Analyses of Saliva Metabolome Reveal Patterns of Metabolites That Differentiate SARS-CoV-2 Infection and COVID-19 Disease Severity
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Collection and Processing of Saliva Samples for Untargeted Metabolomics
2.3. Data Preprocessing and Quality Control
2.4. Statistical Analysis, Variable Selection, and Performance Assessment
2.5. Data Availability
3. Results
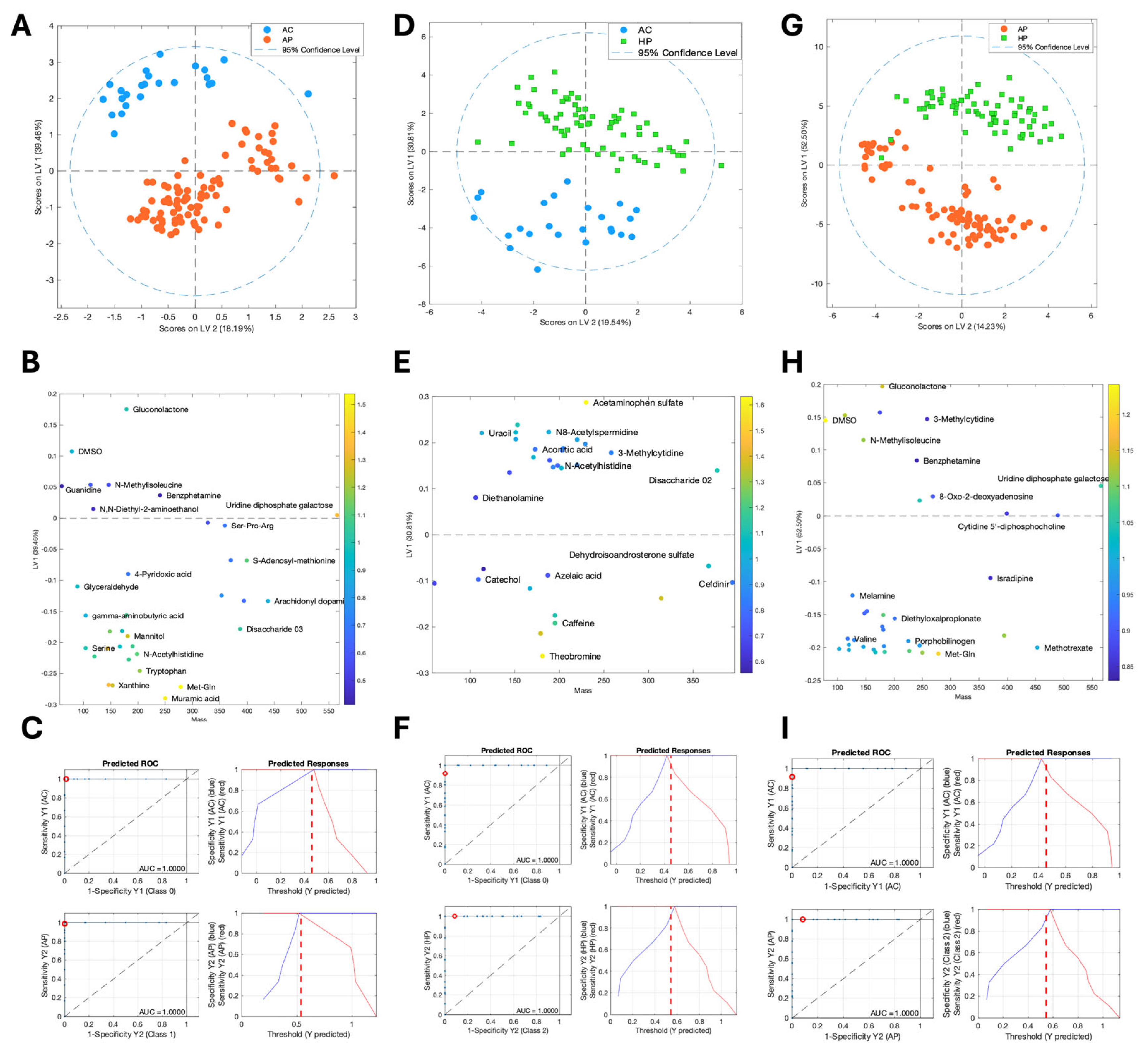
3.1. The Composition of Saliva Metabolites Clearly Distinguished the Clinical Groups
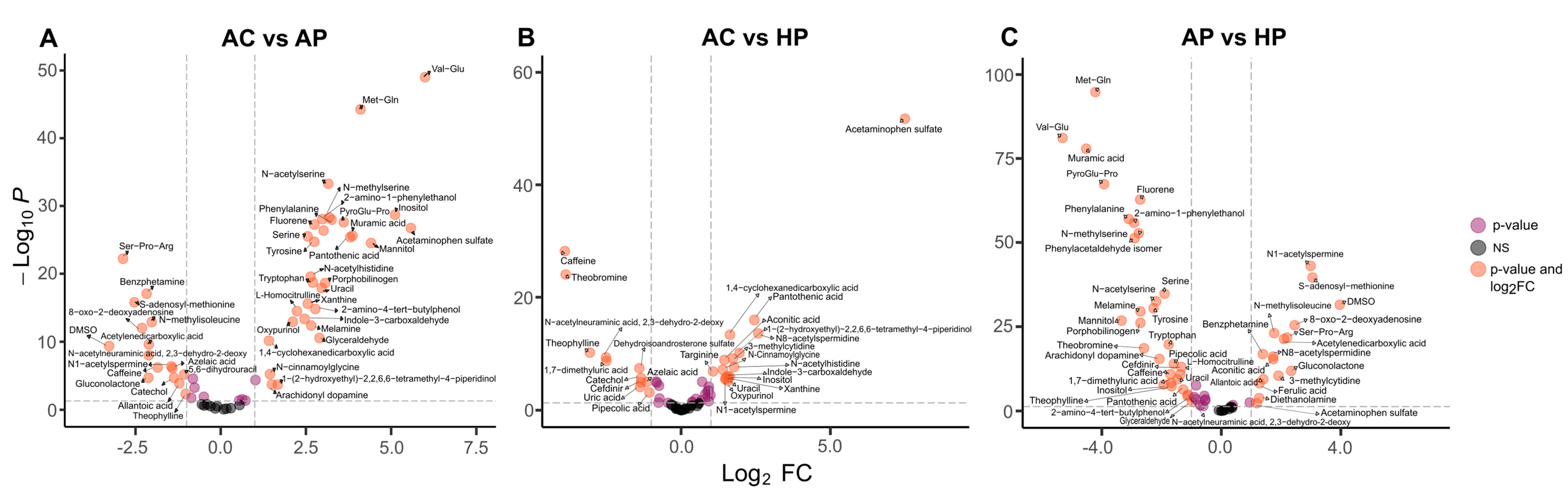
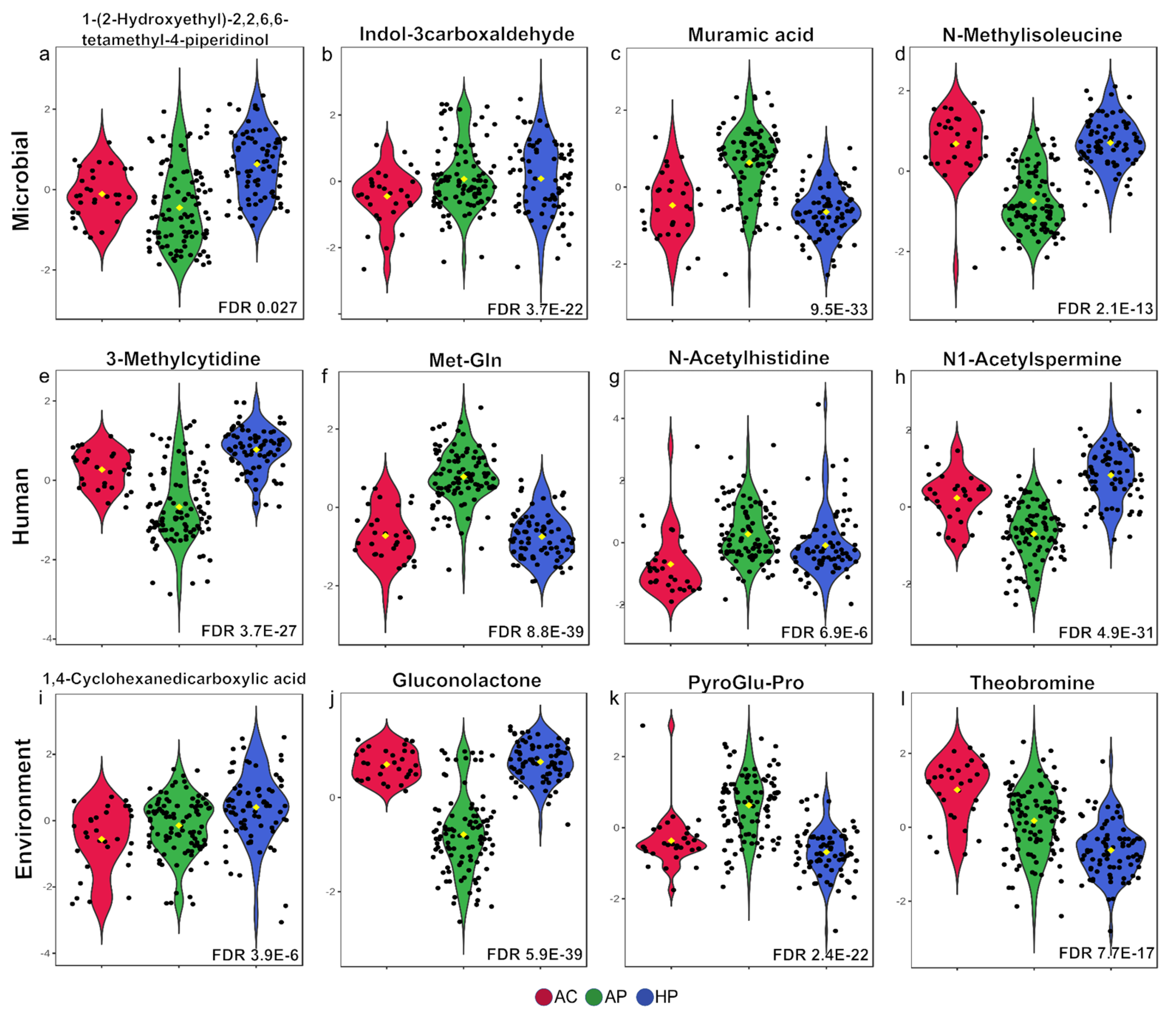
3.2. Amino Acid Metabolites Were Strongly Associated with Moderate COVID Cases and Bacterial Metabolites Were Srongly Associated with Severe Cases
3.3. Some Metabolites Distinguish Deceased from Severe-Hospitalized Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- COVID-19 Epidemiological Update—13 February 2025. Available online: https://www.who.int/publications/m/item/covid-19-epidemiological-update-edition-176 (accessed on 20 February 2025).
