Integrated Transcriptomic and Metabolomic Insights into Flavor-Related Metabolism in Grape Berries Across Cultivars and Developmental Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Metabolite Profiling and DAM Selection
2.3. Metabolite Data Analysis
2.4. Transcriptomic Data Analysis
2.5. Identification of Transcription Factors and Co-Expression Network
3. Results
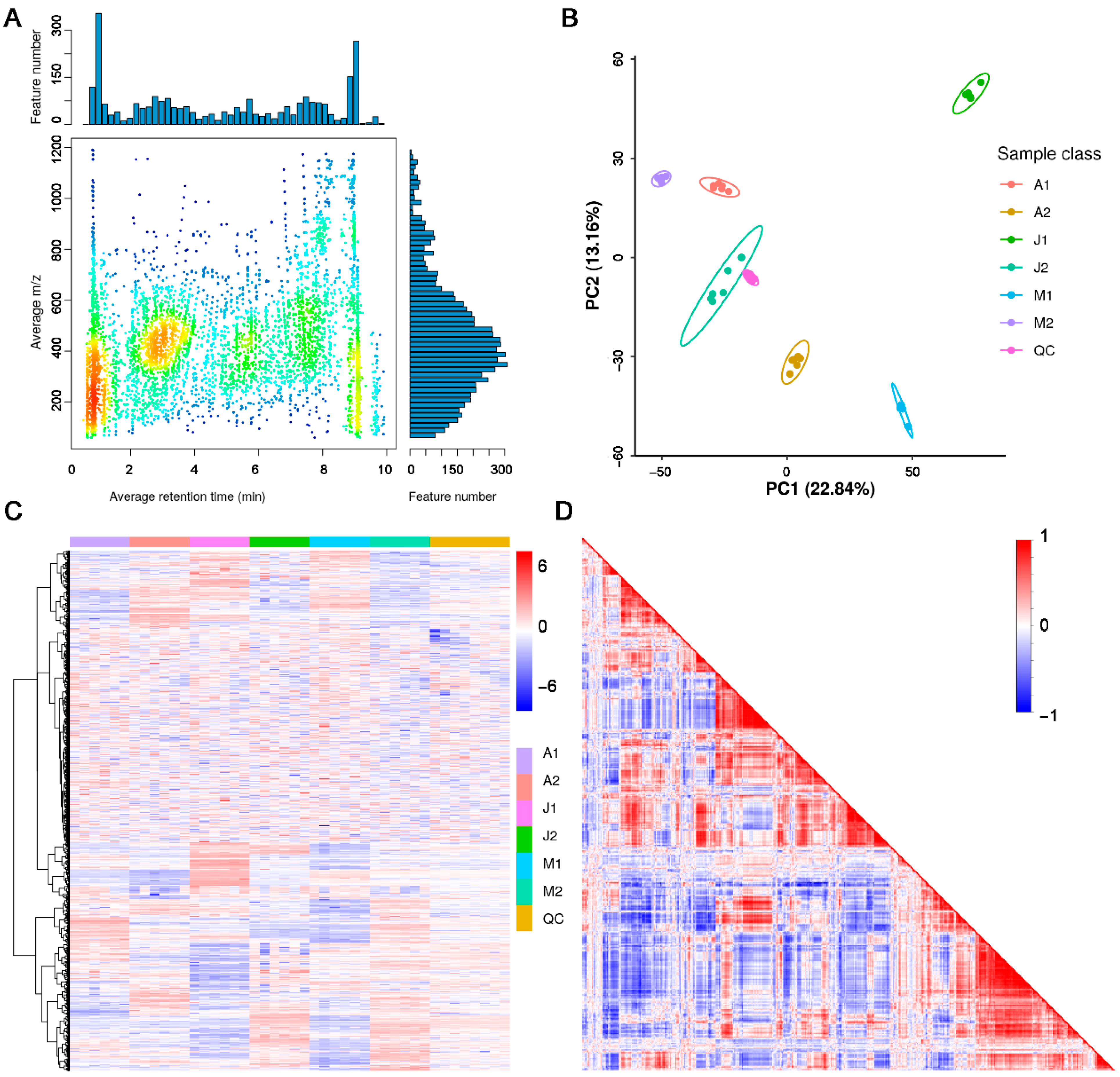
3.1. Overview of LC–MS Metabolomic Profiles
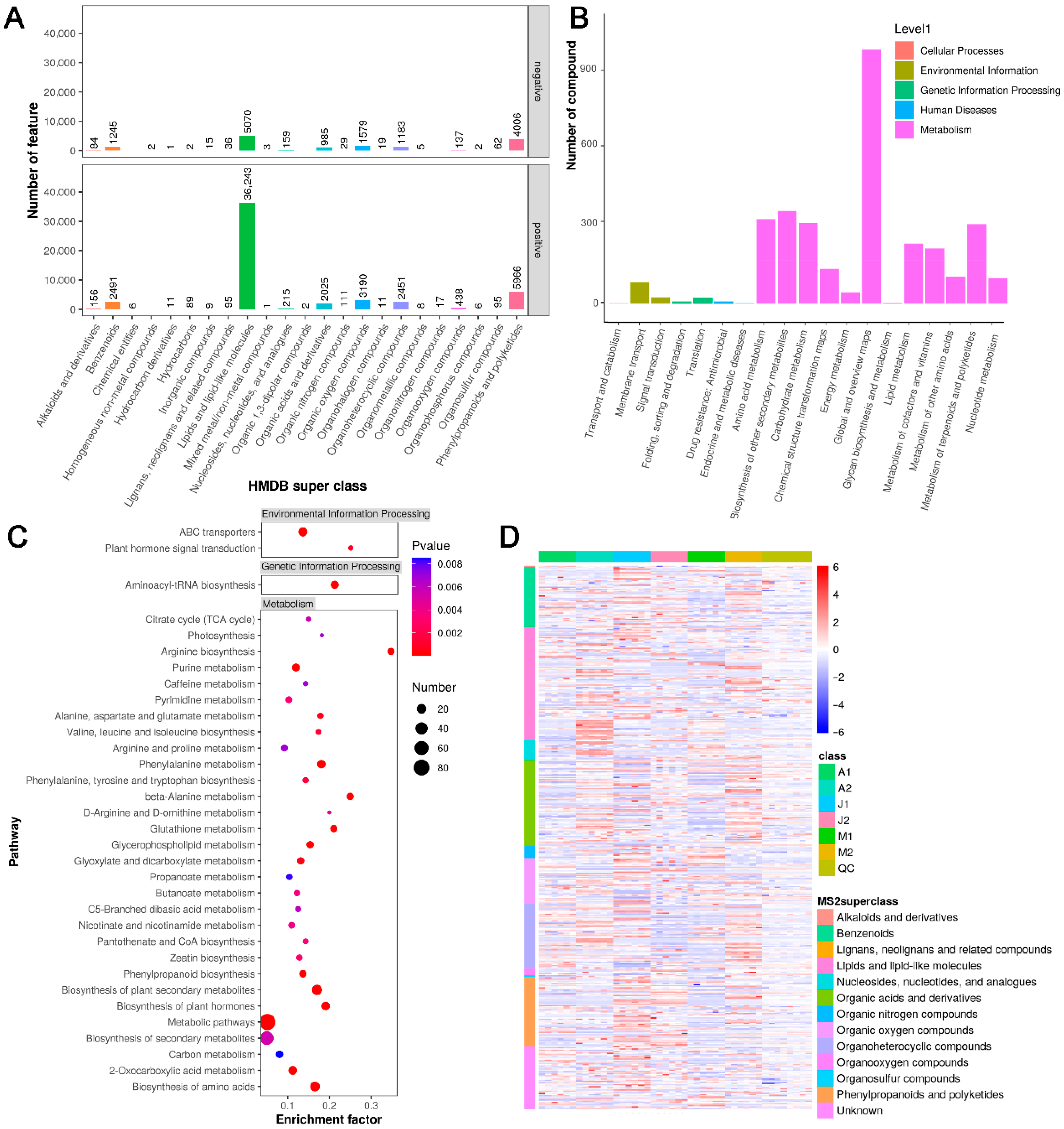
3.2. Functional Classification and Pathway Enrichment of MS2-Annotated Metabolites
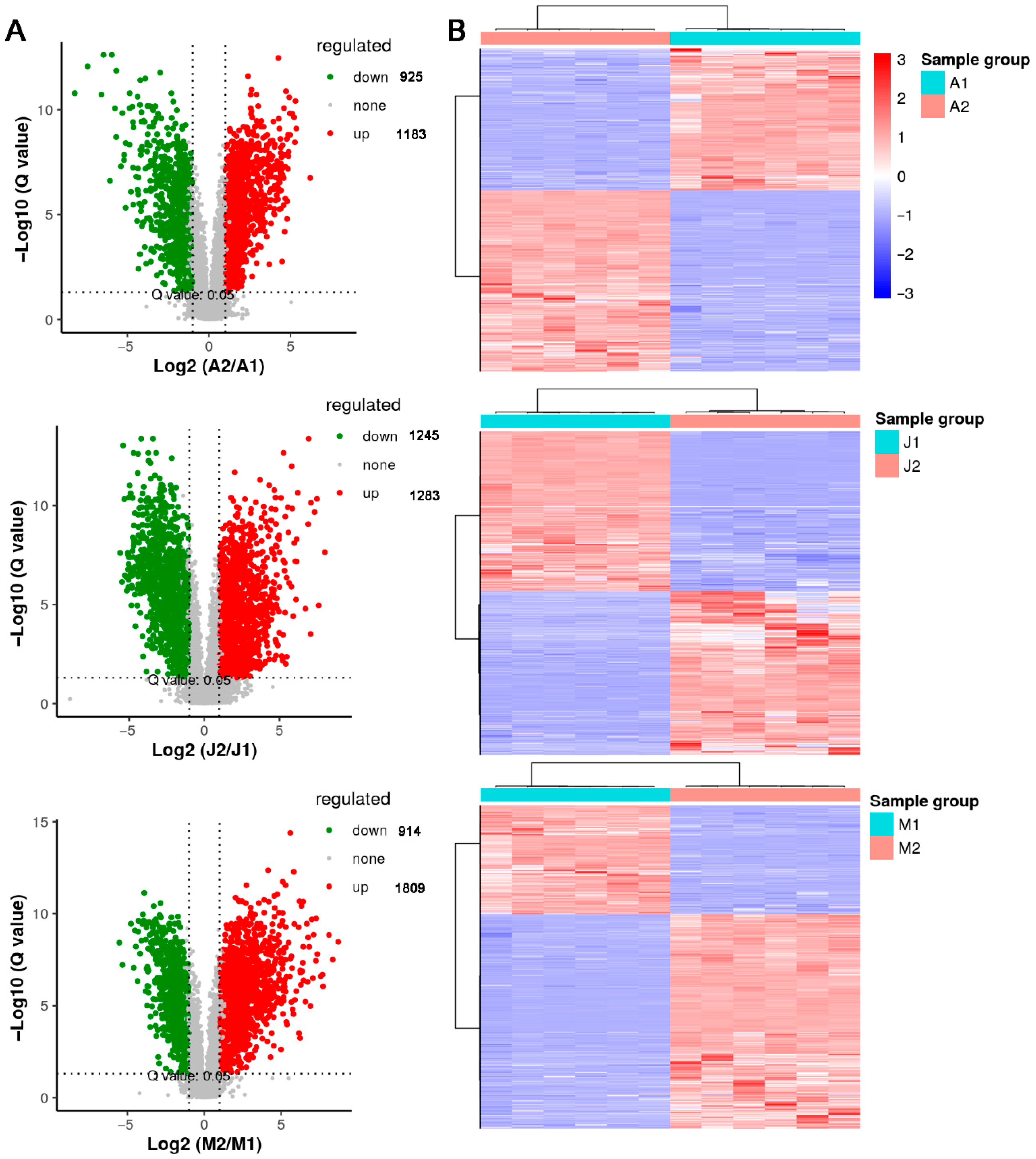
3.3. DAMs Among Cultivars and Developmental Stages
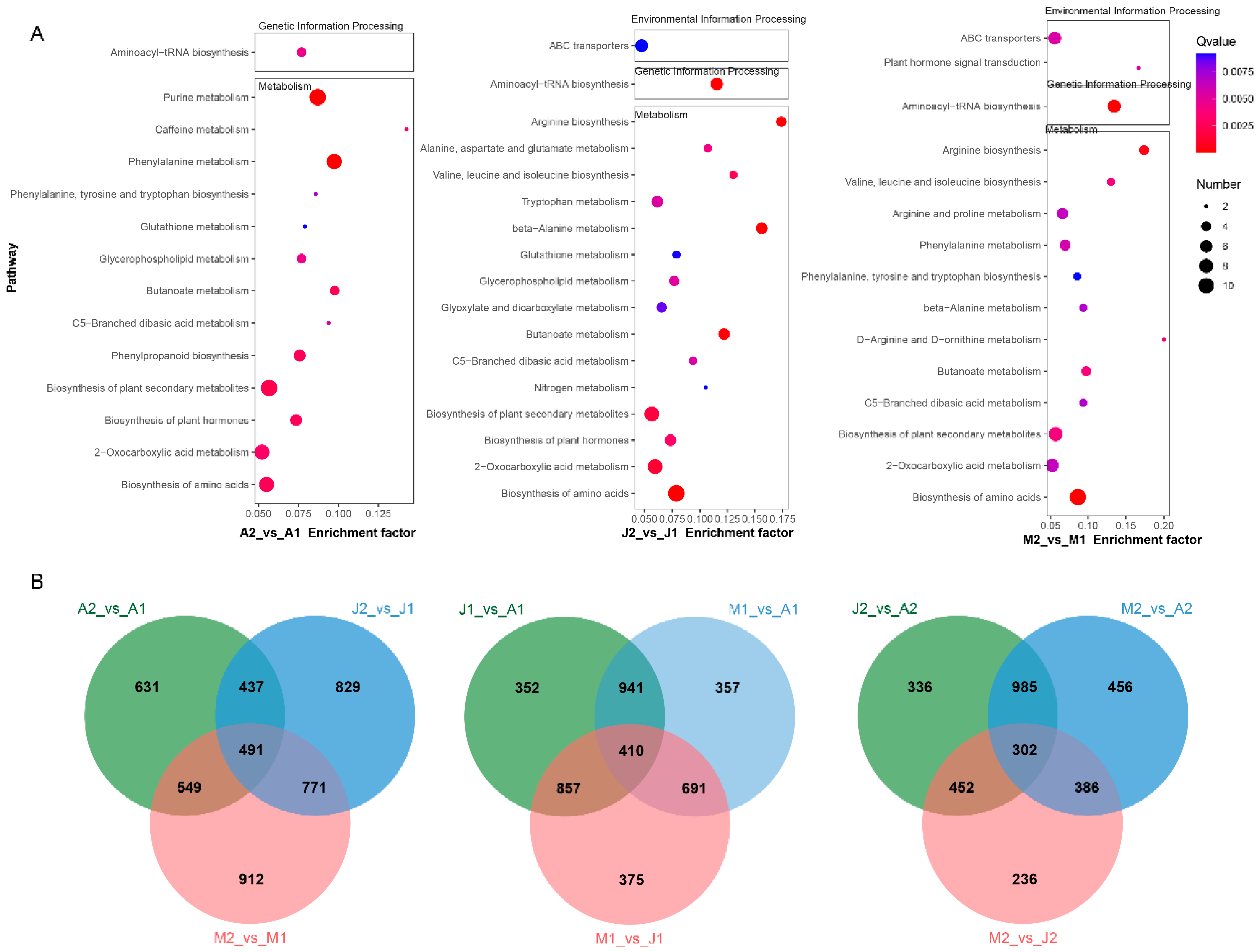
3.4. Functional Classification and Shared Features of DAMs
3.5. Identification of Differentially Expressed Transcription Factors (DETFs)
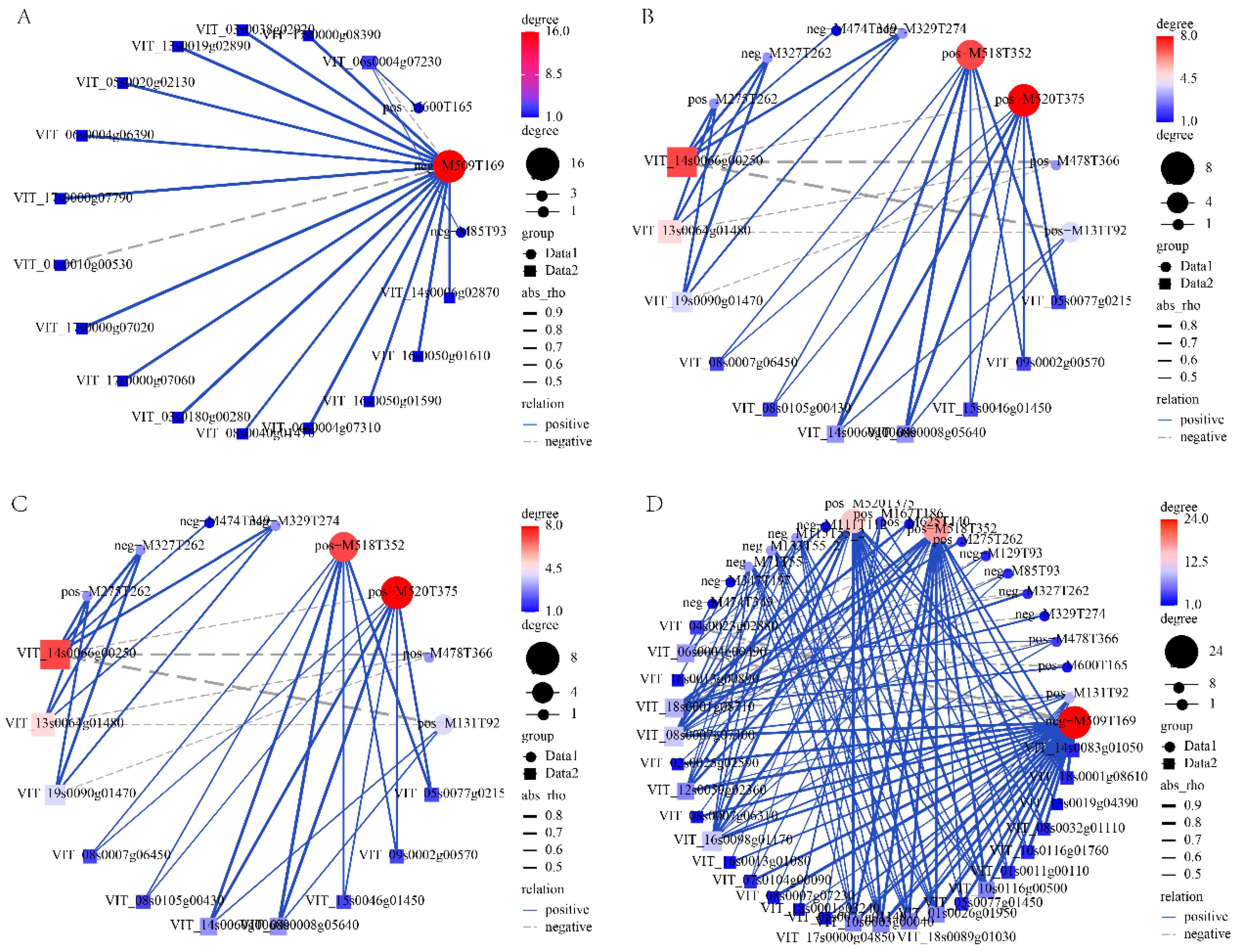
3.6. Correlation Analysis DETFs and Key Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DAMs | Differentially accumulated metabolites |
| DEGs | Differentially expressed genes |
| DETFs | Differentially expressed transcription factors |
| GC–MS | Gas chromatography–mass spectrometry |
| HCA | Hierarchical cluster analysis |
| HMDB | Human Metabolome Database |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC–MS | Liquid chromatography–mass spectrometry |
| OPLS–DA | Orthogonal partial least squares–discriminant analysis |
