Polymorphisms Involved in Insulin Resistance and Metabolic Inflammation: Influence of Nutrients and Dietary Interventions
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
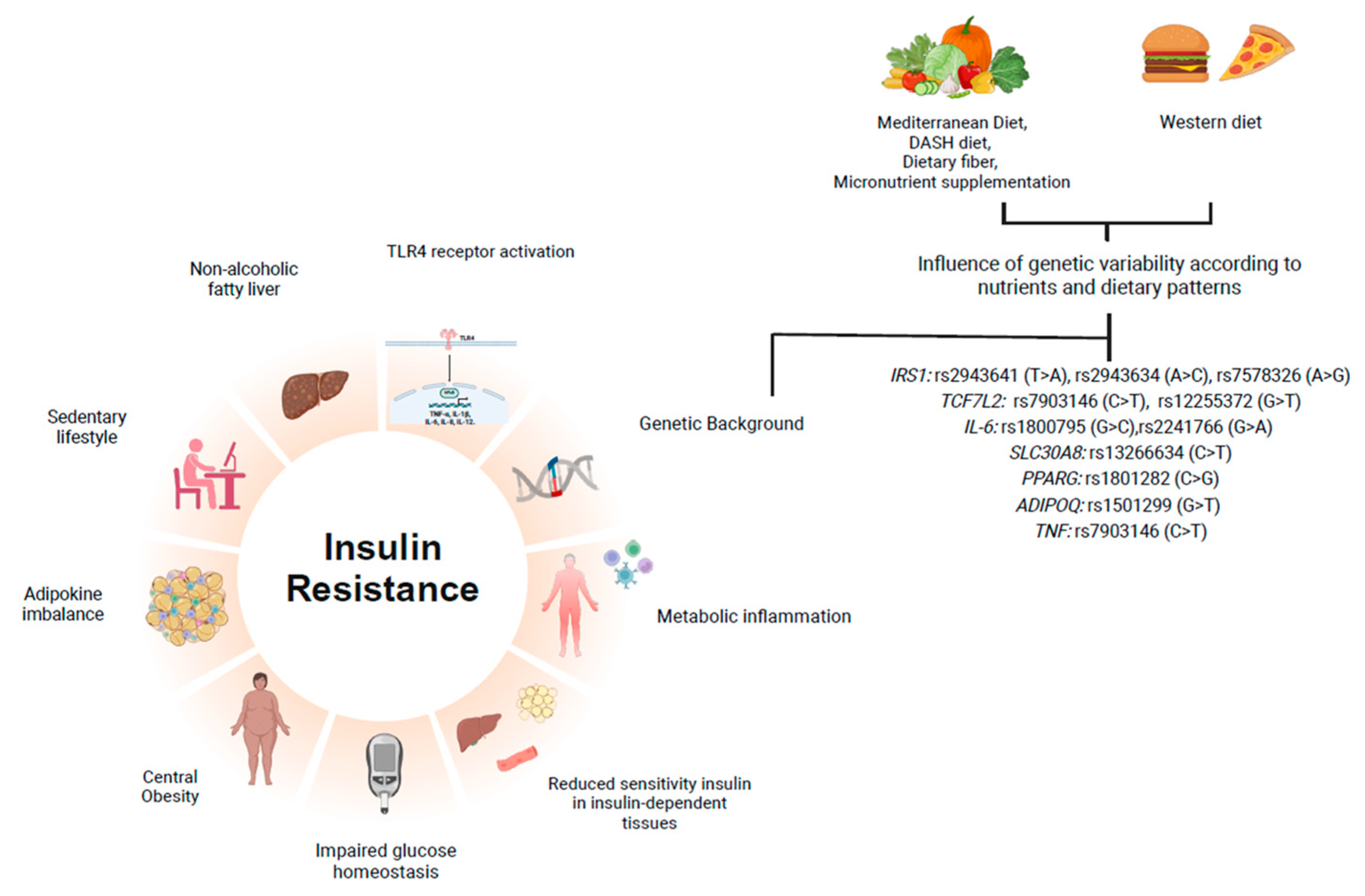
3.1. IR and Metabolic Inflammation
3.2. Gene–Nutrient Interactions in IR: Evidence of Functional Consequences of Genetic Polymorphism
3.2.1. TCF7L2
3.2.2. SLC30A8 (ZnT8)
3.2.3. IRS-1
3.2.4. TNF-α
3.2.5. TLR-4
3.2.6. IL-6
3.2.7. PPAR-γ
3.2.8. Adiponectin
3.3. Effects of Nutritional Interventions on Polygenic Risk Scores Affecting IR Biomarkers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADIPOQ: | Adiponectin |
| APOA-1: | Apolipoprotein A-1 |
| BMI: | Body Mass Index |
| DASH: | Dietary Approaches to Stop Hypertension |
| DNL: | De Novo Lipogenesis |
| FTO: | Fat Mass and Obesity Associated |
| GWAS: | Genome-Wide Association Studies |
| HbA1c: | Glycated Hemoglobin |
| HDL: | High-Density Lipoprotein |
| HOMA-IR: | Homeostasis Model Assessment of Insulin Resistance |
| IL-6: | Interleukin-6 |
| IRS: | Insulin Receptor Substrate |
| IR: | Insulin Resistance |
| JNK: | c-Jun N-terminal Kinase |
| MedDiet: | Mediterranean Diet |
| MC4R: | Melanocortin-4 Receptor |
| MUFA: | Monounsaturated Fatty Acids |
| NF-κB: | Nuclear Factor Kappa B |
| PUFA: | Polyunsaturated Fatty Acids |
| PRS: | Polygenic Risk Score |
| SNP: | Single-Nucleotide Polymorphism |
| SAFA: | Saturated Fatty Acids |
| TCF7L2: | Transcription Factor 7-Like 2 |
| TLR: | Toll-Like Receptor |
| TNF-α: | Tumor Necrosis Factor-Alpha |
| T2DM: | Type 2 Diabetes |
| TyG: | Triglyceride-Glucose Index |
| GRS: | Genetic Risk Score |
| PRS: | Polygenic Risk Score |
References
- Freeman, A.M.; Acevedo, L.A.; Pennings, N. Insulin Resistance. Available online: https://www.ncbi.nlm.nih.gov/books/NBK507839/ (accessed on 4 December 2024).
