Unlocking Plant Resilience: Metabolomic Insights into Abiotic Stress Tolerance in Crops
Abstract
1. Introduction
2. Metabolomics in Plant Research
2.1. Methods Review
2.1.1. Gas Chromatography–Mass Spectrometry (GC-MS)
2.1.2. Liquid Chromatography–Mass Spectrometry (LC-MS)
2.1.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.1.4. Comparison
2.2. Sample Preparation and Extraction Methods
2.3. Expanding the Metabolomic Toolbox: Novel Approaches to Abiotic Stress Analysis
2.4. Standardization and Reproducibility in Metabolomics Across Analytical Platforms
2.5. Analytical Approaches: Targeted vs. Untargeted Metabolomics
2.5.1. Targeted Metabolomics
2.5.2. Untargeted Metabolomics
2.5.3. Complementarity of Targeted and Untargeted Approaches
3. Plant Metabolic Responses and the Critical Role of Secondary Metabolites to Abiotic Stresses
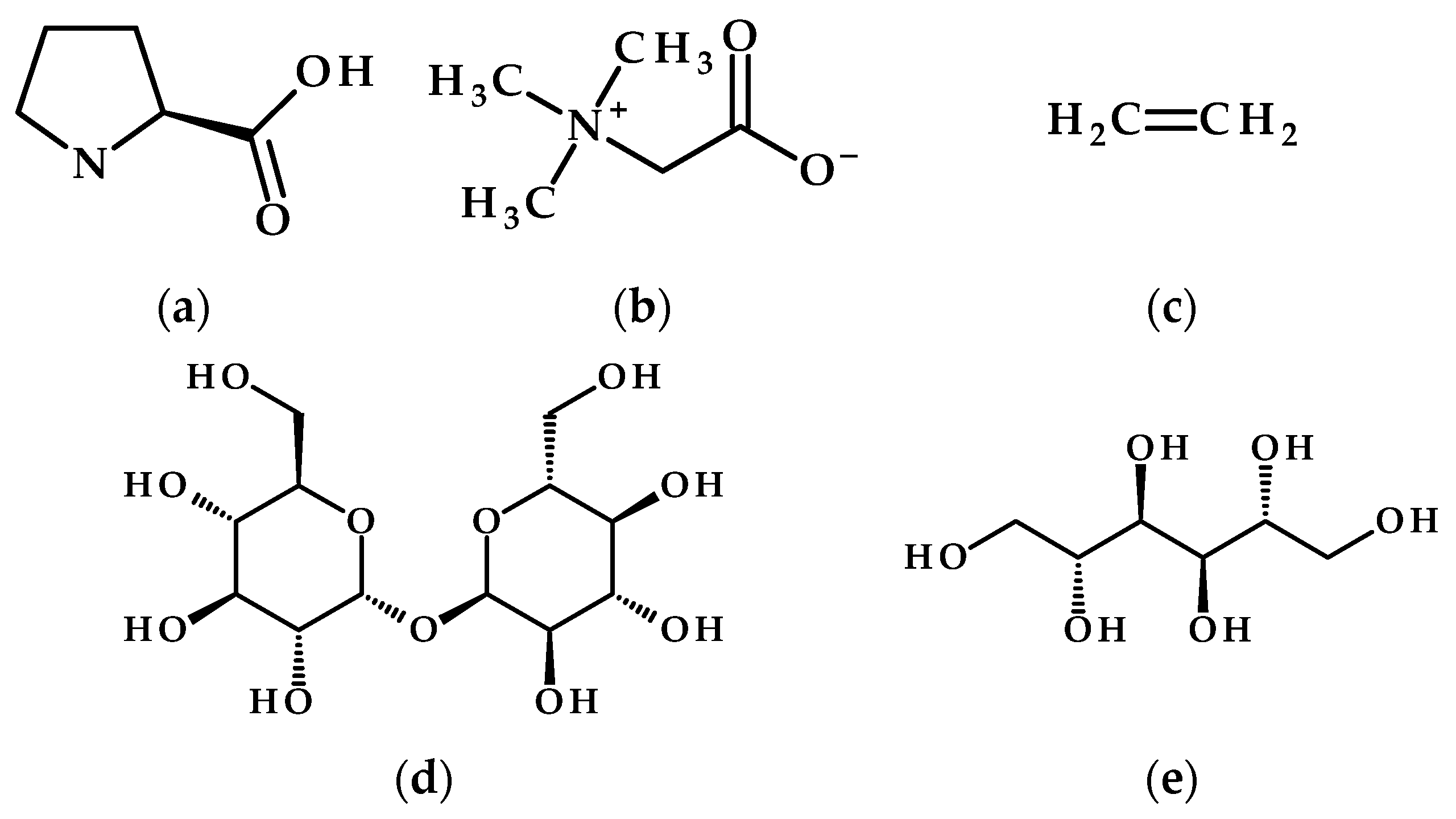
3.1. Salinity Stress
3.2. Drought Stress
3.3. Temperature Stress
3.4. Marker-Assisted Selection in Plant Breeding for Stress Tolerance
4. Integration of Metabolomics with Other “Omics”
4.1. The Integration of Genomics and Metabolomics
4.2. The Integration of Transcriptomics and Metabolomics
4.3. The Integration of Proteomics and Metabolomics
4.4. Benefits and Challenges of Multi-Omics Integration
Methodological Limitations of Multi-Omics Integration
5. Integrative Tools and Platforms for Multi-Omics Data Analysis
6. Future and Practical Applications
6.1. Metabolomics-Assisted Breeding
6.2. Metabolic Engineering
6.3. Understanding Stress Memory and Epigenetic Effects
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| APX | Ascorbate Peroxidase |
| BADH | Betaine-Aldehyde Dehydrogenase |
| BR | Brassinosteroids |
| Cas9 | CRISPR-associated protein 9 |
| CAT | Catalase |
| CE-MS | Capillary electrophoresis–mass spectrometry |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DEGs | Differentially expressed genes |
| DIA | Data-independent acquisition |
| EI | Electron ionization |
| ET | Ethylene |
| F3H | Flavanone 3-hydroxylase |
| FT-IR | Fourier transform infrared spectroscopy |
| GC-MS | Gas chromatography–mass spectrometry |
| GEO | Gene Expression Omnibus |
| GS | Genomic selection |
| HR-MAS | High-resolution magic angle spinning |
| HR-MS | High-Resolution mass spectrometry |
| HSFs | Heat shock factors |
| HSPs | Heat shock proteins |
| IMS-MS | Ion mobility mass spectrometry |
| JA | Jasmonic acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC-MS | Liquid chromatography–mass spectrometry |
| MAS | Marker-assisted selection |
| MDA | Malondialdehyde |
| MOI | Multi-omics integration |
| MSI | Metabolomics Standards Initiative |
| MTBE | Methyl tert-Butyl Ether |
| MSTFA | N-Methyl-N-(trimethylsilyl)trifluoroacetamide |
| MWAS | Metabolome-wide association study |
| NIST | National Institute of Standards and Technology |
| NMR | Nuclear magnetic resonance |
| NO | Nitric oxide |
| PCA | Principal component analysis |
| PLS-DA | Partial least squares discriminant analysis |
| PMN | Plant Metabolic Network |
| PPI | Protein–protein interaction |
| PTMs | Post-translational modifications |
| QC | Quality control |
| QTL | Quantitative trait loci |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SL | Strigolactones |
| SNP | Single nucleotide polymorphism |
| SOD | Superoxide Dismutase |
| SOPs | Standard Operating Procedures |
| SVM | Support vector machine |
| T6P | Trehalose-6-phosphate |
| TSP | Trimethylsilylpropanoic acid |
References
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Masamba, P.; Kappo, A.P. Applications of Metabolomics for the Elucidation of Abiotic Stress Tolerance in Plants: A Special Focus on Osmotic Stress and Heavy Metal Toxicity. Plants 2023, 12, 269. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of Plant Central Metabolism in Response to Abiotic Stresses: A Metabolomics View. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for Plant Stress Response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef]
- Allwood, J.W.; Goodacre, R. An Introduction to Liquid Chromatography-Mass Spectrometry Instrumentation Applied in Plant Metabolomic Analyses. Phytochem. Anal. 2010, 21, 33–47. [Google Scholar] [CrossRef]
- Jorge, T.F.; Rodrigues, J.A.; Caldana, C.; Schmidt, R.; van Dongen, J.T.; Thomas-Oates, J.; António, C. Mass Spectrometry-Based Plant Metabolomics: Metabolite Responses to Abiotic Stress. Mass Spectrom. Rev. 2016, 35, 620–649. [Google Scholar] [CrossRef] [PubMed]
- Roessner, U.; Wagner, C.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Simultaneous Analysis of Metabolites in Potato Tuber by Gas Chromatography–Mass Spectrometry. Plant J. 2000, 23, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Mastovská, K.; Lehotay, S.J. Practical Approaches to Fast Gas Chromatography-Mass Spectrometry. J. Chromatogr. A 2003, 1000, 153–180. [Google Scholar] [CrossRef] [PubMed]