| PCA | Principal component analysis |
| QC | Quality control |
| rOAV | Relative odor activity value |
| TFs | Transcription factors |
| VIP | Variable importance in projection |
References
- Parker, M.; Capone, D.L.; Francis, I.L.; Herderich, M.J. Aroma precursors in grapes and wine: Flavor release during wine production and consumption. J. Agric. Food Chem. 2017, 66, 2281–2286. [Google Scholar] [CrossRef]
- Mele, M.A.; Kang, H.-M.; Lee, Y.-T.; Islam, M.Z. Grape terpenoids: Flavor importance, genetic regulation, and future potential. Crit. Rev. Food Sci. Nutr. 2021, 61, 1429–1447. [Google Scholar] [CrossRef]
- Zheng, T.; Zhang, S.; Leng, X.; Sadeghnezhad, E.; Li, T.; Pervaiz, T.; Liu, F.; Jia, H.; Fang, J. Profiling analysis of volatile and non-volatile compounds in Vitis Vinifera berries (cv. Chardonnay) and spontaneous bud mutation. Front. Nutr. 2021, 8, 715528. [Google Scholar] [CrossRef]
- Feng, T.; Sun, J.; Song, S.; Wang, H.; Yao, L.; Sun, M.; Wang, K.; Chen, D. Geographical differentiation of Molixiang table grapes grown in China based on volatile compounds analysis by HS-GC-IMS coupled with PCA and sensory evaluation of the grapes. Food Chem. X 2022, 15, 100423. [Google Scholar] [CrossRef]
- Chen, X.; Quek, S.Y. Free and glycosidically bound aroma compounds in fruit: Biosynthesis, transformation, and practical control. Crit. Rev. Food Sci. Nutr. 2023, 63, 9052–9073. [Google Scholar] [CrossRef] [PubMed]
