Griffith, C.L.; Ory, J.J.; Doering, T.L. Biosynthesis of UDP-GlcA, a key metabolite for capsular polysaccharide synthesis in the pathogenic fungus Cryptococcus neoformans. Biochem. J. 2004, 381, 131–136. [Google Scholar] [CrossRef]
- Cimini, D.; Carlino, E.; Giovane, A.; Argenzio, O.; Dello Iacono, I.; De Rosa, M.; Schiraldi, C. Engineering a branch of the UDP-precursor biosynthesis pathway enhances the production of capsular polysaccharide in Escherichia coli O5:K4:H4. Biotechnol. J. 2015, 10, 1307–1315. [Google Scholar] [CrossRef]
- Vyas, A.; Nidetzky, B. Energetics of the glycosyl transfer reactions of sucrose phosphorylase. Biochemistry 2023, 62, 1953–1963. [Google Scholar] [CrossRef]
- Corbett, D.; Bennett, H.J.; Askar, H.; Green, J.; Roberts, I.S. SlyA and H-NS regulate transcription of the Escherichia coli K5 capsule gene cluster, and expression of slyA in Escherichia coli is temperature-dependent, positively autoregulated, and independent of H-NS*. J. Biol. Chem. 2007, 282, 33326–33335. [Google Scholar] [CrossRef]
- Yan, H.; Bao, F.; Zhao, L.; Yu, Y.; Tang, J.; Zhou, X. Cyclic AMP (cAMP) receptor protein-cAMP complex regulates heparosan production in Escherichia coli strain Nissle 1917. Appl. Environ. Microbiol. 2015, 81, 7687–7696. [Google Scholar] [CrossRef]
- Kosamia, N.M.; Samavi, M.; Uprety, B.K.; Rakshit, S.K. Valorization of biodiesel byproduct crude glycerol for the production of bioenergy and biochemicals. Catalysts 2020, 10, 609. [Google Scholar] [CrossRef]
- Guo, Y.-S.; Yang, Z.-D.; Huang, J.-S.; Gao, J.-Y.; Chen, X.-P.; Cheng, H.; Zhang, P.-J.; Su, H.-H. Efficient and economical biosynthesis of high-purity isomaltulose from sugar industrial waste molasses using an engineered Corynebacterium glutamicum strain. Green Chem. 2022, 24, 4050–4060. [Google Scholar] [CrossRef]
- Wang, X.; Lu, L.; Liu, Q.; Li, J.; Wang, T.; Wang, J.; Sun, X.; Shen, X.; Yuan, Q. Integration site library for efficient construction of plasmid-free microbial cell factories in Escherichia coli. J. Agric. Food Chem. 2024, 72, 24687–24696. [Google Scholar] [CrossRef]

| Strains/Plasmids | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Wild-type strain for gene cloning | Takara |
| E. coli Nissle 1917 | Wild-type strain for heparosan production | DSMZ * |
| Bacillus subtilis 168 | Wild-type strain for cloning sacA and sacP | DSMZ |
| E. coli EcN05 | Nissle 1917 Δzwf ΔpfkB ΔpfkA Δpgi | This study |
| E. coli EcN05-1 | Nissle 1917 Δzwf ΔpfkB ΔpfkA Δpgi Δpgm | This study |
| Plasmids | ||
| pKD13 | Plasmid harboring KanR and FLP recognition target | Yale CGSC ** |
| pKD46 | λ-Red recombinase expression helper plasmid | Yale CGSC |
| pCP20 | FLP recombinase helper plasmid | Yale CGSC |
| pEC-XK99E | Expression vector, trc promoter, KanR | Sangon |
| pEC-spI | pEC-XK99E derived, harboring spI | This study |
| pEC-sacA | pEC-XK99E derived, harboring sacA | This study |
| pEC-sacP-sacA | pEC-XK99E derived, harboring sacP and sacA | This study |
| pEC-spI-galU | pEC-XK99E derived, harboring spI and galU | This study |
| pEC-spI-kfiD | pEC-XK99E derived, harboring spI and kfiD | This study |
| pEC-spI-glmS | pEC-XK99E derived, harboring spI and glmS | This study |
| pEC-spI-glmM | pEC-XK99E derived, harboring spI and glmM | This study |
| pEC-spI-glmU | pEC-XK99E derived, harboring spI and glmU | This study |
| pEC-IUDM | pEC-XK99E derived, harboring spI, galU, kfiD, and glmM | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wan, Z.; Li, Z.-J. Development of Sucrose-Utilizing Escherichia coli Nissle 1917 for Efficient Heparosan Biosynthesis. Metabolites 2025, 15, 410. https://doi.org/10.3390/metabo15060410
Chen Y, Wan Z, Li Z-J. Development of Sucrose-Utilizing Escherichia coli Nissle 1917 for Efficient Heparosan Biosynthesis. Metabolites. 2025; 15(6):410. https://doi.org/10.3390/metabo15060410
Chicago/Turabian StyleChen, Yaozong, Zihua Wan, and Zheng-Jun Li. 2025. "Development of Sucrose-Utilizing Escherichia coli Nissle 1917 for Efficient Heparosan Biosynthesis" Metabolites 15, no. 6: 410. https://doi.org/10.3390/metabo15060410
APA StyleChen, Y., Wan, Z., & Li, Z.-J. (2025). Development of Sucrose-Utilizing Escherichia coli Nissle 1917 for Efficient Heparosan Biosynthesis. Metabolites, 15(6), 410. https://doi.org/10.3390/metabo15060410