- Mirabelli, M.; Chiefari, E.; Arcidiacono, B.; Corigliano, D.M.; Brunetti, F.S.; Maggisano, V.; Russo, D.; Foti, D.P.; Brunetti, A. Mediterranean Diet Nutrients to Turn the Tide against Insulin Resistance and Related Diseases. Nutrients 2020, 12, 1066. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.O.; Conde, S.V. Impact of Diet Composition on Insulin Resistance. Nutrients 2022, 14, 3716. [Google Scholar] [CrossRef]
- Li, H.; Meng, Y.; He, S.; Tan, X.; Zhang, Y.; Zhang, X.; Wang, L.; Zheng, W. Macrophages, Chronic Inflammation, and Insulin Resistance. Cells 2022, 11, 3001. [Google Scholar] [CrossRef]
- Engin, A.B. Message Transmission Between Adipocyte and Macrophage in Obesity. Adv. Exp. Med. Biol. 2024, 1460, 273–295. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Martinez, M.; Cabail, M.Z. The PI3K/Akt Pathway in Meta-Inflammation. Int. J. Mol. Sci. 2022, 23, 15330. [Google Scholar] [CrossRef] [PubMed]
- Szukiewicz, D. Molecular Mechanisms for the Vicious Cycle between Insulin Resistance and the Inflammatory Response in Obesity. Int. J. Mol. Sci. 2023, 24, 9818. [Google Scholar] [CrossRef]
- Brown, A.E.; Walker, M. Genetics of Insulin Resistance and the Metabolic Syndrome. Curr. Cardiol. Rep. 2016, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bai, R.; Wang, Y.; Qu, M.; Zhou, Y.; Gao, Z.; Wang, Y. The multifaceted function of FoxO1 in pancreatic beta-cell dysfunction and insulin resistance: Therapeutic potential for type 2 diabetes. Life Sci. 2025, 364, 123384. [Google Scholar] [CrossRef]
- Lu, X.; Xie, Q.; Pan, X.; Zhang, R.; Zhang, X.; Peng, G.; Zhang, Y.; Shen, S.; Tong, N. Type 2 diabetes mellitus in adults: Pathogenesis, prevention and therapy. Signal Transduct. Target. Ther. 2024, 9, 262. [Google Scholar] [CrossRef]
- Hosseinpour-Niazi, S.; Mirmiran, P.; Hosseini, S.; Hadaegh, F.; Ainy, E.; Daneshpour, M.S.; Azizi, F. Effect of TCF7L2 on the relationship between lifestyle factors and glycemic parameters: A systematic review. Nutr. J. 2022, 21, 59. [Google Scholar] [CrossRef]
- Dimas, A.S.; Lagou, V.; Barker, A.; Knowles, J.W.; Magi, R.; Hivert, M.F.; Benazzo, A.; Rybin, D.; Jackson, A.U.; Stringham, H.M.; et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes 2014, 63, 2158–2171. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, S.; Costa, V.; Ciccodicola, A.; Aprile, M. PPARgamma and Diabetes: Beyond the Genome and Towards Personalized Medicine. Curr. Diab. Rep. 2021, 21, 18. [Google Scholar] [CrossRef]
- Raskiliene, A.; Smalinskiene, A.; Kriaucioniene, V.; Lesauskaite, V.; Petkeviciene, J. Associations of MC4R, LEP, and LEPR Polymorphisms with Obesity-Related Parameters in Childhood and Adulthood. Genes 2021, 12, 949. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.M. Fat Mass and Obesity Associated (FTO) Gene and Hepatic Glucose and Lipid Metabolism. Nutrients 2018, 10, 1600. [Google Scholar] [CrossRef] [PubMed]
- Jermendy, G.; Horvath, T.; Littvay, L.; Steinbach, R.; Jermendy, A.L.; Tarnoki, A.D.; Tarnoki, D.L.; Metneki, J.; Osztovits, J. Effect of genetic and environmental influences on cardiometabolic risk factors: A twin study. Cardiovasc. Diabetol. 2011, 10, 96. [Google Scholar] [CrossRef]
- de Soysa, A.K.H.; Martins, C.; Langaas, M.; Grill, V.; Mostad, I.L. Exploring Dietary Intake in Adults with Severe Obesity and Associations with the FTO rs9939609 Genotypes. Curr. Dev. Nutr. 2023, 7, 100032. [Google Scholar] [CrossRef]
- Yu, K.; Li, L.; Zhang, L.; Guo, L.; Wang, C. Association between MC4R rs17782313 genotype and obesity: A meta-analysis. Gene 2020, 733, 144372. [Google Scholar] [CrossRef]
- Feng, Y.; Jiang, C.D.; Chang, A.M.; Shi, Y.; Gao, J.; Zhu, L.; Zhang, Z. Interactions among insulin resistance, inflammation factors, obesity-related gene polymorphisms, environmental risk factors, and diet in the development of gestational diabetes mellitus. J. Matern. Fetal Neonatal Med. 2019, 32, 339–347. [Google Scholar] [CrossRef]
- Gardner, C.D.; Trepanowski, J.F.; Del Gobbo, L.C.; Hauser, M.E.; Rigdon, J.; Ioannidis, J.P.A.; Desai, M.; King, A.C. Effect of Low-Fat vs Low-Carbohydrate Diet on 12-Month Weight Loss in Overweight Adults and the Association With Genotype Pattern or Insulin Secretion: The DIETFITS Randomized Clinical Trial. JAMA 2018, 319, 667–679. [Google Scholar] [CrossRef]
- de Luis, D.; Izaola, O.; Primo, D. APOA-5 Genetic Variant rs662799: Role in Lipid Changes and Insulin Resistance after a Mediterranean Diet in Caucasian Obese Subjects. Dis. Markers 2021, 2021, 1257145. [Google Scholar] [CrossRef]
- Vallee Marcotte, B.; Cormier, H.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.C. Polymorphisms in FFAR4 (GPR120) Gene Modulate Insulin Levels and Sensitivity after Fish Oil Supplementation. J. Pers. Med. 2017, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- ElhamKia, M.; Setayesh, L.; Yarizadeh, H.; Pooyan, S.; Veisy, Z.; Aghamohammadi, V.; Casazza, K.; Mirzaei, K. The Interaction Between Dietary Total Antioxidant Capacity and MC4R Gene and HOMA-IR in Metabolically Healthy and Unhealthy Overweight and Obese Women. Nutr. Metab. Insights 2022, 15, 11786388221105984. [Google Scholar] [CrossRef]
- Zhou, L.; Luo, C.; Yin, J.; Zhu, Y.; Li, P.; Chen, S.; Sun, T.; Xie, M.; Shan, Z.; Cao, B.; et al. Diverse Associations of Plasma Selenium Concentrations and SELENOP Gene Polymorphism with Metabolic Syndrome and Its Components. Oxidative Med. Cell. Longev. 2020, 2020, 5343014. [Google Scholar] [CrossRef]