- Šikuten, I.; Štambuk, P.; Tomaz, I.; Marchal, C.; Kontić, J.K.; Lacombe, T.; Maletić, E.; Preiner, D. Discrimination of genetic and geographical groups of grape varieties (Vitis vinifera L.) based on their volatile organic compounds. Front. Plant Sci. 2022, 13, 942148. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, W.; Wang, W.; Nieuwenhuizen, N.J.; Atkinson, R.G.; Gao, L.; Hu, H.; Zhao, W.; Ma, R.; Zheng, H. Integrated transcriptomic and proteomic analysis identifies novel regulatory genes associated with plant growth regulator-induced astringency in grape berries. J. Agric. Food Chem. 2024, 72, 4433–4447. [Google Scholar] [CrossRef]
- Feng, J.; Nieuwenhuizen, N.; Atkinson, R.; Wang, W.; Zeng, J.; Zheng, H.; Tao, J. Comparative study of phenolic compounds reveals a positive relationship between astringency and the phenolic composition in table grape varieties. J. Food Sci. 2023, 88, 447–461. [Google Scholar] [CrossRef]
- Karageçili, H.; İzol, E.; Kireçci, E.; Gülçin, İ. Antioxidant, antidiabetic, antiglaucoma, and anticholinergic effects of Tayfi grape (Vitis vinifera): A phytochemical screening by LC–MS/MS analysis. Open Chem. 2023, 21, 20230120. [Google Scholar] [CrossRef]
- Li, W.; Ye, N.; Xie, P.; Zhang, Z. Regulatory mechanism of methyl jasmonate and methyl dihydrojasmonate in enhancing aroma in ‘Cabernet Gernischt’ via the lipoxygenase pathway. J. Sci. Food. Agric. 2025, 105, 807–815. [Google Scholar] [CrossRef]