- Sievers, B.L.; Cheng, M.T.K.; Csiba, K.; Meng, B.; Gupta, R.K. SARS-CoV-2 and Innate Immunity: The Good, the Bad, and the “Goldilocks”. Cell. Mol. Immunol. 2024, 21, 171–183. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef] [PubMed]
- Mollentze, N.; Streicker, D.G. Viral Zoonotic Risk Is Homogenous among Taxonomic Orders of Mammalian and Avian Reservoir Hosts. Proc. Natl. Acad. Sci. USA 2020, 117, 9423–9430. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Tsang, T.K.; Lee, K.H.; Foxman, B.; Balmaseda, A.; Gresh, L.; Sanchez, N.; Ojeda, S.; Lopez, R.; Yang, Y.; Kuan, G.; et al. Association Between the Respiratory Microbiome and Susceptibility to Influenza Virus Infection. Clin. Infect. Dis. 2020, 71, 1195–1203. [Google Scholar] [CrossRef]
- Larios Serrato, V.; Meza, B.; Gonzalez-Torres, C.; Gaytan-Cervantes, J.; González Ibarra, J.; Santacruz Tinoco, C.E.; Anguiano Hernández, Y.-M.; Martínez Miguel, B.; Cázarez Cortazar, A.; Sarquiz Martínez, B.; et al. Diversity, Composition, and Networking of Saliva Microbiota Distinguish the Severity of COVID-19 Episodes as Revealed by an Analysis of 16S rRNA Variable V1-V3 Region Sequences. mSystems 2023, 8, e0106222. [Google Scholar] [CrossRef]
- Moreno, E.; Ciordia, S.; Fátima, S.M.; Jiménez, D.; Martínez-Sanz, J.; Vizcarra, P.; Ron, R.; Sánchez-Conde, M.; Bargiela, R.; Sanchez-Carrillo, S.; et al. Proteomic Snapshot of Saliva Samples Predicts New Pathways Implicated in SARS-CoV-2 Pathogenesis. Clin. Proteom. 2024, 21, 37. [Google Scholar] [CrossRef]
- Zhang, C.-Z.; Cheng, X.-Q.; Li, J.-Y.; Zhang, P.; Yi, P.; Xu, X.; Zhou, X.-D. Saliva in the Diagnosis of Diseases. Int. J. Oral. Sci. 2016, 8, 133–137. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Sa, M.R.; Ji, J.; Vaniya, A.; Wancewicz, B.; Roberts, B.S.; Torbašinović, H.; Lee, T.; Mehta, S.S.; et al. Structure Annotation of All Mass Spectra in Untargeted Metabolomics. Anal. Chem. 2019, 91, 2155–2162. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Fan, S.; Kind, T.; Cajka, T.; Hazen, S.L.; Tang, W.H.W.; Kaddurah-Daouk, R.; Irvin, M.R.; Arnett, D.K.; Barupal, D.K.; Fiehn, O. Systematic Error Removal Using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 2019, 91, 3590–3596. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 9783319242750. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ansone, L.; Briviba, M.; Silamikelis, I.; Terentjeva, A.; Perkons, I.; Birzniece, L.; Rovite, V.; Rozentale, B.; Viksna, L.; Kolesova, O.; et al. Amino Acid Metabolism Is Significantly Altered at the Time of Admission in Hospital for Severe COVID-19 Patients: Findings from Longitudinal Targeted Metabolomics Analysis. Microbiol. Spectr. 2021, 9, e0033821. [Google Scholar] [CrossRef] [PubMed]
- Bourgin, M.; Derosa, L.; Silva, C.A.C.; Goubet, A.-G.; Dubuisson, A.; Danlos, F.-X.; Grajeda-Iglesias, C.; Cerbone, L.; Geraud, A.; Laparra, A.; et al. Circulating Acetylated Polyamines Correlate with Covid-19 Severity in Cancer Patients. Aging 2021, 13, 20860–20885. [Google Scholar] [CrossRef]
- Frampas, C.F.; Longman, K.; Spick, M.P.; Lewis, H.M.; Costa, C.D.S.; Stewart, A.; Dunn-Walters, D.; Greener, D.; Evetts, G.E.; Skene, D.; et al. Untargeted Saliva Metabolomics Reveals COVID-19 Severity: Saliva Metabolomics for SARS-CoV-2 Prognosis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Pozzi, C.; Levi, R.; Braga, D.; Carli, F.; Darwich, A.; Spadoni, I.; Oresta, B.; Dioguardi, C.C.; Peano, C.; Ubaldi, L.; et al. A “Multiomic” Approach of Saliva Metabolomics, Microbiota, and Serum Biomarkers to Assess the Need of Hospitalization in Coronavirus Disease 2019. Gastro Hep Adv. 2022, 1, 194–209. [Google Scholar] [CrossRef]
- Costa Dos Santos Junior, G.; Pereira, C.M.; Kelly da Silva Fidalgo, T.; Valente, A.P. Saliva NMR-Based Metabolomics in the War Against COVID-19. Anal. Chem. 2020, 92, 15688–15692. [Google Scholar] [CrossRef]
- Thomadaki, K.; Helmerhorst, E.J.; Tian, N.; Sun, X.; Siqueira, W.L.; Walt, D.R.; Oppenheim, F.G. Whole-Saliva Proteolysis and Its Impact on Salivary Diagnostics. J. Dent. Res. 2011, 90, 1325–1330. [Google Scholar] [CrossRef]
- Garreto, L.; Charneau, S.; Mandacaru, S.C.; Nóbrega, O.T.; Motta, F.N.; de Araújo, C.N.; Tonet, A.C.; Modesto, F.M.B.; Paula, L.M.; de Sousa, M.V.; et al. Mapping Salivary Proteases in Sjögren’s Syndrome Patients Reveals Overexpression of Dipeptidyl Peptidase-4/CD26. Front. Immunol. 2021, 12, 686480. [Google Scholar] [CrossRef]
- Turk, B. Targeting Proteases: Successes, Failures and Future Prospects. Nat. Rev. Drug Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef]
- Human Metabolome Database: Showing Metabocard for Valylglutamine (HMDB0029125). Available online: https://hmdb.ca/metabolites/HMDB0029125 (accessed on 25 February 2025).
