Gene Expression-Based Inference of Metabolic Signatures Reveals Distinct Molecular Profiles in Right- and Left-Sided Colon Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Blood Samples
2.2. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) and Gene Enrichment Analysis
2.3. Gene-Metabolite Interaction
2.4. Statistics
3. Results
3.1. Analysis of Sociodemographic and Clinical Parameters
3.2. Gene Expression Analysis in Right and Left Colon Cancer
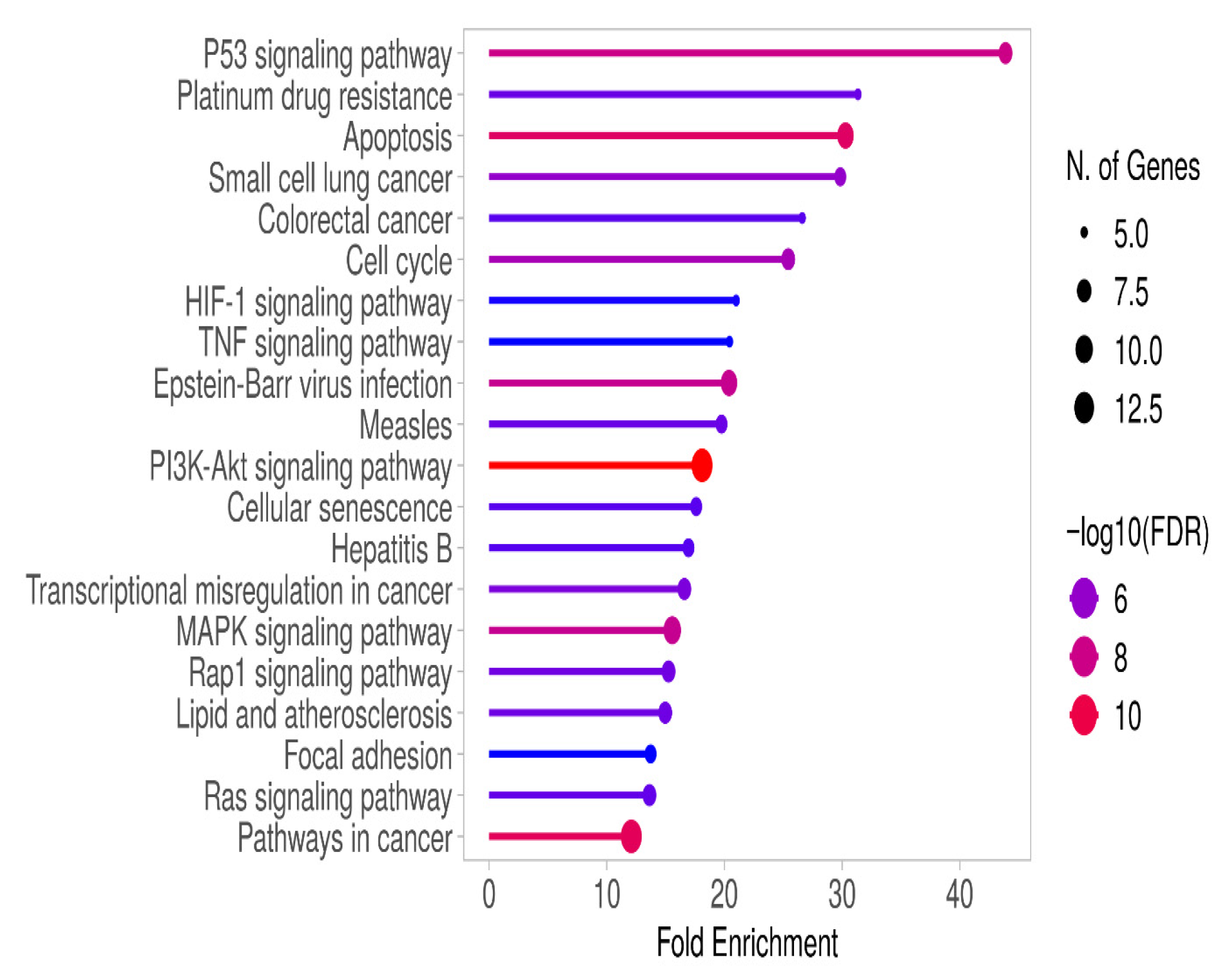
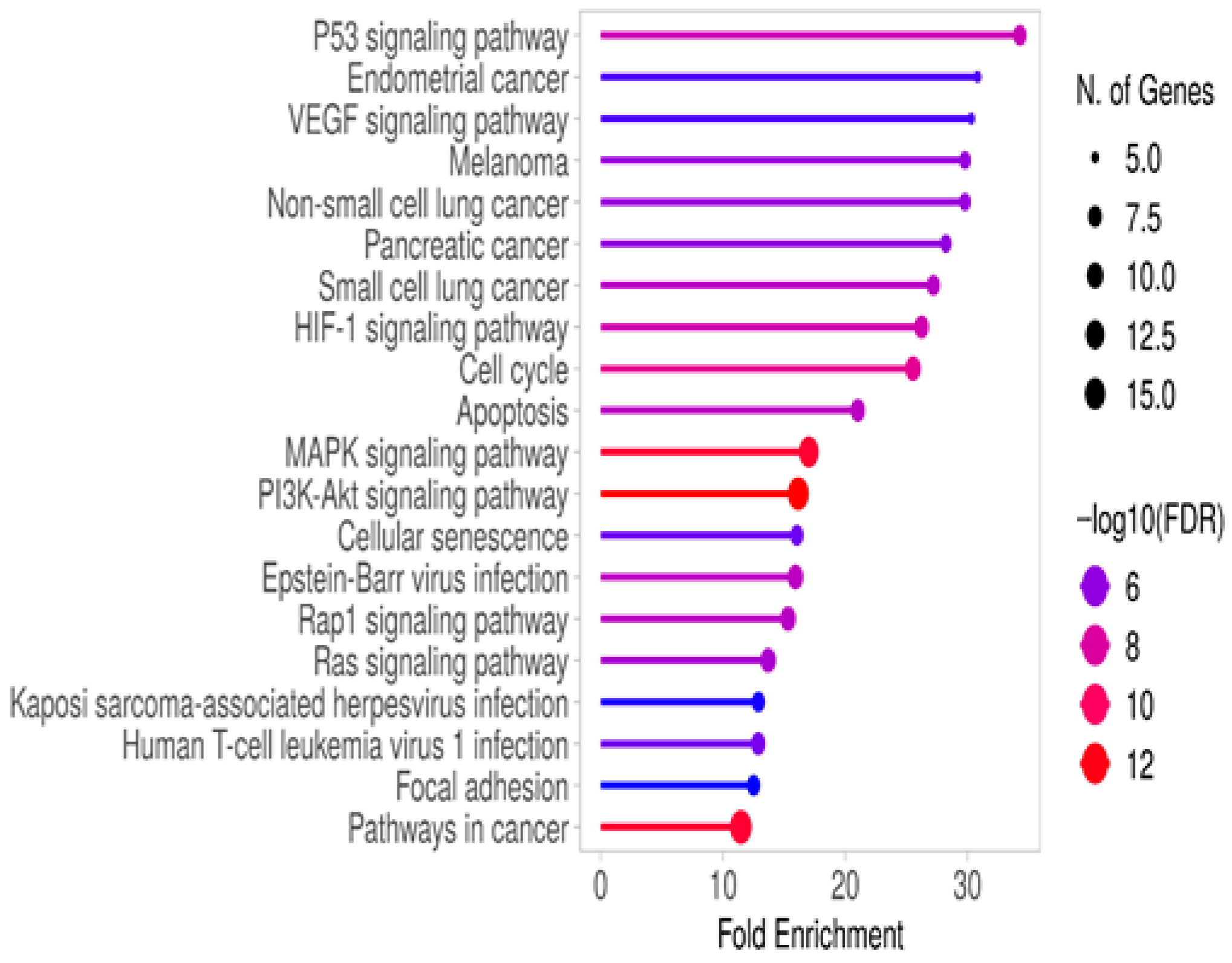
3.3. Gene Enrichment Analysis
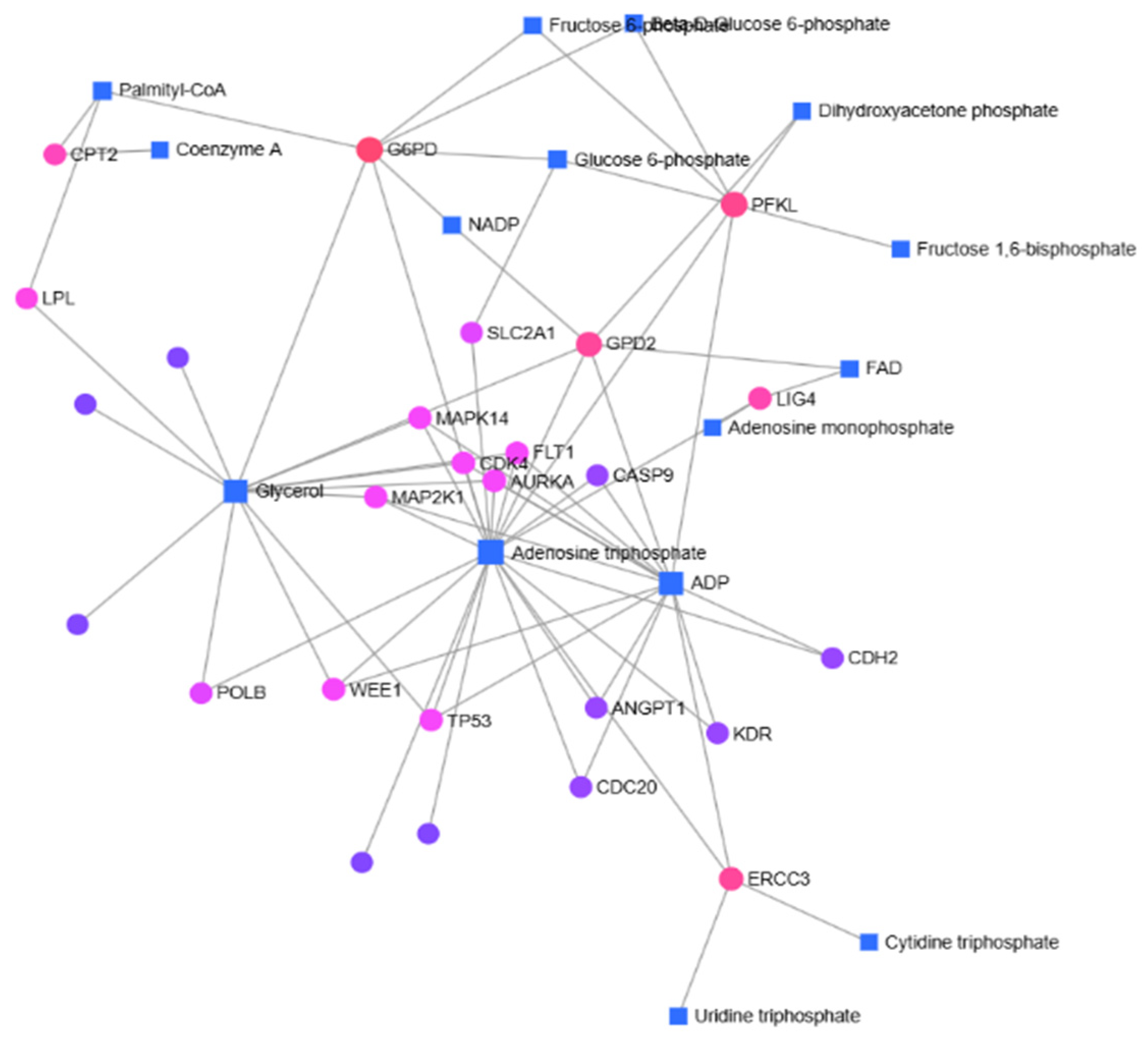
3.4. Gene-Metabolite Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANGPT2 | Angiopoietin-2 |
| APC | Adenomatous polyposis coli |
| AURKA | Aurora kinase A |
| BMI | Body mass index |
| CIN | Chromosomal instability |
| CPT | Carnitine palmitoyltransferase |
| DSP | Desmoplakin |
| EGFR | Epidermal growth factor receptor |
| ERCC3 | Excision repair cross-complementation group 3 |
| FDR | False discovery rate |
| FLT-1 | Fms-related receptor tyrosine kinase 1 (VEGFR1) |
| G6P | Glucose-6-phosphate |
| GPX4 | Glutathione peroxidase 4 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MCM2 | Minichromosome maintenance complex component 2 |
| MMR | Mismatch repair |
| MSI | Microsatellite instability |
References
- Ciepiela, I.; Szczepaniak, M.; Ciepiela, P.; Hińcza-Nowak, K.; Kopczyński, J.; Macek, P.; Kubicka, K.; Chrapek, M.; Tyka, M.; Góźdź, S.; et al. Tumor location matters, next generation sequencing mutation profiling of left-sided, rectal, and right-sided colorectal tumors in 552 patients. Sci. Rep. 2024, 14, 4619. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wen, Y.; Li, H.; Zhang, D.; Zhang, N.; Shi, X.; Jiang, B.; Ma, X.; Yang, P.; Tang, H.; et al. Overexpression of minichromosome maintenance 2 predicts poor prognosis in patients with gastric cancer. Oncol. Rep. 2012, 27, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, S.E.; Helmy, I.M.; Baghdadi, H.M. Maspin and MCM2 immunoprofiling in salivary gland carcinomas. Diagn. Pathol. 2011, 6, 89. [Google Scholar] [CrossRef]
- Yu, J.; Mallon, M.A.; Zhang, W.; Freimuth, R.R.; Marsh, S.; Watson, M.A.; Goodfellow, P.J.; McLeod, H.L. DNA repair pathway profiling and microsatellite instability in colorectal cancer. Clin. Cancer Res. 2006, 12, 5104–5111. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, Y.; Zhang, Y.; Ye, F.; Luo, D.; Li, Y.; Jin, Y.; Han, D.; Wang, Z.; Chen, B.; et al. HSPB1 facilitates chemoresistance through inhibiting ferroptotic cancer cell death and regulating NF-κB signaling pathway in breast cancer. Cell Death Dis. 2023, 14, 434. [Google Scholar] [CrossRef]
- Shimura, T.; Yin, C.; Ma, R.; Zhang, A.; Nagai, Y.; Shiratori, A.; Ozaki, H.; Yamashita, S.; Higashi, K.; Sato, Y.; et al. The prognostic importance of the negative regulators of ferroptosis, GPX4 and HSPB1, in patients with colorectal cancer. Oncol. Lett. 2025, 29, 144. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.E.; Weinberg, B.A.; Xiu, J.; El-Deiry, W.S.; Hwang, J.J.; Gatalica, Z.; Philip, P.A.; Shields, A.F.; Lenz, H.J.; Marshall, J.L. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget 2017, 8, 86356–86368. [Google Scholar] [CrossRef] [PubMed]
- Szostek, J.; Serafin, M.; Mąka, M.; Jabłońska, B.; Mrowiec, S. Right-sided versus left-sided colon cancer—A 5-year single-center observational study. Cancers 2025, 17, 537. [Google Scholar] [CrossRef] [PubMed]