- Wang, W.; Feng, J.; Wei, L.; Khalil-Ur-Rehman, M.; Nieuwenhuizen, N.J.; Yang, L.; Tao, J. Transcriptomics integrated with free and bound terpenoid aroma profiling during “shine muscat”(Vitis labrusca × V. vinifera) grape berry development reveals coordinate regulation of MEP pathway and terpene synthase gene expression. J. Agric. Food Chem. 2021, 69, 1413–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, S.; Gao, W.; Hu, B.; Zhu, B.; Sun, L. Monoterpenoids evolution and MEP pathway gene expression profiles in seven table grape varieties. Plants 2022, 11, 2143. [Google Scholar] [CrossRef]
- Pérez-Navarro, J.; Da Ros, A.; Masuero, D.; Izquierdo-Cañas, P.M.; Hermosín-Gutiérrez, I.; Gómez-Alonso, S.; Mattivi, F.; Vrhovsek, U. LC–MS/MS analysis of free fatty acid composition and other lipids in skins and seeds of Vitis vinifera grape cultivars. Food Res. Int. 2019, 125, 108556. [Google Scholar] [CrossRef]
- Wang, J.; Han, Y.; Chen, C.; Sam, F.E.; Guan, R.; Wang, K.; Zhang, Y.; Zhao, M.; Chen, C.; Liu, X. Influence of Benzothiadiazole on the Accumulation and Metabolism of C6 Compounds in Cabernet Gernischt Grapes (Vitis vinifera L.). Foods 2023, 12, 3710. [Google Scholar] [CrossRef]
