Adapting OptCouple to Identify Strategies with Increased Product Yields in Community Cohorts of E. coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Adapting OptCouple to Community GSMs
2.2. Adapting OptCouple to Maximise the Minimum Possible Product Yield in Community Strains
3. Results
3.1. Coupling 2,3-Butanediol to Community Growth
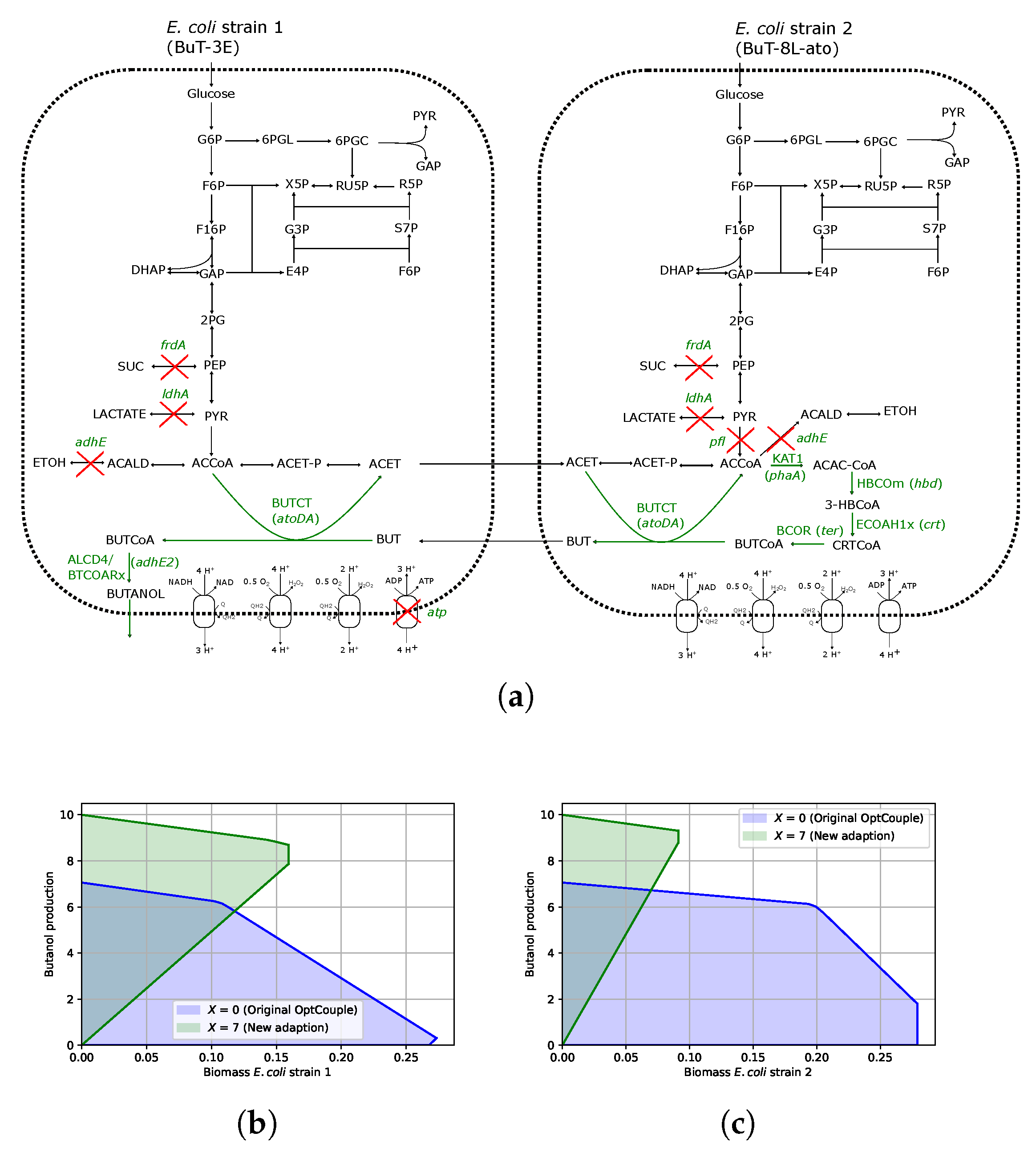
3.2. Coupling Butanol to Community Growth
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MILP | Mixed integer linear program |
| GSM | Genome-scale model |
References
- Matuszyńska, A.; Ebenhöh, O.; Zurbriggen, M.D.; Ducat, D.C.; Axmann, I.M. A new era of synthetic biology—Microbial community design. Synth. Biol. 2024, 9, ysae011. [Google Scholar] [CrossRef]
- Lloyd, C.J.; King, Z.A.; Sandberg, T.E.; Hefner, Y.; Olson, C.A.; Phaneuf, P.V.; O’Brien, E.J.; Sanders, J.G.; Salido, R.A.; Sanders, K.; et al. The genetic basis for adaptation of model-designed syntrophic co-cultures. PLoS Comput. Biol. 2019, 15, e1006213. [Google Scholar] [CrossRef]
- García-Jiménez, B.; Torres-Bacete, J.; Nogales, J. Metabolic modelling approaches for describing and engineering microbial communities. Comput. Struct. Biotechnol. J. 2021, 19, 226–246. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Broeken, V.; Hansen, A.S.L.; Sonnenschein, N.; Herrgård, M.J. OptCouple: Joint simulation of gene knockouts, insertions and medium modifications for prediction of growth-coupled strain designs. Metab. Eng. Commun. 2019, 8, e00087. [Google Scholar] [CrossRef]
- Ji, X.J.; Huang, H.; Zhu, J.G.; Ren, L.J.; Nie, Z.K.; Du, J.; Li, S. Engineering Klebsiella oxytoca for efficient 2,3-butanediol production through insertional inactivation of acetaldehyde dehydrogenase gene. Appl. Microbiol. Biotechnol. 2010, 85, 1751–1758. [Google Scholar] [CrossRef]
- Białkowska, A.M. Strategies for efficient and economical 2, 3-butanediol production: New trends in this field. World J. Microbiol. Biotechnol. 2016, 32, 200. [Google Scholar] [CrossRef]
- Keo-Oudone, C.; Phommachan, K.; Suliya, O.; Nurcholis, M.; Bounphanmy, S.; Kosaka, T.; Yamada, M. Highly efficient production of 2, 3-butanediol from xylose and glucose by newly isolated thermotolerant Cronobacter sakazakii. BMC Microbiol. 2022, 22, 164. [Google Scholar] [CrossRef] [PubMed]