- Minen, R.I.; Thirumalaikumar, V.P.; Skirycz, A. Proteinogenic Dipeptides, an Emerging Class of Small-Molecule Regulators. Curr. Opin. Plant Biol. 2023, 75, 102395. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R. Microbial Tryptophan Catabolites in Health and Disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed]
- Frampas, C.F.; Longman, K.; Spick, M.; Lewis, H.-M.; Costa, C.D.S.; Stewart, A.; Dunn-Walters, D.; Greener, D.; Evetts, G.; Skene, D.J.; et al. Untargeted Saliva Metabolomics by Liquid Chromatography-Mass Spectrometry Reveals Markers of COVID-19 Severity. PLoS ONE 2022, 17, e0274967. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Zheng, Z.; Surapaneni, A.; Yu, B.; Zhou, L.; Zhou, W.; Xie, D.; Shou, H.; Avila-Pacheco, J.; Kalim, S.; et al. Metabolite Profiling of CKD Progression in the Chronic Renal Insufficiency Cohort Study. JCI Insight 2022, 7, e161696. [Google Scholar] [CrossRef]
- Luo, S.; Surapaneni, A.; Zheng, Z.; Rhee, E.P.; Coresh, J.; Hung, A.M.; Nadkarni, G.N.; Yu, B.; Boerwinkle, E.; Tin, A.; et al. Variants, N-Acetylated Amino Acids, and Progression of CKD. Clin. J. Am. Soc. Nephrol. 2020, 16, 37–47. [Google Scholar] [CrossRef]
- Goetzman, E.; Gong, Z.; Rajasundaram, D.; Muzumdar, I.; Goodchild, T.; Lefer, D.; Muzumdar, R. Serum Metabolomics Reveals Distinct Profiles during Ischemia and Reperfusion in a Porcine Model of Myocardial Ischemia-Reperfusion. Int. J. Mol. Sci. 2022, 23, 6711. [Google Scholar] [CrossRef]
- Hammond, T.C.; Xing, X.; Yanckello, L.M.; Stromberg, A.; Chang, Y.-H.; Nelson, P.T.; Lin, A.-L. Human Gray and White Matter Metabolomics to Differentiate APOE and Stage Dependent Changes in Alzheimer’s Disease. J. Cell. Immunol. 2021, 3, 397–412. [Google Scholar]
- Ree, R.; Varland, S.; Arnesen, T. Spotlight on Protein N-Terminal Acetylation. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Strickland, W.N.; Strickland, M.; de Groot, P.C.; Von Holt, C.; Wittmann-Liebold, B. The Primary Structure of Histone H1 from Sperm of the Sea Urchin Parechinus Angulosus. 1. Chemical and Enzymatic Fragmentation of the Protein and the Sequence of Amino Acids in the Four N-Terminal Cyanogen Bromide Peptides. Eur. J. Biochem. 1980, 104, 559–566. [Google Scholar] [CrossRef]
- Pascale, R.M.; Simile, M.M.; Calvisi, D.F.; Feo, C.F.; Feo, F. S-Adenosylmethionine: From the Discovery of Its Inhibition of Tumorigenesis to Its Use as a Therapeutic Agent. Cells 2022, 11, 409. [Google Scholar] [CrossRef] [PubMed]