- Xu, L.; Zang, E.; Sun, S.; Li, M. Main flavor compounds and molecular regulation mechanisms in fruits and vegetables. Crit. Rev. Food Sci. Nutr. 2023, 63, 11859–11879. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhu, Y.; Wang, M.; Xun, Z.; Ma, X.; Zhao, Q. Integrative analysis of transcriptome and metabolome reveals the regulatory network governing aroma formation in grape. Horticulturae 2024, 10, 1159. [Google Scholar] [CrossRef]
- Carmona-Jiménez, Y.; Igartuburu, J.M.; Guillén-Sánchez, D.A.; García-Moreno, M.V. Fatty acid and tocopherol composition of pomace and seed oil from five grape varieties southern Spain. Molecules 2022, 27, 6980. [Google Scholar] [CrossRef]
- Gutiérrez-Escobar, R.; Aliaño-González, M.J.; Cantos-Villar, E. Variety and year: Two key factors on amino acids and biogenic amines content in grapes. Food Res. Int. 2024, 175, 113721. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, J.; Zheng, T.; Gu, W.; Zhang, Y.; Li, W.; Zhou, P.; Fang, Y.; Chen, K. Preharvest application of MeJA enhancing the quality of postharvest grape berries via regulating terpenes biosynthesis and phenylpropanoid metabolisms. Food Chem. 2024, 438, 137958. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhang, G.; Zhang, X.; Huang, L. A Review of Transcriptomics and Metabolomics in Plant Quality and Environmental Response: From Bibliometric Analysis to Science Mapping and Future Trends. Metabolites 2024, 14, 272. [Google Scholar] [CrossRef]