- Sparkman, O.D.; Penton, Z.E.; Kitson, F.G. Gas Chromatography and Mass Spectrometry; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 9780123736284. [Google Scholar]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-Based Metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef]
- Plumb, R.S.; Gethings, L.A.; Rainville, P.D.; Isaac, G.; Trengove, R.; King, A.M.; Wilson, I.D. Advances in High Throughput LC/MS Based Metabolomics: A Review. Trends Analyt. Chem. 2023, 160, 116954. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite Identification and Quantitation in LC-MS/MS-Based Metabolomics. Trends Analyt. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Wang, Y. Mass Spectrometry in Metabolomics. In Molecular Analysis and Genome Discovery; John Wiley & Sons, Ltd.: Chichester, UK, 2011; pp. 271–298. ISBN 9781119977438. [Google Scholar]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-Based Metabolomics: MASS SPECTROMETRY-BASED METABOLOMICS. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted Large-Scale Plant Metabolomics Using Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Weckwerth, W. Metabolomics in Systems Biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef]
- Salem, M.A.; Jüppner, J.; Bajdzienko, K.; Giavalisco, P. Protocol: A Fast, Comprehensive and Reproducible One-Step Extraction Method for the Rapid Preparation of Polar and Semi-Polar Metabolites, Lipids, Proteins, Starch and Cell Wall Polymers from a Single Sample. Plant Methods 2016, 12, 45. [Google Scholar] [CrossRef]
- Putri, S.P.; Yamamoto, S.; Tsugawa, H.; Fukusaki, E. Current Metabolomics: Technological Advances. J. Biosci. Bioeng. 2013, 116, 9–16. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas Chromatography Mass Spectrometry-Based Metabolite Profiling in Plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Ocampos, F.M.M.; de Souza, A.J.B.; Ribeiro, G.H.; Almeida, L.S.; Cônsolo, N.R.B.; Colnago, L.A. NMR-Based Plant Metabolomics Protocols: A Step-by-Step Guide. Front. Nat. Prod. 2024, 3, 1414506. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-Based Plant Metabolomics: Where Do We Stand, Where Do We Go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The Use of Metabolomics to Dissect Plant Responses to Abiotic Stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Soga, T. Advances in Capillary Electrophoresis Mass Spectrometry for Metabolomics. Trends Analyt. Chem. 2023, 158, 116883. [Google Scholar] [CrossRef]
- Serrano-García, I.; Martakos, I.C.; Olmo-García, L.; León, L.; de la Rosa, R.; Gómez-Caravaca, A.M.; Belaj, A.; Serrano, A.; Dasenaki, M.E.; Thomaidis, N.S.; et al. Application of Liquid Chromatography-Ion Mobility Spectrometry-Mass Spectrometry-Based Metabolomics to Investigate the Basal Chemical Profile of Olive Cultivars Differing in Verticillium Dahliae Resistance. J. Agric. Food Chem. 2024, 72, 27561–27574. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Sharma, S.; Kumar, D. Advances of Ion Mobility Platform for Plant Metabolomics. Crit. Rev. Anal. Chem. 2024, 54, 175–191. [Google Scholar] [CrossRef]
- Joshi, R.; Sathasivam, R.; Jayapal, P.K.; Patel, A.K.; Van Nguyen, B.; Faqeerzada, M.A.; Park, S.U.; Lee, S.H.; Kim, M.S.; Baek, I.; et al. Comparative Determination of Phenolic Compounds in Arabidopsis Thaliana Leaf Powder under Distinct Stress Conditions Using Fourier-Transform Infrared (FT-IR) and near-Infrared (FT-NIR) Spectroscopy. Plants 2022, 11, 836. [Google Scholar] [CrossRef]
- López-Ruiz, R.; Maldonado-Reina, A.J.; Marín-Sáez, J.; Romero-González, R.; Martínez-Vidal, J.L.; Garrido Frenich, A. Unravelling Plant Protection Product Analysis: Use of Chromatography Techniques (GC and LC) and High Resolution Mass Spectrometry. Tren. Environ. Anal. Chem. 2023, 37, e00191. [Google Scholar] [CrossRef]
- Mattoli, L.; Gianni, M.; Burico, M. Mass Spectrometry-Based Metabolomic Analysis as a Tool for Quality Control of Natural Complex Products. Mass Spectrom. Rev. 2023, 42, 1358–1396. [Google Scholar] [CrossRef]
- Dias, D.A.; Koal, T. Progress in Metabolomics Standardisation and Its Significance in Future Clinical Laboratory Medicine. EJIFCC 2016, 27, 331–343. [Google Scholar]
- Alseekh, S.; Fernie, A.R. Metabolomics 20 Years on: What Have We Learned and What Hurdles Remain? Plant J. 2018, 94, 933–942. [Google Scholar] [CrossRef]