- Mousa, L.; Salem, M.E.; Mikhail, S. Biomarkers of angiogenesis in colorectal cancer: Supplementary issue: Biomarkers for colon cancer. Biomark. Cancer 2015, 7, 13–19. [Google Scholar] [CrossRef]
- Jary, M.; Hasanova, R.; Vienot, A.; Asgarov, K.; Loyon, R.; Tirole, C.; Bouard, A.; Orillard, E.; Klajer, E.; Kim, S.; et al. Molecular description of ANGPT2 associated colorectal carcinoma. Int. J. Cancer 2020, 147, 2007–2018. [Google Scholar] [CrossRef]
- Jimenez-Luna, C.; González-Flores, E.; Ortiz, R.; Martínez-González, L.J.; Antúnez-Rodríguez, A.; Expósito-Ruiz, M.; Melguizo, C.; Caba, O.; Prados, J. Circulating PTGS2, JAG1, GUCY2C and PGF mRNA in peripheral blood and serum as potential biomarkers for patients with metastatic colon cancer. J. Clin. Med. 2021, 10, 2248. [Google Scholar] [CrossRef]
- Nakada, N.; Mikami, T.; Horie, K.; Nagashio, R.; Sakurai, Y.; Sanoyama, I.; Yoshida, T.; Sada, M.; Kobayashi, K.; Sato, Y.; et al. Expression of CA2 and CA9 carbonic anhydrases in ulcerative colitis and ulcerative colitis-associated colorectal cancer. Pathol. Int. 2020, 70, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Hui, E.P.; Chan, A.T.; Pezzella, F.; Turley, H.; To, K.F.; Poon, T.C.; Zee, B.; Mo, F.; Teo, P.M.; Huang, D.P.; et al. Coexpression of hypoxia-inducible factors 1alpha and 2alpha, carbonic anhydrase IX, and vascular endothelial growth factor in nasopharyngeal carcinoma and relationship to survival. Clin. Cancer Res. 2002, 8, 2595–2604. [Google Scholar]
- Wang, Z.; Ni, F.; Yu, F.; Cui, Z.; Zhu, X.; Chen, J. Prognostic significance of mRNA expression of CASPs in gastric cancer. Oncol. Lett. 2019, 18, 4535–4554. [Google Scholar] [CrossRef]