- Liu, S.; Shan, B.; Zhou, X.; Gao, W.; Liu, Y.; Zhu, B.; Sun, L. Transcriptome and metabolomics integrated analysis reveals terpene synthesis genes controlling linalool synthesis in grape berries. J. Agric. Food Chem. 2022, 70, 9084–9094. [Google Scholar] [CrossRef]
- Li, X.; Yan, Y.; Wang, L.; Li, G.; Wu, Y.; Zhang, Y.; Xu, L.; Wang, S. Integrated transcriptomic and metabolomic analysis revealed abscisic acid-induced regulation of monoterpene biosynthesis in grape berries. Plants 2024, 13, 1862. [Google Scholar] [CrossRef]
- Villano, C.; Demurtas, O.C.; Esposito, S.; Granell, A.; Rambla, J.L.; Piombino, P.; Frusciante, L.; Carputo, D.; Diretto, G.; Aversano, R. Integrative analysis of metabolome and transcriptome profiles to highlight aroma determinants in Aglianico and Falanghina grape berries. BMC Plant Biol. 2023, 23, 241. [Google Scholar] [PubMed]
- Tang, X.; Dong, Z.; Li, X.; Tan, W.; Ma, X.; Zhao, Q.; Wang, M. A new early season tetraploid grape cultivar ‘Meixiangbao’. J. Fruit Sci. 2017, 34, 115–118. [Google Scholar]
- Kuhalskaya, A.; Wijesingha Ahchige, M.; Perez de Souza, L.; Vallarino, J.; Brotman, Y.; Alseekh, S. Network analysis provides insight into tomato lipid metabolism. Metabolites 2020, 10, 152. [Google Scholar] [CrossRef]
- Li, X.; Tieman, D.; Liu, Z.; Chen, K.; Klee, H.J. Identification of a lipase gene with a role in tomato fruit short-chain fatty acid-derived flavor volatiles by genome-wide association. Plant J. 2020, 104, 631–644. [Google Scholar] [CrossRef]