- Furey, A.; Moriarty, M.; Bane, V.; Kinsella, B.; Lehane, M. Ion Suppression; a Critical Review on Causes, Evaluation, Prevention and Applications. Talanta 2013, 115, 104–122. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Spicer, R.A.; Salek, R.; Steinbeck, C. A Decade after the Metabolomics Standards Initiative It’s Time for a Revision. Sci. Data 2017, 4, 170138. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and Considerations for the Use of System Suitability and Quality Control Samples in Mass Spectrometry Assays Applied in Untargeted Clinical Metabolomic Studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI): Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of Recent Developments in GC-MS Approaches to Metabolomics-Based Research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a Tool to Investigate Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Allwood, J.W.; Williams, A.; Uthe, H.; van Dam, N.M.; Mur, L.A.J.; Grant, M.R.; Pétriacq, P. Unravelling Plant Responses to Stress-the Importance of Targeted and Untargeted Metabolomics. Metabolites 2021, 11, 558. [Google Scholar] [CrossRef]
- Patel, M.K.; Pandey, S.; Kumar, M.; Haque, M.I.; Pal, S.; Yadav, N.S. Plants Metabolome Study: Emerging Tools and Techniques. Plants 2021, 10, 2409. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef]
- Shen, S.; Zhan, C.; Yang, C.; Fernie, A.R.; Luo, J. Metabolomics-Centered Mining of Plant Metabolic Diversity and Function: Past Decade and Future Perspectives. Mol. Plant 2023, 16, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Hagel, J.M.; Facchini, P.J. Plant Metabolomics: Analytical Platforms and Integration with Functional Genomics. Phytochem. Rev. 2008, 7, 479–497. [Google Scholar] [CrossRef]
- Han, W.; Ward, J.L.; Kong, Y.; Li, X. Editorial: Targeted and Untargeted Metabolomics for the Evaluation of Plant Metabolites in Response to the Environment. Front. Plant Sci. 2023, 14, 1167513. [Google Scholar] [CrossRef]
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic Stress in Crop Production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef]
- El Moukhtari, A.; Cabassa-Hourton, C.; Farissi, M.; Savouré, A. How Does Proline Treatment Promote Salt Stress Tolerance during Crop Plant Development? Front. Plant Sci. 2020, 11, 1127. [Google Scholar] [CrossRef]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ashraf, M. Proline Alleviates Abiotic Stress Induced Oxidative Stress in Plants. J. Plant Growth Regul. 2023, 42, 4629–4651. [Google Scholar] [CrossRef]
- Naliwajski, M.; Skłodowska, M. The Relationship between the Antioxidant System and Proline Metabolism in the Leaves of Cucumber Plants Acclimated to Salt Stress. Cells 2021, 10, 609. [Google Scholar] [CrossRef]
- Cárdenas-Pérez, S.; Piernik, A.; Ludwiczak, A.; Duszyn, M.; Szmidt-Jaworska, A.; Chanona-Pérez, J.J. Image and Fractal Analysis as a Tool for Evaluating Salinity Growth Response between Two Salicornia Europaea Populations. BMC Plant Biol. 2020, 20, 467. [Google Scholar] [CrossRef]
- Chourasia, K.N.; Lal, M.K.; Tiwari, R.K.; Dev, D.; Kardile, H.B.; Patil, V.U.; Kumar, A.; Vanishree, G.; Kumar, D.; Bhardwaj, V.; et al. Salinity Stress in Potato: Understanding Physiological, Biochemical and Molecular Responses. Life 2021, 11, 545. [Google Scholar] [CrossRef]
- Dustgeer, Z.; Seleiman, M.F.; Khan, I.; Chattha, M.U.; Ali, E.F.; Alhammad, B.A.; Jalal, R.S.; Refay, Y.; Hassan, M.U. Glycine-Betaine Induced Salinity Tolerance in Maize by Regulating the Physiological Attributes, Antioxidant Defense System and Ionic Homeostasis. Not. Bot. Horti Agrobot. Cluj Napoca 2021, 49, 12248. [Google Scholar] [CrossRef]
- Sofy, M.R.; Elhawat, N.; Alshaal, T. Glycine Betaine Counters Salinity Stress by Maintaining High K+/Na+ Ratio and Antioxidant Defense via Limiting Na+ Uptake in Common Bean (Phaseolus vulgaris L.). Ecotoxicol. Environ. Saf. 2020, 200, 110732. [Google Scholar] [CrossRef]
- Reddikavitha, E.; Sathiavelu, A. Role of Quaternary Aminoacid Derivatives and Ion Homeostasis in Amelioration of Salinity Stress. Res. J. Biotechnol. 2024, 19, 124–131. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Delorge, I.; Janiak, M.; Carpentier, S.; Van Dijck, P. Fine Tuning of Trehalose Biosynthesis and Hydrolysis as Novel Tools for the Generation of Abiotic Stress Tolerant Plants. Front. Plant Sci. 2014, 5, 147. [Google Scholar] [CrossRef]
- Nawaz, M.; Hassan, M.U.; Chattha, M.U.; Mahmood, A.; Shah, A.N.; Hashem, M.; Alamri, S.; Batool, M.; Rasheed, A.; Thabit, M.A.; et al. Trehalose: A Promising Osmo-Protectant against Salinity Stress-Physiological and Molecular Mechanisms and Future Prospective. Mol. Biol. Rep. 2022, 49, 11255–11271. [Google Scholar] [CrossRef]
- Alhudhaibi, A.M.; Ibrahim, M.A.R.; Abd-Elaziz, S.M.S.; Farag, H.R.M.; Elsayed, S.M.; Ibrahim, H.A.; Hossain, A.S.; Alharbi, B.M.; Haouala, F.; Elkelish, A.; et al. Enhancing Salt Stress Tolerance in Wheat (Triticum aestivum) Seedlings: Insights from Trehalose and Mannitol. BMC Plant Biol. 2024, 24, 472. [Google Scholar] [CrossRef]