- Feng, J.; Wang, Q.; Yang, W.; Liu, J.; Gao, M.Q. Omega-3 polyunsaturated fatty acids ameliorated inflammatory response of mammary epithelial cells and mammary gland induced by lipopolysaccharide. Acta. Biochim. Biophys. Sin. 2021, 53, 1142–1153. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 Fatty Acids and Inflammatory Processes. Nutrients 2010, 2, 355–374. [Google Scholar] [CrossRef]
- Zheng, J.S.; Arnett, D.K.; Parnell, L.D.; Smith, C.E.; Li, D.; Borecki, I.B.; Tucker, K.L.; Ordovas, J.M.; Lai, C.Q. Modulation by dietary fat and carbohydrate of IRS1 association with type 2 diabetes traits in two populations of different ancestries. Diabetes. Care 2013, 36, 2621–2627. [Google Scholar] [CrossRef]
- Shen, L.; Liu, J.; Zhao, X.; Wang, A.; Hu, X. Association between insulin receptor substrate 1 gene polymorphism rs1801278 and gestational diabetes mellitus: An updated meta- analysis. Diabetol. Metab. Syndr. 2024, 16, 62. [Google Scholar] [CrossRef]
- Zhang, Z.; Ji, G.; Li, M. Glucokinase regulatory protein: A balancing act between glucose and lipid metabolism in NAFLD. Front. Endocrinol. 2023, 14, 1247611. [Google Scholar] [CrossRef]
- James, D.E.; Stockli, J.; Birnbaum, M.J. The aetiology and molecular landscape of insulin resistance. Nat. Rev. Mol. Cell. Biol. 2021, 22, 751–771. [Google Scholar] [CrossRef]
- Yaribeygi, H.; Farrokhi, F.R.; Butler, A.E.; Sahebkar, A. Insulin resistance: Review of the underlying molecular mechanisms. J. Cell. Physiol. 2019, 234, 8152–8161. [Google Scholar] [CrossRef]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 1–25. [Google Scholar]
- Shulman, G.I. Cellular mechanisms of insulin resistance. J. Clin. Investig. 2000, 106, 171–176. [Google Scholar] [CrossRef]
- Petersen, M.C.; Shulman, G.I. Mechanisms of Insulin Action and Insulin Resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef] [PubMed]
- Suren Garg, S.; Kushwaha, K.; Dubey, R.; Gupta, J. Association between obesity, inflammation and insulin resistance: Insights into signaling pathways and therapeutic interventions. Diabetes Res. Clin. Pract. 2023, 200, 110691. [Google Scholar] [CrossRef]
- Ye, J. Mechanisms of insulin resistance in obesity. Front. Med. 2013, 7, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Tahapary, D.L.; Pratisthita, L.B.; Fitri, N.A.; Marcella, C.; Wafa, S.; Kurniawan, F.; Rizka, A.; Tarigan, T.J.E.; Harbuwono, D.S.; Purnamasari, D.; et al. Challenges in the diagnosis of insulin resistance: Focusing on the role of HOMA-IR and Tryglyceride/glucose index. Diabetes Metab. Syndr. 2022, 16, 102581. [Google Scholar] [CrossRef]
- Zhao, X.; An, X.; Yang, C.; Sun, W.; Ji, H.; Lian, F. The crucial role and mechanism of insulin resistance in metabolic disease. Front. Endocrinol. 2023, 14, 1149239. [Google Scholar] [CrossRef]
- Russo, S.; Kwiatkowski, M.; Govorukhina, N.; Bischoff, R.; Melgert, B.N. Meta-Inflammation and Metabolic Reprogramming of Macrophages in Diabetes and Obesity: The Importance of Metabolites. Front. Immunol. 2021, 12, 746151. [Google Scholar] [CrossRef]
- Abd El-Kader, S.M.; Al-Jiffri, O.H. Impact of weight reduction on insulin resistance, adhesive molecules and adipokines dysregulation among obese type 2 diabetic patients. Afr. Health Sci. 2018, 18, 873–883. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation, metaflammation and immunometabolic disorders. Nature 2017, 542, 177–185. [Google Scholar] [CrossRef]
- Quintanilha, B.J.; Reis, B.Z.; Duarte, G.B.S.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, 9, 1168. [Google Scholar] [CrossRef] [PubMed]
- Rogero, M.M.; Calder, P.C. Obesity, Inflammation, Toll-Like Receptor 4 and Fatty Acids. Nutrients 2018, 10, 432. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Moschen, A.R. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol. Metab. 2008, 19, 371–379. [Google Scholar] [CrossRef]
- Elashi, A.A.; Toor, S.M.; Umlai, U.I.; Al-Sarraj, Y.A.; Taheri, S.; Suhre, K.; Abou-Samra, A.B.; Albagha, O.M.E. Genome-wide association study and trans-ethnic meta-analysis identify novel susceptibility loci for type 2 diabetes mellitus. BMC Med. Genom. 2024, 17, 115. [Google Scholar] [CrossRef]
- Ortega-Contreras, B.; Armella, A.; Appel, J.; Mennickent, D.; Araya, J.; Gonzalez, M.; Castro, E.; Obregon, A.M.; Lamperti, L.; Gutierrez, J.; et al. Pathophysiological Role of Genetic Factors Associated With Gestational Diabetes Mellitus. Front. Physiol. 2022, 13, 769924. [Google Scholar] [CrossRef]
- Nguyen-Tu, M.S.; da Silva Xavier, G.; Leclerc, I.; Rutter, G.A. Transcription factor-7-like 2 (TCF7L2) gene acts downstream of the Lkb1/Stk11 kinase to control mTOR signaling, beta cell growth, and insulin secretion. J. Biol. Chem. 2018, 293, 14178–14189. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Xu, L.; Zhang, L.; Han, Z.; Jiang, Q.; Wang, Z.; Jin, S. Meta-analysis of association between TCF7L2 polymorphism rs7903146 and type 2 diabetes mellitus. BMC Med. Genet. 2018, 19, 38. [Google Scholar] [CrossRef]
- Sladek, R.; Rocheleau, G.; Rung, J.; Dina, C.; Shen, L.; Serre, D.; Boutin, P.; Vincent, D.; Belisle, A.; Hadjadj, S.; et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007, 445, 881–885. [Google Scholar] [CrossRef]
- Assmann, T.S.; Duarte, G.C.; Rheinheimer, J.; Cruz, L.A.; Canani, L.H.; Crispim, D. The TCF7L2 rs7903146 (C/T) polymorphism is associated with risk to type 2 diabetes mellitus in Southern-Brazil. Arq. Bras. Endocrinol. E Metabol. 2014, 58, 918–925. [Google Scholar] [CrossRef]
- Lin, P.C.; Lin, W.T.; Yeh, Y.H.; Wung, S.F. Transcription Factor 7-Like 2 (TCF7L2) rs7903146 Polymorphism as a Risk Factor for Gestational Diabetes Mellitus: A Meta-Analysis. PLoS ONE 2016, 11, e0153044. [Google Scholar] [CrossRef]