- Rivero Meza, S.L.; de Castro Tobaruela, E.; Benedetti Pascoal, G.; Louro Massaretto, I.; Purgatto, E. Post-harvest treatment with methyl jasmonate impacts lipid metabolism in tomato pericarp (Solanum lycopersicum L. cv. Grape) at different ripening stages. Foods 2021, 10, 877. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.-B.; Xiang, X.-F.; Yang, W.-X.; Zhu, B.-Q.; Pu, H.-T.; Duan, C.-Q. Characterization of free and glycosidically bound volatile compounds, fatty acids, and amino acids in Vitis davidii Foex grape species native to China. Food Sci. Biotechnol. 2020, 29, 1641–1653. [Google Scholar] [CrossRef]
- Wang, Z.; Fu, M.; Yang, H.; Sun, F.; Qu, X.; Liu, J.; Chen, Q.; Yan, Y. (E)-2-Hexenyl acetate improved carotenoid synthesis in tomato fruits by reducing (E)-2-hexenal biosynthesis and activating jasmonic acid pathway. N. Z. J. Crop Hortic. Sci. 2025, 53, 2827–2842. [Google Scholar] [CrossRef]
- Tawfik, M.M.; Yamato, K.T.; Kohchi, T.; Koeduka, T.; Matsui, K. n-Hexanal and (Z)-3-hexenal are generated from arachidonic acid and linolenic acid by a lipoxygenase in Marchantia polymorpha L. Biosci. Biotechnol. Biochem. 2017, 81, 1148–1155. [Google Scholar] [CrossRef]