- Pujni, D.; Chaudhary, A.; Rajam, M.V. Increased Tolerance to Salinity and Drought in Transgenic Indica Rice by Mannitol Accumulation. J. Plant Biochem. Biotechnol. 2007, 16, 1–7. [Google Scholar] [CrossRef]
- Dąbrowska, J.; Menéndez Orellana, A.E.; Kilian, W.; Moryl, A.; Cielecka, N.; Michałowska, K.; Policht-Latawiec, A.; Michalski, A.; Bednarek, A.; Włóka, A. Between Flood and Drought: How Cities Are Facing Water Surplus and Scarcity. J. Environ. Manag. 2023, 345, 118557. [Google Scholar] [CrossRef]
- Yadav, B.; Jogawat, A.; Rahman, M.S.; Narayan, O.P. Secondary Metabolites in the Drought Stress Tolerance of Crop Plants: A Review. Gene Rep. 2021, 23, 101040. [Google Scholar] [CrossRef]
- Jogawat, A.; Yadav, B.; Chhaya; Lakra, N.; Singh, A.K.; Narayan, O.P. Crosstalk between Phytohormones and Secondary Metabolites in the Drought Stress Tolerance of Crop Plants: A Review. Physiol. Plant. 2021, 172, 1106–1132. [Google Scholar] [CrossRef] [PubMed]
- Białecka-Florjańczyk, E.; Fabiszewska, A.; Zieniuk, B. Phenolic Acids Derivatives—Biotechnological Methods of Synthesis and Bioactivity. Curr. Pharm. Biotechnol. 2018, 19, 1098–1113. [Google Scholar] [CrossRef] [PubMed]
- Quan, N.; Anh, L.; Khang, D.; Tuyen, P.; Toan, N.; Minh, T.; Minh, L.; Bach, D.; Ha, P.; Elzaawely, A.; et al. Involvement of Secondary Metabolites in Response to Drought Stress of Rice (Oryza sativa L.). Agriculture 2016, 6, 23. [Google Scholar] [CrossRef]
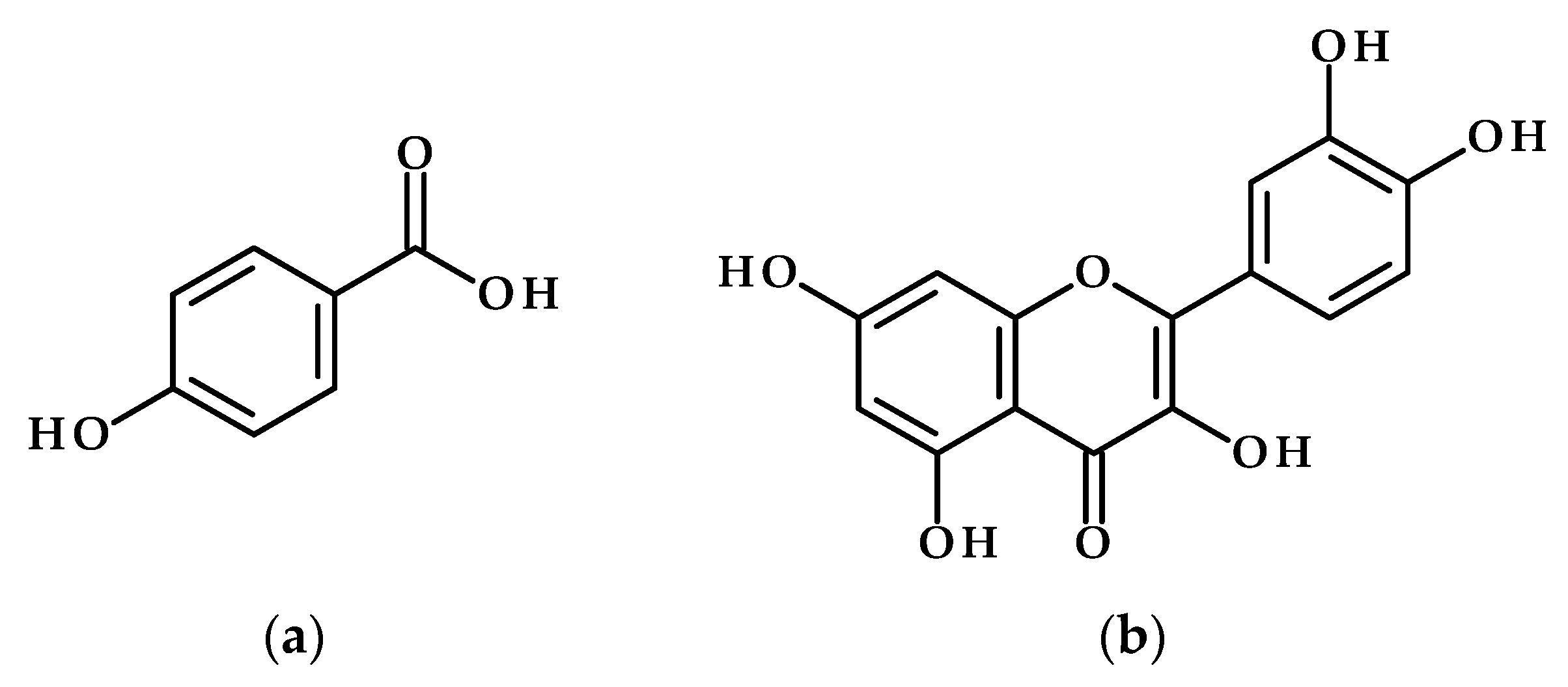
- Jan, R.; Khan, M.-A.; Asaf, S.; Lubna; Waqas, M.; Park, J.-R.; Asif, S.; Kim, N.; Lee, I.-J.; Kim, K.-M. Drought and UV Radiation Stress Tolerance in Rice Is Improved by Overaccumulation of Non-Enzymatic Antioxidant Flavonoids. Antioxidants 2022, 11, 917. [Google Scholar] [CrossRef]
- Ma, D.; Sun, D.; Wang, C.; Li, Y.; Guo, T. Expression of Flavonoid Biosynthesis Genes and Accumulation of Flavonoid in Wheat Leaves in Response to Drought Stress. Plant Physiol. Biochem. 2014, 80, 60–66. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Li, B.; Fan, R.; Sun, G.; Sun, T.; Fan, Y.; Bai, S.; Guo, S.; Huang, S.; Liu, J.; Zhang, H.; et al. Flavonoids Improve Drought Tolerance of Maize Seedlings by Regulating the Homeostasis of Reactive Oxygen Species. Plant Soil 2021, 461, 389–405. [Google Scholar] [CrossRef]
- Ahmed, U.; Rao, M.J.; Qi, C.; Xie, Q.; Noushahi, H.A.; Yaseen, M.; Shi, X.; Zheng, B. Expression Profiling of Flavonoid Biosynthesis Genes and Secondary Metabolites Accumulation in Populus under Drought Stress. Molecules 2021, 26, 5546. [Google Scholar] [CrossRef]
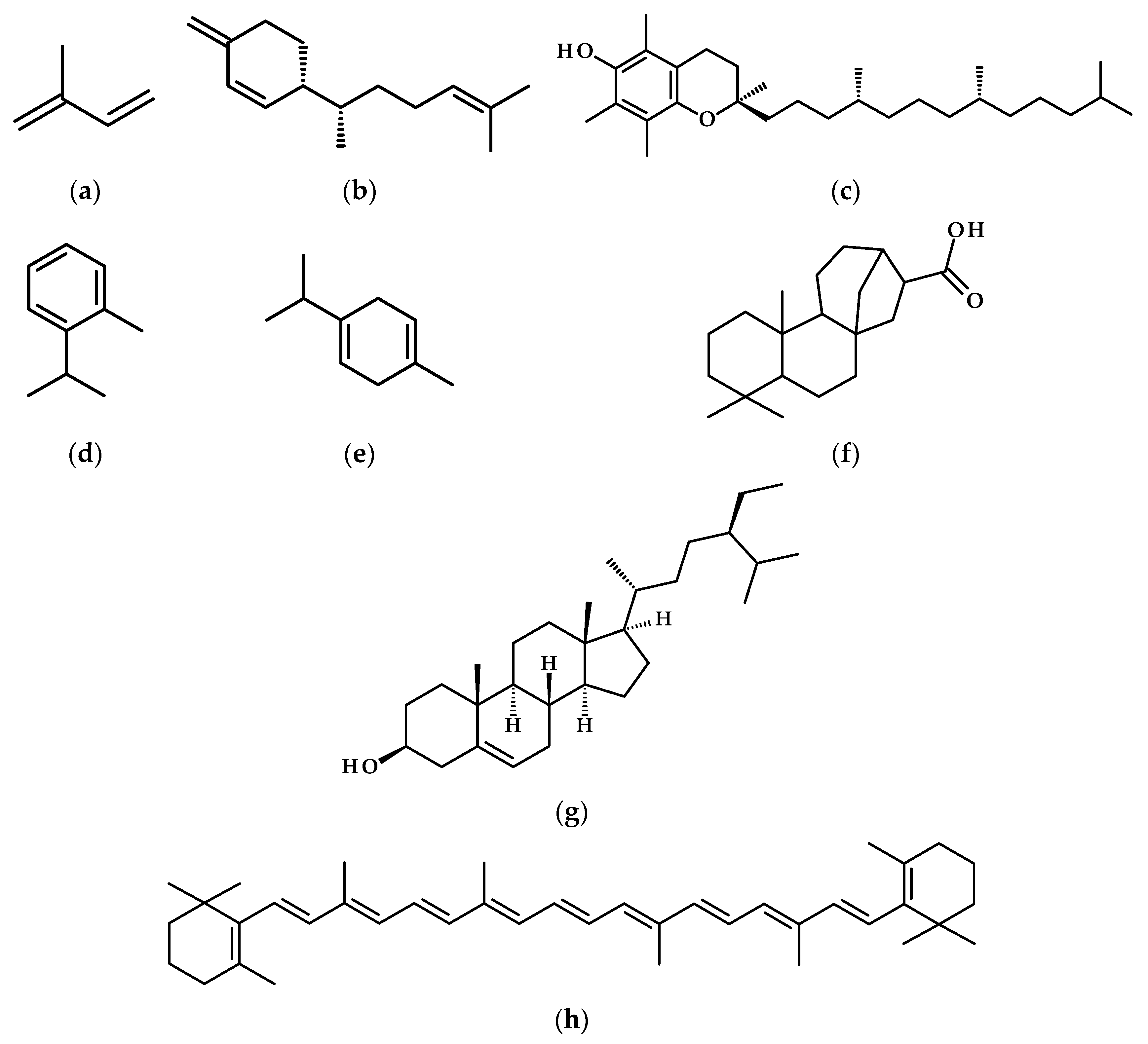
- Boncan, D.A.T.; Tsang, S.S.K.; Li, C.; Lee, I.H.T.; Lam, H.-M.; Chan, T.-F.; Hui, J.H.L. Terpenes and Terpenoids in Plants: Interactions with Environment and Insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef]
- Kleine, S.; Müller, C. Drought Stress and Leaf Herbivory Affect Root Terpenoid Concentrations and Growth of Tanacetum vulgare. J. Chem. Ecol. 2014, 40, 1115–1125. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Changes in Carotenoids, Tocopherols and Diterpenes during Drought and Recovery, and the Biological Significance of Chlorophyll Loss in Rosmarinus officinalis Plants. Planta 2000, 210, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, A.; Moradi, P.; Mastinu, A. Variation in Terpene Profiles of Thymus vulgaris in Water Deficit Stress Response. Molecules 2020, 25, 1091. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.M.; Christensen, S.; Schmelz, E.A.; Huffaker, A.; McAuslane, H.J.; Alborn, H.T.; Romero, M.; Allen, L.H.; Teal, P.E.A. Accumulation of Terpenoid Phytoalexins in Maize Roots Is Associated with Drought Tolerance: Maize Root Phytoalexins Play a Role in Drought Tolerance. Plant Cell Environ. 2015, 38, 2195–2207. [Google Scholar] [CrossRef]
- Loyola, J.; Verdugo, I.; González, E.; Casaretto, J.A.; Ruiz-Lara, S. Plastidic Isoprenoid Biosynthesis in Tomato: Physiological and Molecular Analysis in Genotypes Resistant and Sensitive to Drought Stress: Isoprenoid Pathway in Tomato. Plant Biol. 2012, 14, 149–156. [Google Scholar] [CrossRef]
- Patanè, C.; Siah, S.; Pellegrino, A.; Cosentino, S.L.; Siracusa, L. Fruit Yield, Polyphenols, and Carotenoids in Long Shelf-Life Tomatoes in Response to Drought Stress and Rewatering. Agronomy 2021, 11, 1943. [Google Scholar] [CrossRef]
- Kumar, M.S.S.; Ali, K.; Dahuja, A.; Tyagi, A. Role of Phytosterols in Drought Stress Tolerance in Rice. Plant Physiol. Biochem. 2015, 96, 83–89. [Google Scholar] [CrossRef]