- Ouhaibi-Djellouli, H.; Mediene-Benchekor, S.; Lardjam-Hetraf, S.A.; Hamani-Medjaoui, I.; Meroufel, D.N.; Boulenouar, H.; Hermant, X.; Saidi-Mehtar, N.; Amouyel, P.; Houti, L.; et al. The TCF7L2 rs7903146 polymorphism, dietary intakes and type 2 diabetes risk in an Algerian population. BMC Genet. 2014, 15, 134. [Google Scholar] [CrossRef]
- Phillips, C.M.; Goumidi, L.; Bertrais, S.; Field, M.R.; McManus, R.; Hercberg, S.; Lairon, D.; Planells, R.; Roche, H.M. Dietary saturated fat, gender and genetic variation at the TCF7L2 locus predict the development of metabolic syndrome. J. Nutr. Biochem. 2012, 23, 239–244. [Google Scholar] [CrossRef]
- Fisher, E.; Boeing, H.; Fritsche, A.; Doering, F.; Joost, H.G.; Schulze, M.B. Whole-grain consumption and transcription factor-7-like 2 (TCF7L2) rs7903146: Gene-diet interaction in modulating type 2 diabetes risk. Br. J. Nutr. 2009, 101, 478–481. [Google Scholar] [CrossRef]
- Corella, D.; Carrasco, P.; Sorli, J.V.; Estruch, R.; Rico-Sanz, J.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Covas, M.I.; Coltell, O.; Aros, F.; et al. Mediterranean diet reduces the adverse effect of the TCF7L2-rs7903146 polymorphism on cardiovascular risk factors and stroke incidence: A randomized controlled trial in a high-cardiovascular-risk population. Diabetes Care 2013, 36, 3803–3811. [Google Scholar] [CrossRef]
- Sotos-Prieto, M.; Smith, C.E.; Lai, C.Q.; Tucker, K.L.; Ordovas, J.M.; Mattei, J. Mediterranean Diet Adherence Modulates Anthropometric Measures by TCF7L2 Genotypes among Puerto Rican Adults. J. Nutr. 2020, 150, 167–175. [Google Scholar] [CrossRef]
- Barabash, A.; Valerio, J.D.; Garcia de la Torre, N.; Jimenez, I.; del Valle, L.; Melero, V.; Assaf-Balut, C.; Fuentes, M.; Bordiu, E.; Durán, A.; et al. TCF7L2 rs7903146 polymorphism modulates the association between adherence to a Mediterranean diet and the risk of gestational diabetes mellitus. Metab. Open 2020, 8, 100069. [Google Scholar] [CrossRef]
- Hosseinpour-Niazi, S.; Mirmiran, P.; Hadaegh, F.; Daneshpour, M.S.; Hedayati, M.; Azizi, F. The effect of TCF7L2 polymorphisms on inflammatory markers after 16 weeks of legume-based dietary approach to stop hypertension (DASH) diet versus a standard DASH diet: A randomised controlled trial. Nutr. Metab. 2022, 19, 35. [Google Scholar] [CrossRef]
- Vardatsikos, G.; Pandey, N.R.; Srivastava, A.K. Insulino-mimetic and anti-diabetic effects of zinc. J. Inorg. Biochem. 2013, 120, 8–17. [Google Scholar] [CrossRef]
- Cruz, K.J.C.; de Oliveira, A.R.S.; Morais, J.B.S.; Severo, J.S.; Mendes, P.M.V.; de Sousa Melo, S.R.; de Sousa, G.S.; Marreiro, D.D.N. Zinc and Insulin Resistance: Biochemical and Molecular Aspects. Biol. Trace Elem. Res. 2018, 186, 407–412. [Google Scholar] [CrossRef]
- Norouzi, S.; Adulcikas, J.; Sohal, S.S.; Myers, S. Zinc transporters and insulin resistance: Therapeutic implications for type 2 diabetes and metabolic disease. J. Biomed. Sci. 2017, 24, 87. [Google Scholar] [CrossRef]
- Huang, Q.; Du, J.; Merriman, C.; Gong, Z. Genetic, Functional, and Immunological Study of ZnT8 in Diabetes. Int. J. Endocrinol. 2019, 2019, 1524905. [Google Scholar] [CrossRef] [PubMed]
- Cauchi, S.; Del Guerra, S.; Choquet, H.; D’Aleo, V.; Groves, C.J.; Lupi, R.; McCarthy, M.I.; Froguel, P.; Marchetti, P. Meta-analysis and functional effects of the SLC30A8 rs13266634 polymorphism on isolated human pancreatic islets. Mol. Genet. Metab. 2010, 100, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Krentz, N.A.J.; Gloyn, A.L. Insights into pancreatic islet cell dysfunction from type 2 diabetes mellitus genetics. Nat. Rev. Endocrinol. 2020, 16, 202–212. [Google Scholar] [CrossRef]
- Drake, I.; Hindy, G.; Ericson, U.; Orho-Melander, M. A prospective study of dietary and supplemental zinc intake and risk of type 2 diabetes depending on genetic variation in SLC30A8. Genes Nutr. 2017, 12, 30. [Google Scholar] [CrossRef]
- Maruthur, N.M.; Clark, J.M.; Fu, M.; Linda Kao, W.; Shuldiner, A.R. Effect of zinc supplementation on insulin secretion: Interaction between zinc and SLC30A8 genotype in Old Order Amish. Diabetologia 2015, 58, 295–303. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Y.E.; Kirschke, C.P.; Stephensen, C.B.; Newman, J.W.; Keim, N.L.; Cai, Y.; Huang, L. Effects of a genetic variant rs13266634 in the zinc transporter 8 gene (SLC30A8) on insulin and lipid levels before and after a high-fat mixed macronutrient tolerance test in U.S. adults. J. Trace Elem. Med. Biol. 2023, 77, 127142. [Google Scholar] [CrossRef]
- Kanoni, S.; Nettleton, J.A.; Hivert, M.F.; Ye, Z.; van Rooij, F.J.; Shungin, D.; Sonestedt, E.; Ngwa, J.S.; Wojczynski, M.K.; Lemaitre, R.N.; et al. Total zinc intake may modify the glucose-raising effect of a zinc transporter (SLC30A8) variant: A 14-cohort meta-analysis. Diabetes 2011, 60, 2407–2416. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Pranto, S.; Ahammad, I.; Chowdhury, Z.M.; Juliana, F.M.; Das, K.C.; Keya, C.A.; Salimullah, M. High risk genetic variants of human insulin receptor substrate 1(IRS1) infer structural instability and functional interference. J. Biomol. Struct. Dyn. 2023, 41, 15150–15164. [Google Scholar] [CrossRef]
- Li, Q.; Qiao, Y.; Wang, C.; Zhang, G.; Zhang, X.; Xu, L. Associations between two single-nucleotide polymorphisms (rs1801278 and rs2943641) of insulin receptor substrate 1 gene and type 2 diabetes susceptibility: A meta-analysis. Endocrine 2016, 51, 52–62. [Google Scholar] [CrossRef]