- Hong, W.; Gu, Y.; Guan, R.; Xie, D.; Zhou, H.; Yu, M. Pan-cancer analysis of the CASP gene family in relation to survival, tumor-infiltrating immune cells and therapeutic targets. Genomics 2020, 112, 4304–4315. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, X.; Yan, H.; Wu, J.; Yang, Y.; He, J.; Chen, J.; Jiang, Z.; Wu, F.; Jiang, Z. Downregulation of CPT2 promotes proliferation and inhibits apoptosis through p53 pathway in colorectal cancer. Cell. Signal. 2022, 92, 110267. [Google Scholar] [CrossRef]
- Dai, G.; Wang, D.; Ma, S.; Hong, S.; Ding, K.; Tan, X.; Ju, W. ACSL4 promotes colorectal cancer and is a potential therapeutic target of emodin. Phytomedicine 2022, 102, 154149. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Xiao, Q.; Li, Y.; Peng, Y.; Gan, Y.; Shu, G.; Yi, H.; Yin, G. Identification of CPT2 as a prognostic biomarker by integrating the metabolism-associated gene signature in colorectal cancer. BMC Cancer 2022, 22, 1038. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, C.; Bao, N. Molecular mechanism of palmitic acid and its derivatives in tumor progression. Front. Oncol. 2023, 13, 1224125. [Google Scholar] [CrossRef]
- Baxter, B.A.; Parker, K.D.; Nosler, M.J.; Rao, S.; Craig, R.; Seiler, C.; Ryan, E.P. Metabolite profile comparisons between ascending and descending colon tissue in healthy adults. World J. Gastroenterol. 2020, 26, 335–352. [Google Scholar] [CrossRef]


| Parameters | Total (n = 18) | Left Colon (n = 6) | Right Colon (n = 6) | Healthy (n = 6) |
|---|---|---|---|---|
| Age | ||||
| Mean ± SD | 58.77 ± 11.17 | 63.83 ± 10.57 | 63.16 ± 8.01 | 49.33 ± 9.37 |