- Zhang, R.-R.; Wang, Y.-H.; Li, T.; Tan, G.-F.; Tao, J.-P.; Su, X.-J.; Xu, Z.-S.; Tian, Y.-S.; Xiong, A.-S. Effects of Simulated Drought Stress on Carotenoid Contents and Expression of Related Genes in Carrot Taproots. Protoplasma 2021, 258, 379–390. [Google Scholar] [CrossRef]
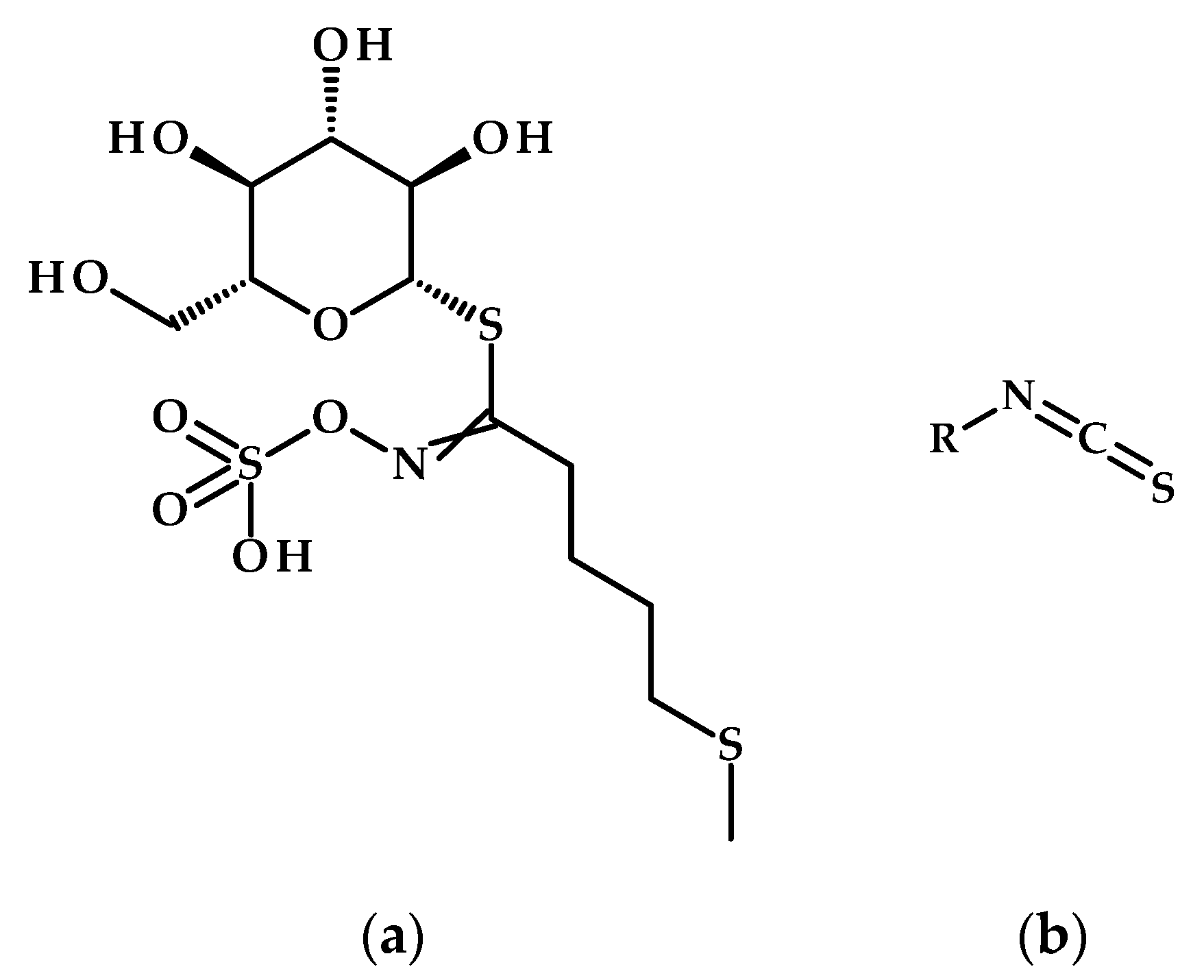
- Eom, S.H.; Baek, S.-A.; Kim, J.K.; Hyun, T.K. Transcriptome Analysis in Chinese Cabbage (Brassica rapa ssp. pekinensis) Provides the Role of Glucosinolate Metabolism in Response to Drought Stress. Molecules 2018, 23, 1186. [Google Scholar] [CrossRef]
- Ben Ammar, H.; Arena, D.; Treccarichi, S.; Di Bella, M.C.; Marghali, S.; Ficcadenti, N.; Lo Scalzo, R.; Branca, F. The Effect of Water Stress on the Glucosinolate Content and Profile: A Comparative Study on Roots and Leaves of Brassica oleracea L. Crops. Agronomy 2023, 13, 579. [Google Scholar] [CrossRef]
- Del Carmen Martínez-Ballesta, M.; Moreno, D.A.; Carvajal, M. The Physiological Importance of Glucosinolates on Plant Response to Abiotic Stress in Brassica. Int. J. Mol. Sci. 2013, 14, 11607–11625. [Google Scholar] [CrossRef]
- Tiwari, Y.K.; Yadav, S.K. High Temperature Stress Tolerance in Maize (Zea mays L.): Physiological and Molecular Mechanisms. J. Plant Biol. 2019, 62, 93–102. [Google Scholar] [CrossRef]
- Abasi, F.; Raja, N.I.; Mashwani, Z.-U.-R.; Ehsan, M.; Ali, H.; Shahbaz, M. Heat and Wheat: Adaptation Strategies with Respect to Heat Shock Proteins and Antioxidant Potential; an Era of Climate Change. Int. J. Biol. Macromol. 2024, 256, 128379. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Boro, P.; Karmakar, K.; Pradhan, P.; Saha Chowdhury, R.; Das, B.; Mandal, R.; Kumar, D. Advances in the Understanding of Heat Shock Proteins and Their Functions in Reducing Abiotic Stress in Plants. J. Plant Biochem. Biotechnol. 2024, 33, 474–491. [Google Scholar] [CrossRef]
- Manasa S, L.; Panigrahy, M.; Panigrahi, K.C.S.; Rout, G.R. Overview of Cold Stress Regulation in Plants. Bot. Rev. 2022, 88, 359–387. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Sung, D.Y.; Guy, C.L. Exploring the Temperature-Stress Metabolome of Arabidopsis. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef]
- Shao, J.; Huang, K.; Batool, M.; Idrees, F.; Afzal, R.; Haroon, M.; Noushahi, H.A.; Wu, W.; Hu, Q.; Lu, X.; et al. Versatile Roles of Polyamines in Improving Abiotic Stress Tolerance of Plants. Front. Plant Sci. 2022, 13, 1003155. [Google Scholar] [CrossRef]
- Tyagi, A.; Ali, S.; Ramakrishna, G.; Singh, A.; Park, S.; Mahmoudi, H.; Bae, H. Revisiting the Role of Polyamines in Plant Growth and Abiotic Stress Resilience: Mechanisms, Crosstalk, and Future Perspectives. J. Plant Growth Regul. 2023, 42, 5074–5098. [Google Scholar] [CrossRef]
- Jing, J.; Guo, S.; Li, Y.; Li, W. The Alleviating Effect of Exogenous Polyamines on Heat Stress Susceptibility of Different Heat Resistant Wheat (Triticum aestivum L.) Varieties. Sci. Rep. 2020, 10, 7467. [Google Scholar] [CrossRef]
- Shen, W.; Nada, K.; Tachibana, S. Involvement of Polyamines in the Chilling Tolerance of Cucumber Cultivars. Plant Physiol. 2000, 124, 431–439. [Google Scholar] [CrossRef]
- Paupière, M.J.; Müller, F.; Li, H.; Rieu, I.; Tikunov, Y.M.; Visser, R.G.F.; Bovy, A.G. Untargeted Metabolomic Analysis of Tomato Pollen Development and Heat Stress Response. Plant Reprod. 2017, 30, 81–94. [Google Scholar] [CrossRef]
- Henkrar, F.; Udupa, S. Marker Assisted Selection in Plant Breeding. Moroc. J. Agric. Sci. 2020, 1, 237–247. Available online: https://www.techagro.org/index.php/MJAS/article/view/865/930 (accessed on 5 June 2025).
- Latif, M.A.; Kayess, O.; Hasan, R.; Rahman, L. Molecular Marker Assisted Gene Stacking for Multiple Diseases Resistance in an Elite Rice Cultivar, BRRI Dhan48. Plant Gene 2025, 42, 100505. [Google Scholar] [CrossRef]
- Kumar, P.; Choudhary, M.; Halder, T.; Prakash, N.R.; Singh, V.; V, V.T.; Sheoran, S.; T, R.K.; Longmei, N.; Rakshit, S.; et al. Salinity Stress Tolerance and Omics Approaches: Revisiting the Progress and Achievements in Major Cereal Crops. Heredity 2022, 128, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Boros, A.; Szólik, E.; Desalegn, G.; Tőzsér, D. A Systematic Review of Opportunities and Limitations of Innovative Practices in Sustainable Agriculture. Agronomy 2024, 15, 76. [Google Scholar] [CrossRef]
- Garg, R.; Varshney, R.K.; Jain, M. Molecular Genetics and Genomics of Abiotic Stress Responses. Front. Plant Sci. 2014, 5, 398. [Google Scholar] [CrossRef][Green Version]
- Roychoudhury, A.; Datta, K.; Datta, S.K. Abiotic Stress in Plants: From Genomics to Metabolomics. In Omics and Plant Abiotic Stress Tolerance; Roychoudhury, A., Datta, K., Datta, S.K., Eds.; Bentham Science Publishers: Sharjah, United Arab Emirates, 2011; pp. 91–120. ISBN 9781608050581. [Google Scholar]