- Ericson, U.; Rukh, G.; Stojkovic, I.; Sonestedt, E.; Gullberg, B.; Wirfalt, E.; Wallstrom, P.; Orho-Melander, M. Sex-specific interactions between the IRS1 polymorphism and intakes of carbohydrates and fat on incident type 2 diabetes. Am. J. Clin. Nutr. 2013, 97, 208–216. [Google Scholar] [CrossRef]
- Fajardo, C.M.; Cerda, A.; Bortolin, R.H.; de Oliveira, R.; Stefani, T.I.M.; Dos Santos, M.A.; Braga, A.A.; Dorea, E.L.; Bernik, M.M.S.; Bastos, G.M.; et al. Influence of polymorphisms in IRS1, IRS2, MC3R, and MC4R on metabolic and inflammatory status and food intake in Brazilian adults: An exploratory pilot study. Nutr. Res. 2023, 119, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Cormier, H.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.C. Expression and Sequence Variants of Inflammatory Genes; Effects on Plasma Inflammation Biomarkers Following a 6-Week Supplementation with Fish Oil. Int. J. Mol. Sci. 2016, 17, 375. [Google Scholar] [CrossRef]
- Oki, E.; Norde, M.N.; Carioca, A.A.F.; Souza, J.M.P.; Castro, I.A.; Marchioni, D.M.L.; Fisberg, R.M.; Rogero, M.M. Polymorphisms of the TNF-alpha gene interact with plasma fatty acids on inflammatory biomarker profile: A population-based, cross-sectional study in Sao Paulo, Brazil. Br. J. Nutr. 2017, 117, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Ghareeb, D.; Abdelazem, A.S.; Hussein, E.M.; Al-Karamany, A.S. Association of TNF-alpha-308 G>A (rs1800629) polymorphism with susceptibility of metabolic syndrome. J. Diabetes Metab. Disord. 2021, 20, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.X.; Zhang, L.; Zhang, D.L.; Zhou, J.P.; Jiang, X.J.; Jin, Y.L.; Chang, W.W. Association between TNF-alpha G-308A (rs1800629) polymorphism and susceptibility to chronic periodontitis and type 2 diabetes mellitus: A meta-analysis. J. Periodontal Res. 2021, 56, 226–235. [Google Scholar] [CrossRef]
- Gomez-Delgado, F.; Alcala-Diaz, J.F.; Garcia-Rios, A.; Delgado-Lista, J.; Ortiz-Morales, A.; Rangel-Zuñiga, O.; Tinahones, F.J.; Gonzalez-Guardia, L.; Malagon, M.M.; Bellido-Muñoz, E.; et al. Polymorphism at the TNF-alpha gene interacts with Mediterranean diet to influence triglyceride metabolism and inflammation status in metabolic syndrome patients: From the CORDIOPREV clinical trial. Mol. Nutr. Food Res. 2014, 58, 1519–1527. [Google Scholar] [CrossRef]
- Ter Horst, K.W.; Serlie, M.J. Fructose Consumption, Lipogenesis, and Non-Alcoholic Fatty Liver Disease. Nutrients 2017, 9, 981. [Google Scholar] [CrossRef]
- Geidl-Flueck, B.; Gerber, P.A. Fructose drives de novo lipogenesis affecting metabolic health. J. Endocrinol. 2023, 257, e220270. [Google Scholar] [CrossRef]
- Sharif, E.; Al-Wakeel, M.; Mohamed, A.; Kerkadi, A.; Rizk, N. TLR4 Receptor D299G/T399I Haplotype Polymorphism Is Associated with Insulin Resistance in Obese Female Subjects. Genes 2020, 11, 814. [Google Scholar] [CrossRef]
- Chait, A.; den Hartigh, L.J. Adipose Tissue Distribution, Inflammation and Its Metabolic Consequences, Including Diabetes and Cardiovascular Disease. Front. Cardiovasc. Med. 2020, 7, 22. [Google Scholar] [CrossRef]
- Renovato-Martins, M.; Moreira-Nunes, C.; Atella, G.C.; Barja-Fidalgo, C.; Moraes, J.A. Obese Adipose Tissue Secretion Induces Inflammation in Preadipocytes: Role of Toll-Like Receptor-4. Nutrients 2020, 12, 2828. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Garcia, R.; Vazquez-Villamar, M.; Aparicio-Ozores, G.; Parra-Rojas, I.; Radilla-Vazquez, R.B.; Castro-Alarcon, N. TLR4 polymorphism and haplotype are associated with obesity and lipid profile in young population: A pilot study. J. Endocrinol. Investig. 2023, 46, 903–913. [Google Scholar] [CrossRef]
- Abbas, S.A.; Raza, S.T.; Mir, S.S.; Siddiqi, Z.; Zaidi, A.; Zaidi, Z.H.; Mahdi, F. Role of variants rs5030717 and rs5030718 of TLR4 in the risk prediction of nephropathy, hypertension and dyslipidaemia in type 2 diabetes mellitus. Br. J. Biomed. Sci. 2018, 75, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Cuda, C.; Badawi, A.; Karmali, M.; El-Sohemy, A. Polymorphisms in Toll-like receptor 4 are associated with factors of the metabolic syndrome and modify the association between dietary saturated fat and fasting high-density lipoprotein cholesterol. Metab. Clin. Exp. 2011, 60, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Al-Mansoori, L.; Al-Jaber, H.; Prince, M.S.; Elrayess, M.A. Role of Inflammatory Cytokines, Growth Factors and Adipokines in Adipogenesis and Insulin Resistance. Inflammation 2022, 45, 31–44. [Google Scholar] [CrossRef]
- Cheng, H.; Zhu, W.; Zhu, M.; Sun, Y.; Sun, X.; Jia, D.; Yang, C.; Yu, H.; Zhang, C. Meta-analysis: Interleukin 6 gene -174G/C polymorphism associated with type 2 diabetes mellitus and interleukin 6 changes. J. Cell. Mol. Med. 2021, 25, 5628–5639. [Google Scholar] [CrossRef]
- Ali, Y.B.M.; El-Gahel, H.E.; Abdel-Hakem, N.E.; Gadalla, M.E.; El-Hefnawy, M.H.; El-Shahat, M. Association between IL-18 and IL-6 gene polymorphisms and the risk of T1D in Egyptian children. J. Diabetes Metab. Disord. 2021, 20, 439–446. [Google Scholar] [CrossRef]
- Obirikorang, C.; Lokpo, S.Y.; Owiredu, W.; Ahenkorah-Fondjo, L.; Osei-Yeboah, J.; Duedu, K.O.; Adejumo, E.N.; Ametepe, S.; Asamoah, E.A.; Coffie, S.A.; et al. Association between Interleukin-6 Gene Polymorphism (rs1800795 and rs1800796) and Type 2 Diabetes Mellitus in a Ghanaian Population: A Case-Control Study in the Ho Municipality. Biomed. Res. Int. 2024, 2024, 3610879. [Google Scholar] [CrossRef]