- Aboud, O.; Liu, Y.; Dahabiyeh, L.; Abuaisheh, A.; Li, F.; Aboubechara, J.P.; Riess, J.; Bloch, O.; Hodeify, R.; Tagkopoulos, I.; et al. Profile Characterization of Biogenic Amines in Glioblastoma Patients Undergoing Standard-of-Care Treatment. Biomedicines 2023, 11, 2261. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, S.; Ravcheev, D.; de Crécy-Lagard, V.; Thiele, I. Systematic Genome Assessment of B-Vitamin Biosynthesis Suggests Co-Operation among Gut Microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, J. Indole as an Intercellular Signal in Microbial Communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Wallace, H.M.; Fraser, A.V.; Hughes, A. A Perspective of Polyamine Metabolism. Biochem. J. 2003, 376, 1–14. [Google Scholar] [CrossRef]
- Nayak, A.; Liu, C.; Mehta, A.; Ko, Y.-A.; Tahhan, A.S.; Dhindsa, D.S.; Uppal, K.; Jones, D.P.; Butler, J.; Morris, A.A.; et al. N8-Acetylspermidine: A Polyamine Biomarker in Ischemic Cardiomyopathy With Reduced Ejection Fraction. J. Am. Heart Assoc. 2020, 9, e016055. [Google Scholar] [CrossRef]
- Qu, Y.; Eguchi, A.; Ma, L.; Wan, X.; Mori, C.; Hashimoto, K. Role of the Gut-Brain Axis via the Subdiaphragmatic Vagus Nerve in Stress Resilience of 3,4-Methylenedioxymethamphetamine in Mice Exposed to Chronic Restrain Stress. Neurobiol. Dis. 2023, 189, 106348. [Google Scholar] [CrossRef]
- Wenzel, A.G.; Reiner, J.L.; Kohno, S.; Wolf, B.J.; Brock, J.W.; Cruze, L.; Newman, R.B.; Kucklick, J.R. Biomonitoring of Emerging DINCH Metabolites in Pregnant Women in Charleston, SC: 2011–2014. Chemosphere 2021, 262, 128369. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Kleiber, N.; Lemus-Diaz, N.; Bohnsack, M.T. Roles and Dynamics of 3-Methylcytidine in Cellular RNAs. Trends Biochem. Sci. 2022, 47, 596–608. [Google Scholar] [CrossRef]
- Aarsand, A.K.; Petersen, P.H.; Sandberg, S. Estimation and Application of Biological Variation of Urinary Delta-Aminolevulinic Acid and Porphobilinogen in Healthy Individuals and in Patients with Acute Intermittent Porphyria. Clin. Chem. 2006, 52, 650–656. [Google Scholar] [CrossRef]
| Group | Number of Cases | Description |
|---|---|---|
| Asymptomatic cases (ACs) | 30 | Asymptomatic cases, negative for SARS-CoV-2 infection |
| Ambulatory patients (APs) | 102 | Ambulatory patients, PCR+ for SARS-CoV-2 |
| Hospitalized patients (HPs) | 61 | Patients that required hospitalization because of severe symptoms, PCR+ for SARS-CoV-2 |
| Total studied | 210 |
| Microbial | Human | Environment | Drugs | |
|---|---|---|---|---|
| AC vs. AP | N-Methylisoleucine | N-Acetylserine | Val-Glu | S-Adenosyl-methionine |
| Indole-3-carboxaldehyde | N-Methylserine | Ser-Pro-Arg | N,N Diethyl-2-aminoethanol | |
| Muramic acid | gamma-aminobutyric acid | Serine | DMSO | |
| Guanidine | Phenylalanine | Isradipine | ||
| Inositol | Tyrosine | |||
| L-Homocitrulline | Gluconolactone | Xanthine | ||
| N-Acetylhistidine | 1,4-Cyclohexanedicarboxylic acid | Mannitol | ||
| Met-Gln | Fluorene | Cefdinir | ||
| Arachidonyl dopamine | Tryptophan | Benzphetamine | ||
| Acetylenedicarboxylic acid | 2-Amino-4-tert-butylphenol | |||
| Uridine diphosphate galactose | ||||