- Dawid, C.; Hille, K. Functional Metabolomics—A Useful Tool to Characterize Stress-Induced Metabolome Alterations Opening New Avenues towards Tailoring Food Crop Quality. Agronomy 2018, 8, 138. [Google Scholar] [CrossRef]
- Karan, R. Omics Study for Abiotic Stress Responses in Plants. Adv. Plants Agric. Res. 2015, 2, 28–34. [Google Scholar] [CrossRef]
- Yadav, P.; Li, G.; Mulet, J.M. Editorial: Transcriptome & Metabolic Profiling: An Insight into the Abiotic Stress Response Crosstalk in Plants. Front. Plant Sci. 2024, 15, 1370817. [Google Scholar] [CrossRef]
- Dickinson, E.; Rusilowicz, M.J.; Dickinson, M.; Charlton, A.J.; Bechtold, U.; Mullineaux, P.M.; Wilson, J. Integrating Transcriptomic Techniques and K-Means Clustering in Metabolomics to Identify Markers of Abiotic and Biotic Stress in Medicago Truncatula. Metabolomics 2018, 14, 126. [Google Scholar] [CrossRef]
- Ahmad, P.; Abdel Latef, A.A.H.; Rasool, S.; Akram, N.A.; Ashraf, M.; Gucel, S. Role of Proteomics in Crop Stress Tolerance. Front. Plant Sci. 2016, 7, 1336. [Google Scholar] [CrossRef]
- Gong, F.; Hu, X.; Wang, W. Proteomic Analysis of Crop Plants under Abiotic Stress Conditions: Where to Focus Our Research? Front. Plant Sci. 2015, 6, 418. [Google Scholar] [CrossRef] [PubMed]
- Moshood, A.Y.; Abdulraheem, M.I.; Li, L.; Zhang, Y.; Raghavan, V.; Hu, J. Deciphering Nutrient Stress in Plants: Integrative Insight from Metabolomics and Proteomics. Funct. Integr. Genom. 2025, 25, 38. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, R.; Ahmad, F.; Kaya, C.; Upadhyay, S.K.; Muneer, S.; Kumar, V.; Meena, M.; Liu, H.; Upadhyaya, H.; Seth, C.S. Decrypting Proteomics, Transcriptomics, Genomics, and Integrated Omics for Augmenting the Abiotic, Biotic, and Climate Change Stress Resilience in Plants. J. Plant Physiol. 2025, 305, 154430. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses. Genes 2023, 14, 1281. [Google Scholar] [CrossRef]
- Varadharajan, V.; Rajendran, R.; Muthuramalingam, P.; Runthala, A.; Madhesh, V.; Swaminathan, G.; Murugan, P.; Srinivasan, H.; Park, Y.; Shin, H.; et al. Multi-Omics Approaches against Abiotic and Biotic Stress-A Review. Plants 2025, 14, 865. [Google Scholar] [CrossRef]
- Mahmood, U.; Li, X.; Fan, Y.; Chang, W.; Niu, Y.; Li, J.; Qu, C.; Lu, K. Multi-Omics Revolution to Promote Plant Breeding Efficiency. Front. Plant Sci. 2022, 13, 1062952. [Google Scholar] [CrossRef]
- Bisht, A.; Saini, D.K.; Kaur, B.; Batra, R.; Kaur, S.; Kaur, I.; Jindal, S.; Malik, P.; Sandhu, P.K.; Kaur, A.; et al. Multi-Omics Assisted Breeding for Biotic Stress Resistance in Soybean. Mol. Biol. Rep. 2023, 50, 3787–3814. [Google Scholar] [CrossRef]
- Razzaq, A.; Sadia, B.; Raza, A.; Khalid Hameed, M.; Saleem, F. Metabolomics: A Way Forward for Crop Improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef]
- Suharti, W.S.; Nose, A.; Zheng, S.-H. Metabolomic Study of Two Rice Lines Infected by Rhizoctonia Solani in Negative Ion Mode by CE/TOF-MS. J. Plant Physiol. 2016, 206, 13–24. [Google Scholar] [CrossRef]
- Kajrolkar, A. Integrating Multi-Omics Data for Plant Stress Response: Current Advances and Future Directions. Prem. J. Plant Biol. 2025, 3, 100012. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, C.; Yu, C.; Dong, J.; Hu, J. Integration of Multi-Omics Technologies for Crop Improvement: Status and Prospects. Front. Bioinform. 2022, 2, 1027457. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated Omics Approaches in Plant Systems Biology. Curr. Opin. Chem. Biol. 2009, 13, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, C. Development and Applications of Metabolic Models in Plant Multi-Omics Research. Front. Plant Sci. 2024, 15, 1361183. [Google Scholar] [CrossRef]
- Zhang, J. Emerging Trends in Multi-Omics Data Integration: Challenges and Future Directions. Comput. Mol. Biol. 2024, 14, 64–75. [Google Scholar] [CrossRef]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and Metabolomic Data Integration. Brief. Bioinform. 2016, 17, 891–901. [Google Scholar] [CrossRef]