- Martinez-Ramirez, O.C.; Salazar-Pina, D.A.; de Lorena, R.M.; Castro-Hernandez, C.; Casas-Avila, L.; Portillo-Jacobo, J.A.; Rubio, J. Association of NFkappabeta, TNFalpha, IL-6, IL-1beta, and LPL Polymorphisms with Type 2 Diabetes Mellitus and Biochemical Parameters in a Mexican Population. Biochem. Genet. 2021, 59, 940–965. [Google Scholar] [CrossRef]
- Halvatsiotis, P.; Tsokaki, T.; Tsitsis, V.; Palaiodimou, L.; Tsivgoulis, G.; Tsangaris, I.; Panagiotou, M.O.; Houhoula, D. IL-6 Polymorphism as a Predisposing Genetic Factor for Gestational Diabetes or Preeclampsia Development in Pregnancy with Obesity in Relation to VEGF and VEGFF Receptor Gene Expression Modalities. Diagn 2024, 14, 1206. [Google Scholar] [CrossRef]
- Rana, B.K.; Flatt, S.W.; Health, D.D.; Pakiz, B.; Quintana, E.L.; Natarajan, L.; Rock, C.L. The IL-6 Gene Promoter SNP and Plasma IL-6 in Response to Diet Intervention. Nutrients 2017, 9, 552. [Google Scholar] [CrossRef]
- Janani, C.; Ranjitha Kumari, B.D. PPAR gamma gene--a review. Diabetes Metab. Syndr. 2015, 9, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Sarhangi, N.; Sharifi, F.; Hashemian, L.; Hassani Doabsari, M.; Heshmatzad, K.; Rahbaran, M.; Jamaldini, S.H.; Aghaei Meybodi, H.R.; Hasanzad, M. PPARG (Pro12Ala) genetic variant and risk of T2DM: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 12764. [Google Scholar] [CrossRef]
- Bhushan, R.; Haque, S.; Gupta, R.K.; Rani, A.; Diwakar, A.; Agarwal, S.; Tripathi, A.; Dubey, P.K. Genetic variants related to insulin metabolism are associated with gestational diabetes mellitus. Gene 2024, 927, 148704. [Google Scholar] [CrossRef]
- Chmurzynska, A.; Muzsik, A.; Krzyzanowska-Jankowska, P.; Madry, E.; Walkowiak, J.; Bajerska, J. PPARG and FTO polymorphism can modulate the outcomes of a central European diet and a Mediterranean diet in centrally obese postmenopausal women. Nutr. Res. 2019, 69, 94–100. [Google Scholar] [CrossRef]
- Garaulet, M.; Smith, C.E.; Hernández-González, T.; Lee, Y.C.; Ordovás, J.M. PPARγ Pro12Ala interacts with fat intake for obesity and weight loss in a behavioural treatment based on the Mediterranean diet. Mol. Nutr. Food Res. 2011, 55, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Koohdani, F.; Sotoudeh, G.; Kalantar, Z.; Mansoori, A. PPARγ Pro12Ala polymorphism influences the relationship between dietary fat intake, adiposity and lipid profile in patients with type 2 diabetes. Int. J. Vitam. Nutr. Res. 2018, 88, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Pihlajamäki, J.; Schwab, U.; Kaminska, D.; Ågren, J.; Kuusisto, J.; Kolehmainen, M.; Paananen, J.; Laakso, M.; Uusitupa, M. Dietary polyunsaturated fatty acids and the Pro12Ala polymorphisms of PPARG regulate serum lipids through divergent pathways: A randomized crossover clinical trial. Genes. Nutr. 2015, 10, 1–9. [Google Scholar]
- Rodrigues, A.P.S.; Rosa, L.P.S.; Silveira, E.A. PPARG2 Pro12Ala polymorphism influences body composition changes in severely obese patients consuming extra virgin olive oil: A randomized clinical trial. Nutr. Metab. 2018, 15, 52. [Google Scholar] [CrossRef]
- Mansoori, A.; Sotoudeh, G.; Djalali, M.; Eshraghian, M.R.; Keramatipour, M.; Nasli-Esfahani, E.; Shidfar, F.; Alvandi, E.; Toupchian, O.; Koohdani, F. Docosahexaenoic acid-rich fish oil supplementation improves body composition without influence of the PPARγ Pro12Ala polymorphism in patients with type 2 diabetes: A randomized, double-blind, placebo-controlled clinical trial. J. Nutr. Nutr. 2016, 8, 195–204. [Google Scholar] [CrossRef]
- Katsiki, N.; Mantzoros, C.; Mikhailidis, D.P. Adiponectin, lipids and atherosclerosis. Curr. Opin. Lipidol. 2017, 28, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Achari, A.E.; Jain, S.K. Adiponectin, a Therapeutic Target for Obesity, Diabetes, and Endothelial Dysfunction. Int. J. Mol. Sci. 2017, 18, 1321. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.S.; Han, J.W.; Lee, S.B.; You, J.H.; Kim, M.J.; Kang, S.; Park, J.S.; Ahn, C.W. Calpain-10 and Adiponectin Gene Polymorphisms in Korean Type 2 Diabetes Patients. Endocrinol. Metab. 2018, 33, 364–371. [Google Scholar] [CrossRef]
- Sun, P.; Liu, L.; Chen, J.; Chen, Y.; Shi, L.; Imam, M.U.; Chen, Y.; Pei, X.; Xu, Y.; Guo, Y.; et al. The polymorphism of rs266729 in adiponectin gene and type 2 diabetes mellitus: A Meta-Analysis. Medicine 2017, 96, e8745. [Google Scholar] [CrossRef]
- Alfaqih, M.A.; Al-Hawamdeh, A.; Amarin, Z.O.; Khader, Y.S.; Mhedat, K.; Allouh, M.Z. Single Nucleotide Polymorphism in the ADIPOQ Gene Modifies Adiponectin Levels and Glycemic Control in Type Two Diabetes Mellitus Patients. Biomed. Res. Int. 2022, 2022, 6632442. [Google Scholar] [CrossRef]
- Sanchez, M.P.; Prieto, C.; Mujica, E.; Vergara, K.; Valencia, E.; Villalobos, E.; Medina, M.; Parra, M.; D’Addosio, R.; Hoedebecke, K.; et al. Association between +45T>G adiponectin polymorphism gene and type 2 diabetes mellitus and metabolic syndrome in a Venezuelan population. F1000Research 2019, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Park, J.E.; Choi, Y.J.; Huh, K.B.; Chang, N.; Kim, W.Y. Carbohydrate intake interacts with SNP276G>T polymorphism in the adiponectin gene to affect fasting blood glucose, HbA1C, and HDL cholesterol in Korean patients with type 2 diabetes. J. Am. Coll. Nutr. 2013, 32, 143–150. [Google Scholar] [CrossRef]
- de Luis Roman, D.A.; Primo, D.; Izaola, O.; Gomez, E.; Lopez, J.J. Adiponectin Gene Variant rs3774261, Effects on Lipid Profile and Adiponectin Levels after a High Polyunsaturated Fat Hypocaloric Diet with Mediterranean Pattern. Nutrients 2021, 13, 1811. [Google Scholar] [CrossRef]