- Gawal, P.M.; Subudhi, S. Advances and challenges in bio-based 2,3-BD downstream purification: A comprehensive review. Bioresour. Technol. Rep. 2023, 24, 101638. [Google Scholar] [CrossRef]
- Xu, Y.; Chu, H.; Gao, C.; Tao, F.; Zhou, Z.; Li, K.; Li, L.; Ma, C.; Xu, P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab. Eng. 2014, 23, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Fleming, R.M.T.; Palsson, B. Reconstruction and Use of Microbial Metabolic Networks: The Core Escherichia coli Metabolic Model as an Educational Guide. EcoSal Plus 2010, 4, 1–47. [Google Scholar] [CrossRef] [PubMed]
- E. coli Core Model. 2016. Available online: http://bigg.ucsd.edu/models/e_coli_core (accessed on 1 April 2025).
- Tan, Z.; Chen, J.; Zhang, X. Systematic engineering of pentose phosphate pathway improves Escherichia coli succinate production. Biotechnol. Biofuels 2016, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Saini, M.; Chen, M.H.; Chiang, C.J.; Chao, Y.P. Potential production platform of n-butanol in Escherichia coli. Metab. Eng. 2015, 27, 76–82. [Google Scholar] [CrossRef] [PubMed]

| Product | Knockouts | Cross-Feeding | Production Rate | Yield |
|---|---|---|---|---|
| 2,3-butanediol | ||||
| Butanol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pearcy, N.; Twycross, J. Adapting OptCouple to Identify Strategies with Increased Product Yields in Community Cohorts of E. coli. Metabolites 2025, 15, 309. https://doi.org/10.3390/metabo15050309
Pearcy N, Twycross J. Adapting OptCouple to Identify Strategies with Increased Product Yields in Community Cohorts of E. coli. Metabolites. 2025; 15(5):309. https://doi.org/10.3390/metabo15050309
Chicago/Turabian StylePearcy, Nicole, and Jamie Twycross. 2025. "Adapting OptCouple to Identify Strategies with Increased Product Yields in Community Cohorts of E. coli" Metabolites 15, no. 5: 309. https://doi.org/10.3390/metabo15050309
APA StylePearcy, N., & Twycross, J. (2025). Adapting OptCouple to Identify Strategies with Increased Product Yields in Community Cohorts of E. coli. Metabolites, 15(5), 309. https://doi.org/10.3390/metabo15050309