- Jamil, I.N.; Remali, J.; Azizan, K.A.; Nor Muhammad, N.A.; Arita, M.; Goh, H.-H.; Aizat, W.M. Systematic Multi-Omics Integration (MOI) Approach in Plant Systems Biology. Front. Plant Sci. 2020, 11, 944. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-Omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Reska, D.; Czajkowski, M.; Jurczuk, K.; Boldak, C.; Kwedlo, W.; Bauer, W.; Koszelew, J.; Kretowski, M. Integration of Solutions and Services for Multi-Omics Data Analysis towards Personalized Medicine. Biocybern. Biomed. Eng. 2021, 41, 1646–1663. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Lê Cao, K.-A. DIABLO: An Integrative Approach for Identifying Key Molecular Drivers from Multi-Omics Assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef] [PubMed]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis-a Framework for Unsupervised Integration of Multi-Omics Data Sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Nagel, A.; Thimm, O.; Redestig, H.; Blaesing, O.E.; Palacios-Rojas, N.; Selbig, J.; Hannemann, J.; Piques, M.C.; Steinhauser, D.; et al. Extension of the Visualization Tool MapMan to Allow Statistical Analysis of Arrays, Display of Corresponding Genes, and Comparison with Known Responses. Plant Physiol. 2005, 138, 1195–1204. [Google Scholar] [CrossRef]
- Rico-Chávez, A.K.; Franco, J.A.; Fernandez-Jaramillo, A.A.; Contreras-Medina, L.M.; Guevara-González, R.G.; Hernandez-Escobedo, Q. Machine Learning for Plant Stress Modeling: A Perspective towards Hormesis Management. Plants 2022, 11, 970. [Google Scholar] [CrossRef]
- Cembrowska-Lech, D.; Krzemińska, A.; Miller, T.; Nowakowska, A.; Adamski, C.; Radaczyńska, M.; Mikiciuk, G.; Mikiciuk, M. An Integrated Multi-Omics and Artificial Intelligence Framework for Advance Plant Phenotyping in Horticulture. Biology 2023, 12, 1298. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.-P.; Bodein, A.; Périn, O.; Droit, A. Integration Strategies of Multi-Omics Data for Machine Learning Analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Zhou, Y.; Luo, K.; Liang, L.; Chen, M.; He, X. A New Bayesian Factor Analysis Method Improves Detection of Genes and Biological Processes Affected by Perturbations in Single-Cell CRISPR Screening. Nat. Methods 2023, 20, 1693–1703. [Google Scholar] [CrossRef]
- Razzaq, A.; Wishart, D.S.; Wani, S.H.; Hameed, M.K.; Mubin, M.; Saleem, F. Advances in Metabolomics-Driven Diagnostic Breeding and Crop Improvement. Metabolites 2022, 12, 511. [Google Scholar] [CrossRef]
- Salam, U.; Ullah, S.; Tang, Z.-H.; Elateeq, A.A.; Khan, Y.; Khan, J.; Khan, A.; Ali, S. Plant Metabolomics: An Overview of the Role of Primary and Secondary Metabolites against Different Environmental Stress Factors. Life 2023, 13, 706. [Google Scholar] [CrossRef]
- Song, Y.; John Martin, J.J.; Liu, X.; Li, X.; Hou, M.; Zhang, R.; Xu, W.; Li, W.; Cao, H. Unraveling the Response of Secondary Metabolites to Cold Tolerance in Oil Palm by Integration of Physiology and Metabolomic Analyses. BMC Plant Biol. 2025, 25, 279. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, J.; Sade, N.; Wu, S.; Egbaria, A.; Fernie, A.R.; Yan, J.; Qin, F.; Chen, W.; Brotman, Y.; et al. Genomic Basis Underlying the Metabolome-Mediated Drought Adaptation of Maize. Genome Biol. 2021, 22, 260. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, X.; Shi, T.; Yin, H.; Sun, D.; Hao, Y.; Xia, X.; Luo, J.; Fernie, A.R.; He, Z.; et al. Metabolite-Based Genome-Wide Association Study Enables Dissection of the Flavonoid Decoration Pathway of Wheat Kernels. Plant Biotechnol. J. 2020, 18, 1722–1735. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Han, T.; Wu, Y.; Lyu, L.; Wu, W.; Li, W. Quality Analysis and Metabolomic Profiling of the Effects of Exogenous Abscisic Acid on Rabbiteye Blueberry. Front. Plant Sci. 2023, 14, 1224245. [Google Scholar] [CrossRef] [PubMed]
- Tinte, M.M.; Chele, K.H.; van der Hooft, J.J.J.; Tugizimana, F. Metabolomics-Guided Elucidation of Plant Abiotic Stress Responses in the 4IR Era: An Overview. Metabolites 2021, 11, 445. [Google Scholar] [CrossRef]
- Parida, A.K.; Panda, A.; Rangani, J. Metabolomics-Guided Elucidation of Abiotic Stress Tolerance Mechanisms in Plants. In Plant Metabolites and Regulation Under Environmental Stress; Elsevier: Amsterdam, The Netherlands, 2018; pp. 89–131. ISBN 9780128126899. [Google Scholar]
- Kumar, M.; Prusty, M.R.; Pandey, M.K.; Singh, P.K.; Bohra, A.; Guo, B.; Varshney, R.K. Application of CRISPR/Cas9-Mediated Gene Editing for Abiotic Stress Management in Crop Plants. Front. Plant Sci. 2023, 14, 1157678. [Google Scholar] [CrossRef]
- Nascimento, F.d.S.; Rocha, A.d.J.; Soares, J.M.d.S.; Mascarenhas, M.S.; Ferreira, M.d.S.; Morais Lino, L.S.; Ramos, A.P.d.S.; Diniz, L.E.C.; Mendes, T.A.d.O.; Ferreira, C.F.; et al. Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review. Plants 2023, 12, 305. [Google Scholar] [CrossRef]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, F.; Wang, F.; Le, L.; Pu, L. Synthetic Biology and Artificial Intelligence in Crop Improvement. Plant Commun. 2025, 6, 101220. [Google Scholar] [CrossRef]
- Lv, X.; Hueso-Gil, A.; Bi, X.; Wu, Y.; Liu, Y.; Liu, L.; Ledesma-Amaro, R. New Synthetic Biology Tools for Metabolic Control. Curr. Opin. Biotechnol. 2022, 76, 102724. [Google Scholar] [CrossRef]