- AlSaleh, A.; D O’Dell, S.; Frost, G.S.; Griffin, B.A.; Lovegrove, J.A.; Jebb, S.A.; Sanders, T.A.; Group, R.S. Single nucleotide polymorphisms at the ADIPOQ gene locus interact with age and dietary intake of fat to determine serum adiponectin in subjects at risk of the metabolic syndrome. Am. J. Clin. Nutr. 2011, 94, 262–269. [Google Scholar] [CrossRef]
- Sugrue, L.P.; Desikan, R.S. What Are Polygenic Scores and Why Are They Important? JAMA 2019, 321, 1820–1821. [Google Scholar] [CrossRef]
- Collister, J.A.; Liu, X.; Clifton, L. Calculating Polygenic Risk Scores (PRS) in UK Biobank: A Practical Guide for Epidemiologists. Front. Genet. 2022, 13, 818574. [Google Scholar] [CrossRef]
- Alsulami, S.; Cruvinel, N.T.; da Silva, N.R.; Antoneli, A.C.; Lovegrove, J.A.; Horst, M.A.; Vimaleswaran, K.S. Effect of dietary fat intake and genetic risk on glucose and insulin-related traits in Brazilian young adults. J. Diabetes Metab. Disord. 2021, 20, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Alsulami, S.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Tasrif, N.; Yani, F.F.; Lovegrove, J.A.; Sudji, I.R.; Lipoeto, N.I.; Vimaleswaran, K.S. Interaction between the genetic risk score and dietary protein intake on cardiometabolic traits in Southeast Asian. Genes. Nutr. 2020, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Sekar, P.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Tasrif, N.; Yani, F.F.; Lovegrove, J.A.; Sudji, I.R.; Lipoeto, N.I.; Vimaleswaran, K.S. A Novel Interaction between a 23-SNP Genetic Risk Score and Monounsaturated Fatty Acid Intake on HbA1c Levels in Southeast Asian Women. Nutrients 2024, 16, 3022. [Google Scholar] [CrossRef] [PubMed]
- Franck, M.; de Toro-Martin, J.; Guenard, F.; Rudkowska, I.; Lemieux, S.; Lamarche, B.; Couture, P.; Vohl, M.C. Prevention of Potential Adverse Metabolic Effects of a Supplementation with Omega-3 Fatty Acids Using a Genetic Score Approach. Lifestyle Genom. 2020, 13, 32–42. [Google Scholar] [CrossRef]
- Seral-Cortes, M.; Sabroso-Lasa, S.; De Miguel-Etayo, P.; Gonzalez-Gross, M.; Gesteiro, E.; Molina-Hidalgo, C.; De Henauw, S.; Erhardt, E.; Censi, L.; Manios, Y.; et al. Interaction Effect of the Mediterranean Diet and an Obesity Genetic Risk Score on Adiposity and Metabolic Syndrome in Adolescents: The HELENA Study. Nutrients 2020, 12, 3841. [Google Scholar] [CrossRef]
- Watanabe, D.; Kuranuki, S.; Sunto, A.; Matsumoto, N.; Nakamura, T. Daily yogurt consumption improves glucose metabolism and insulin sensitivity in young nondiabetic Japanese subjects with type-2 diabetes risk alleles. Nutrients 2018, 10, 1834. [Google Scholar] [CrossRef]
- Lopez-Portillo, M.L.; Huidobro, A.; Tobar-Calfucoy, E.; Yanez, C.; Retamales-Ortega, R.; Garrido-Tapia, M.; Acevedo, J.; Paredes, F.; Cid-Ossandon, V.; Ferreccio, C.; et al. The Association between Fasting Glucose and Sugar Sweetened Beverages Intake Is Greater in Latin Americans with a High Polygenic Risk Score for Type 2 Diabetes Mellitus. Nutrients 2021, 14, 69. [Google Scholar] [CrossRef]

| Gene | SNPs | Nutrients and/or Nutritional Intervention | Main Findings |
|---|---|---|---|
| TCF7L2 | rs7903146 (C > T) and rs12255372 (G > T) | Mediterranean Diet | (1) T allele for both SNPs: individuals with high adherence to the Mediterranean diet presented ↓ weight, BMI, WC [56], and lower risk for GDM [57]; (2) individuals with low adherence had higher fasting glucose [55]. |
| rs7903146 (C > T) | Quantitative evaluation of dairy products (>200 mL/day), processed sweets (>30 g/day), and refined grains (≥5 portions/week) | T allele: individuals with higher consumption of desserts and milk had ↑ risk for T2D [52]. | |
| Evaluation of habitual intake of SAFA (low intake ≤ 12% TEV and high intake ≥ 15%TEV) | T allele: ↑ intake of SAFA associated with impaired insulin sensitivity [53]. | ||
| Evaluation of intake of whole grains (>50 g/day vs. <30 g/day) | GG genotype: 14% less risk of developing T2D [54]. | ||
| DASH diet or legume-based DASH diet | No effects of the SNP on inflammatory biomarkers or oxidative stress indicator [58]. | ||
| SLC30A8 rs13266634 (C > T) | Zinc intake | TT genotype: zinc supplement users had a more pronounced reduction in T2D risk [65]. | |
| Supplementation of oral zinc acetate | (1) CT/TT genotypes: ↑ fasting insulin. (2) No differences between genotypes for serum zinc concentration [66]. | ||
| MMTT test with high lipids content (60% lipids; 25% carbohydrates; 15% proteins) | (1) CC genotype: ↓ NEFA in men. (2) TT genotype carriers: ↑ insulin basal levels in women and attenuated lipid response with no gender-specific effect [67]. | ||
| IRS1 | rs2943641 (T > A) | Macronutrients intake | T allele: ↓ T2D risk in lower tertiles of carbohydrate intake for women and in the lower tertiles of fat intake for men [71]. |
| rs2943634 (A > C) | Counseling program: reduction in refined carbohydrates (<45% TEV), emphasis on dietary fiber (>25 g/day) | A allele: associated with ↓ carbohydrate intake [72]. | |
| rs2943641 (T > A) and rs7578326 (A > G) | Dietary analysis of SAFA: carbohydrates (Group 1: Ratio ≤0.24 and Group 2: Ratio >0.24) and MUFA intake | (1) haplotype G-T: ↓ risk of IR and MetS with low dietary SAFA-to-carbohydrate ratio. (2) G allele: ↓ MetS risk when dietary MUFA was lower than median intake [27]. | |
| TNF | rs7903146 (C > T) | Mediterranean Diet | GG genotype: improvement of TG and plasma hsCRP after 1 year of intervention [77]. |