| Median (min-max) | 60.0 (36–72) | 65.5 (44–76) | 63.0 (55–73) | 51.0 (36–63) |
| Sex | ||||
| Female | 13 (72.2%) | 5 (83.3%) | 4 (66.7%) | 4 (66.7%) |
| Male | 5 (27.8%) | 1 (16.7%) | 2 (33.3%) | 2 (33.3%) |
| Height | ||||
| Mean ± SD | 166.72 ± 9.56 | 172.0 ± 4.85 | 164.00 ± 12.63 | 164.16 ± 8.81 |
| Median (min-max) | 167.0 (149–185) | 173.0 (163–177) | 165.0 (149–185) | 163.5 (155–175) |
| Weight | ||||
| Mean ± SD | 75.72 ± 13.25 | 81.83 ± 9.28 | 76.33 ± 17.94 | 69.00 ± 9.46 |
| Median (min-max) | 75.0 (55–110) | 82.0 (70–95) | 70.5 (62–110) | 72.0 (55–78) |
| BMI | ||||
| Mean ± SD | 27.2 ± 3.7 | 27.72 ± 2.93 | 28.25 ± 4.64 | 25.62 ± 3.44 |
| Median (min-max) | 27.2 (22.1–34.1) | 27.5 (22.8–31.7) | 28.6 (22.1–34.1) | 24.4 (22.8–32.1) |
| Diagnosis | ||||
| Cecum tumor | 1 (5.6%) | 1 (16.7%) | ||
| Ascending colon tumor | 3 (16.7%) | 3 (50.0%) | ||
| Hepatic flux tumor | 1 (5.6%) | 1 (16.7%) | ||
| Normal | 6 (33.3%) | 6 (100.0%) | ||
| Rectosigmoid tumor | 1 (5.6%) | 1 (16.7%) | ||
| Rectum tumor | 1 (5.6%) | 1 (16.7%) | ||
| Right colon tumor | 1 (5.6%) | 1 (16.7%) | ||
| Sigmoid colon tumor | 4 (22.2%) | 4 (66.7%) | ||
| Tumor location | ||||
| Cecum | 1 (8.3%) | 1 (16.7%) | ||
| Ileocecal valve | 2 (16.6%) | 2 (33.3%) | ||
| Rectosigmoid colon | 2 (16.6%) | 2 (33.3%) | ||
| Right colon | 3 (25.0%) | 3 (50.0%) | ||
| Sigmoid colon | 3 (25.0%) | 3 (50.0%) | ||
| Left colon | 1 (8.3%) | 1 (16.7%) | ||
| Smoking | ||||
| No | 16 (88.9%) | 5 (83.3%) | 5 (83.3%) | 6 (100.0%) |
| Yes | 2 (11.1%) | 1 (16.7%) | 1 (16.7%) | 0 (0.0%) |
| Alcohol consumption | ||||
| No | 16 (88.9%) | 5 (83.3%) | 5 (83.3%) | 6 (100.0%) |