- Kambona, C.M.; Koua, P.A.; Léon, J.; Ballvora, A. Stress Memory and Its Regulation in Plants Experiencing Recurrent Drought Conditions. Züchter Genet. Breed. Res. 2023, 136, 26. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Zhang, Z.; Mullasseri, S.; Kalendar, R.; Ahmad, Z.; Sharma, A.; Liu, G.; Zhou, M.; Wei, Q. Epigenetic Stress Memory: A New Approach to Study Cold and Heat Stress Responses in Plants. Front. Plant Sci. 2022, 13, 1075279. [Google Scholar] [CrossRef] [PubMed]
- Romero, H.; Pott, D.M.; Vallarino, J.G.; Osorio, S. Metabolomics-Based Evaluation of Crop Quality Changes as a Consequence of Climate Change. Metabolites 2021, 11, 461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, S.; Li, B.; Chen, W.; Li, Y.; He, X.; Wang, N. Responses of Spring Leaf Phenological and Functional Traits of Two Urban Tree Species to Air Warming and/or Elevated Ozone. Plant Physiol. Biochem. 2022, 179, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Wakchaure, G.C.; Kumar, D.; Kumar, S.; Gawhale, B.J.; Meena, K.K.; Sawant, C.P.; Singh, D.K.; Paramasivam, S.K.; Minhas, P.S. Grafting Wild Rootstocks as a Climate-Resilient Strategy to Enhance Productivity, Quality and Tolerance in Eggplant under Variable Water Stress Induced by Deficit Irrigation. Agric. Water Manag. 2025, 314, 109492. [Google Scholar] [CrossRef]

| Feature | GC-MS | LC-MS | NMR Spectroscopy |
|---|---|---|---|
| Sensitivity | High | Very high | Moderate |
| Reproducibility | High | Moderate–high | Very high |
| Sample preparation | Requires derivatization | No derivatization needed | Minimal, especially for HR-MAS |
| Metabolite coverage | Volatile, polar metabolites | Broad (polar and non-polar) | Limited, mostly abundant metabolites |
| Quantification | Relative or absolute (with standards) | Relative or absolute (with standards) | Absolute without standards |
| Structural elucidation | Limited | Limited to fragmentation data | Strong (direct molecular structure) |
| Destructive analysis | Yes | Yes | No |
| Throughput | Moderate | High | Moderate |
| Cost (instrument and maintenance) | Moderate–high | High | Very high |
| Common applications | Sugars, amino acids, and organic acids | Secondary metabolites, lipids, phenolics | Structural ID, metabolite fingerprinting |
| Step | Purpose | Methods/Techniques | Practical Notes |
|---|---|---|---|
| Ensure biological relevance and statistical robustness | Define replicates, randomization, and sample size | Standardize environmental/growth conditions |
| Preserve metabolic state at harvest | Rapid harvesting, consistent timing | Avoid contamination; handle with gloves/tools |
| Halt metabolic activity immediately | Flash-freezing in liquid nitrogen, cold solvents (methanol, dry ice–ethanol) | Store at −80 °C; transport on dry ice |
| Disrupt tissue for uniform extraction | Cryogenic grinding (mortar and pestle or bead beater) | Prevent thawing; use precooled tools |
| Isolate metabolites from the plant matrix | Solvent systems (methanol:water, chloroform:methanol:water, MTBE, etc.) | Select a solvent based on target metabolites and platform |
| Remove solids and reduce matrix effects | Centrifugation, filtration (0.2 μm), Solid Phase Extraction | Avoid contamination, work on ice |
| Improve volatility and stability | Oximation + silylation (e.g., MSTFA) | Required for GC-MS; not needed for LC-MS or NMR |
| Prepare a sample for analysis | Reconstitute in LC/NMR-compatible solvents (e.g., acetonitrile:water, D2O) | Use appropriate internal standards |
| Ensure analytical reproducibility | Use pooled QC samples, internal standards, and randomized injection order | Monitor instrument drift and variability |
| Maintain sample integrity | Freeze at −80 °C; minimize freeze-thaw cycles | Aliquot samples for repeatability |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głuchowska, A.; Zieniuk, B.; Pawełkowicz, M. Unlocking Plant Resilience: Metabolomic Insights into Abiotic Stress Tolerance in Crops. Metabolites 2025, 15, 384. https://doi.org/10.3390/metabo15060384
Głuchowska A, Zieniuk B, Pawełkowicz M. Unlocking Plant Resilience: Metabolomic Insights into Abiotic Stress Tolerance in Crops. Metabolites. 2025; 15(6):384. https://doi.org/10.3390/metabo15060384
Chicago/Turabian StyleGłuchowska, Agata, Bartłomiej Zieniuk, and Magdalena Pawełkowicz. 2025. "Unlocking Plant Resilience: Metabolomic Insights into Abiotic Stress Tolerance in Crops" Metabolites 15, no. 6: 384. https://doi.org/10.3390/metabo15060384
APA StyleGłuchowska, A., Zieniuk, B., & Pawełkowicz, M. (2025). Unlocking Plant Resilience: Metabolomic Insights into Abiotic Stress Tolerance in Crops. Metabolites, 15(6), 384. https://doi.org/10.3390/metabo15060384