| Glyceraldehyde | ||||
| 4-Pyridoxic acid | ||||
| AC vs. HP | Pantothenic acid | N8-Acetylspermidine | Theobromine | Acetaminophen sulfate |
| 1-(2-Hydroxyethyl)-2,2,6,6-tetramethyl-4-piperidinol | 3-Methylcytidine | Theophylline | Ornidazole | |
| Indole-3-carboxaldehyde | 1,7 Dimethyluric acid | Caffeine | Xanthine | |
| N-Acetylneuraminic acid | Aconitic acid | Cefdinir | ||
| N-Cinnamoylglycine | Dehydroisoandrosterone sulfate | Diethanolamine | ||
| Oxypurinol | 1,4-Cyclohexanedicarboxylic acid | |||
| N-Acetylhistidine | Ferulic acid | |||
| Targinine | Catechol | |||
| Uracil | ||||
| 5,6-Dihydrouracil | ||||
| Uric acid | ||||
| AP vs. HP | Muramic acid | N1-Acetylspermine | Ser-Pro-Arg | S-Adenosyl-methionine |
| 2-Amino-1-phenylethanol | 8-Oxo-2-deoxyadenosine | Met-Gln | DMSO | |
| Allantoic acid | N-Methylisoleucine | Val-Glu | Cefdinir | |
| 3-Hydroxyanthranilic acid | Acetylenedicarboxylic acid | Phenylalanine | Benzphetamine | |
| N8-Acetylspermidine | PyroGlu-Pro | Mannitol | ||
| N-Methylserine | Tyrosine | Methotrexate | ||
| Serine | Phenylacetaldehyde isomer | Isradipine | ||
| Porphobilinogen | Gluconolactone | |||
| 3-Methylcytidine | Theobromine | |||
| Diethyloxalpropionate | Theophylline | |||
| N-Acetylserine | ||||
| Uridine diphosphate galactose | Fluorene | |||
| Cytidine 5′-diphosphocholine | 2-Amino-4-tert-butylphenol | |||
| Pipecolic acid | ||||
| Melamine | ||||
| Valine |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larios-Serrato, V.; Vázquez-Manjarrez, N.; Resendis-Antonio, O.; Rios-Sarabia, N.; Meza, B.; Fiehn, O.; Torres, J. Analyses of Saliva Metabolome Reveal Patterns of Metabolites That Differentiate SARS-CoV-2 Infection and COVID-19 Disease Severity. Metabolites 2025, 15, 192. https://doi.org/10.3390/metabo15030192
Larios-Serrato V, Vázquez-Manjarrez N, Resendis-Antonio O, Rios-Sarabia N, Meza B, Fiehn O, Torres J. Analyses of Saliva Metabolome Reveal Patterns of Metabolites That Differentiate SARS-CoV-2 Infection and COVID-19 Disease Severity. Metabolites. 2025; 15(3):192. https://doi.org/10.3390/metabo15030192
Chicago/Turabian StyleLarios-Serrato, Violeta, Natalia Vázquez-Manjarrez, Osbaldo Resendis-Antonio, Nora Rios-Sarabia, Beatriz Meza, Oliver Fiehn, and Javier Torres. 2025. "Analyses of Saliva Metabolome Reveal Patterns of Metabolites That Differentiate SARS-CoV-2 Infection and COVID-19 Disease Severity" Metabolites 15, no. 3: 192. https://doi.org/10.3390/metabo15030192
APA StyleLarios-Serrato, V., Vázquez-Manjarrez, N., Resendis-Antonio, O., Rios-Sarabia, N., Meza, B., Fiehn, O., & Torres, J. (2025). Analyses of Saliva Metabolome Reveal Patterns of Metabolites That Differentiate SARS-CoV-2 Infection and COVID-19 Disease Severity. Metabolites, 15(3), 192. https://doi.org/10.3390/metabo15030192