| Minimal | 2 (11.1%) | 1 (16.7%) | 1 (16.7%) | 0 (0.0%) |
| Surgery performed | ||||
| Anterior resection | 1 (5.6%) | 1 (16.7%) | ||
| Extended right hemicolectomy | 1 (5.6%) | 1 (16.7%) | ||
| Low anterior resection (LAR) | 5 (27.8%) | 5 (83.3%) | ||
| Right hemicolectomy | 5 (27.8%) | 5 (83.3%) | ||
| None | 6 (33.3%) | 6 (100.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ertuğrul, I.; Çelik, A.B.; Al, M.; Duman, M.; Altuntaş, Y.E.; Polat, E.; Ertuğrul, Y.E.; Küçük, H.F.; Tutar, Y. Gene Expression-Based Inference of Metabolic Signatures Reveals Distinct Molecular Profiles in Right- and Left-Sided Colon Cancer. Metabolites 2025, 15, 768. https://doi.org/10.3390/metabo15120768
Ertuğrul I, Çelik AB, Al M, Duman M, Altuntaş YE, Polat E, Ertuğrul YE, Küçük HF, Tutar Y. Gene Expression-Based Inference of Metabolic Signatures Reveals Distinct Molecular Profiles in Right- and Left-Sided Colon Cancer. Metabolites. 2025; 15(12):768. https://doi.org/10.3390/metabo15120768
Chicago/Turabian StyleErtuğrul, Ismail, Ayşe Büşranur Çelik, Mervenur Al, Mustafa Duman, Yunus Emre Altuntaş, Erdal Polat, Yunus Emre Ertuğrul, Hasan Fehmi Küçük, and Yusuf Tutar. 2025. "Gene Expression-Based Inference of Metabolic Signatures Reveals Distinct Molecular Profiles in Right- and Left-Sided Colon Cancer" Metabolites 15, no. 12: 768. https://doi.org/10.3390/metabo15120768
APA StyleErtuğrul, I., Çelik, A. B., Al, M., Duman, M., Altuntaş, Y. E., Polat, E., Ertuğrul, Y. E., Küçük, H. F., & Tutar, Y. (2025). Gene Expression-Based Inference of Metabolic Signatures Reveals Distinct Molecular Profiles in Right- and Left-Sided Colon Cancer. Metabolites, 15(12), 768. https://doi.org/10.3390/metabo15120768