| Fatty acids intake evaluation | A allele: ↑ inflammation— ↑ plasma stearic acid and SAFA [74]. | ||
| TLR4 | rs4986790 (Asp299Gly) and rs5030718 (G > A) | Dietary fat intake evaluation | (1) Gly allele: ↑ insulin, HOMA-IR, and HOMA-β. (2) G allele: SFA intake -> inversely associated with HDL-c among GG individuals and a positive relationship for GA [85] |
| IL6 | rs1800795 (G > C) | 3 intervention groups: (a) lower fat and higher carbohydrate; (b) lower carbohydrate and higher fat; or (c) a walnut-rich diet, with higher fat and lower carbohydrate. | CC genotype: ↓ IL-6 levels before adjustment for BMI [92]. |
| PPARG | rs1801282 (C > G | Mediterranean Diet or Centro-European diet | (1) Central European diet—G allele: ↓ weight, lean mass, and HDL-c. (2) Mediterranean Diet—G allele: ↓ abdominal fat [96]. |
| 2-year behavioral weight-loss program based on Mediterranean Diet | (1) CG/GG genotype: baseline—↓ insulin and HOMA-IR; (2) G allele: after intervention—↑ % of weight loss and body fat, especially in those with high MUFA intake [97]. | ||
| High SAFA content or high PUFA intake | (1) CC genotype: a positive association between total fat intake and WC [98]; (2) PUFA diet—↑ HDL-c and APOA [99]. | ||
| 3 interventional groups: extra virgin olive oil supplementation (50 mL/day), a traditional Brazilian diet (TBD), or a TBD with extra virgin olive oil supplementation. | TBD with extra virgin olive oil supplementation: weight loss and improvement body composition independently of genotype [100]. | ||
| ↓ body weight, fasting glucose, and HOMA-IR independent of genotype [101]. | |||
| Daily DHA-rich fish oil supplementation | |||
| ADIPOQ | rs1501299 (G > T) | Carbohydrate intake evaluation | T allele carriers influence the degree to which fasting glucose, HbA1C e HDL-c are affected by the amount of carbohydrate intake [108]. |
| rs2241766 (G > A) | High-monounsaturated diet | A allele: ↓ serum adiponectin [110]. | |
| PRS/GRS Composition | Nutrients and/or Nutritional Intervention | Nutrients and/or Nutritional Intervention |
|---|---|---|
| 10 SNPs associated with glucose and insulin metabolism, and obesity (rs12255372, rs7903146, rs17782313, rs8050136, rs10163409, rs2237892, rs2237895, rs10811661, rs5030952, rs1801282) | Macronutrients intake | Individuals within the high-fat intake category and with ≥5 risk alleles: ↑ fasting insulin, insulin-glucose ratio, HOMA-β, and HOMA-IR than those with < 5 risk allele [113]. |
| 16 SNPs associated with metabolic traits (rs3792267, rs5030952, rs9939609, rs10163409, rs8050136, rs17782313, rs2229616, rs12255372, rs7903146, rs2237895, rs2237892, rs10811661, rs1801282, s266729, rs17846866) | Protein intake evaluation | No influence of PRS on dietary protein intake and glucose concentration, insulin, and HbA1C [114]. |
| 23 SNPs associated with metabolic traits (rs1801133, rs7903146, rs12255372, rs8050136, rs9939609, rs17782313, rs1801282, rs5219, rs2237895, rs2237892, rs10741567, rs12794714, rs12785878, rs6013897, rs2282679, rs6680429, rs266729, rs10811661, rs1801725, rs5030952, rs3742801, rs2270655, rs778805) | MUFA intake evaluation | Women with ↓ MUFA intake (<7.0 g/day) and a higher PRS (>13 risk allele) had elevated HbA1C [115] |
| 8 SNPs associated with changes in the HOMA-IR after n-3 PUFA supplementation (rs72723587, rs77850702, rs72703546, rs17174795, rs12437986, rs35621498, rs55842940, rs6001872) | Omega-3 PUFA supplementation (5 g/day of fish oil) | ↑ PRS increased HOMA-IR after dietary intervention [116] |
| 21 SNPS associated with obesity (rs2010899, rs4135275, rs4912905, rs9355296, rs1524107, rs2183013, rs1019731, rs1568400, rs4246444, rs7701443, rs13182800, rs3211867, rs2515362, rs1800497, rs9939609, rs4783961, rs8068149, rs7502966, rs1044250, rs17373080, rs2143511). | Mediterranean Diet | Women with adherence to the intervention diet and ↑ PRS: improvement of IR [117] |
| 5 SNPs associated with T2D risk (rs2237892, rs2206734, rs2383208, rs6780569, rs1470579) | Intake of 150 g of yogurt | ↑ PRS group: improvement of postprandial plasma glucose, insulin, and HOMA-IR after the intervention [118] |
| 16 SNPs associated with T2D (rs3923113, rs1801282, rs11717195, rs4402960, rs6878122, rs7756992, rs849135, rs516946, rs17791513, rs2796441, rs11257655, rs7903146, rs10830963, rs7403531, rs7202877, rs5945326, rs188827514) | Sugar-sweetened beverages (SSB) intake evaluation | SSB intake: increased glucose levels in individuals with ↑ PRS [119] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, G.B.S.; Pascoal, G.d.F.L.; Rogero, M.M. Polymorphisms Involved in Insulin Resistance and Metabolic Inflammation: Influence of Nutrients and Dietary Interventions. Metabolites 2025, 15, 245. https://doi.org/10.3390/metabo15040245
Duarte GBS, Pascoal GdFL, Rogero MM. Polymorphisms Involved in Insulin Resistance and Metabolic Inflammation: Influence of Nutrients and Dietary Interventions. Metabolites. 2025; 15(4):245. https://doi.org/10.3390/metabo15040245
Chicago/Turabian StyleDuarte, Graziela Biude Silva, Gabriela de Freitas Laiber Pascoal, and Marcelo Macedo Rogero. 2025. "Polymorphisms Involved in Insulin Resistance and Metabolic Inflammation: Influence of Nutrients and Dietary Interventions" Metabolites 15, no. 4: 245. https://doi.org/10.3390/metabo15040245
APA StyleDuarte, G. B. S., Pascoal, G. d. F. L., & Rogero, M. M. (2025). Polymorphisms Involved in Insulin Resistance and Metabolic Inflammation: Influence of Nutrients and Dietary Interventions. Metabolites, 15(4), 245. https://doi.org/10.3390/metabo15040245