- Liscombe, D.K.; Kamiyoshihara, Y.; Ghironzi, J.; Kempthorne, C.J.; Hooton, K.; Bulot, B.; Kanellis, V.; McNulty, J.; Lam, N.B.; Nadeau, L.F.; et al. A flavin-dependent monooxygenase produces nitrogenous tomato aroma volatiles using cysteine as a nitrogen source. Proc. Natl. Acad. Sci. USA 2022, 119, e2118676119. [Google Scholar] [CrossRef]
- Walker, R.P.; Famiani, F. Organic acids in fruits: Metabolism, functions and contents. Hortic. Rev. 2018, 45, 371–430. [Google Scholar]
- Jia, D.; Xu, Z.; Chen, L.; Huang, Q.; Huang, C.; Tao, J.; Qu, X.; Xu, X. Analysis of organic acid metabolism reveals citric acid and malic acid play major roles in determining acid quality during the development of kiwifruit (Actinidia eriantha). J. Sci. Food Agric. 2023, 103, 6055–6069. [Google Scholar] [CrossRef]
- Yang, G.; Zhao, S.; Gong, J.; Huang, M.; Yu, W.; Zhang, K.; Hu, D. Dissipation and the effects of thidiazuron on antioxidant enzyme activity and malondialdehyde content in strawberry. J. Sci. Food Agric. 2019, 99, 4331–4337. [Google Scholar] [CrossRef]
- Wei, C.; Liu, H.; Cao, X.; Zhang, M.; Li, X.; Chen, K.; Zhang, B. Synthesis of flavour-related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol. J. 2021, 19, 2082–2096. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Guo, Z.-H.; Hao, P.-P.; Wang, G.-M.; Jin, Z.-M.; Zhang, S.-L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Li, Z.; Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–20. [Google Scholar] [CrossRef]
- Tirumalai, V.; Swetha, C.; Nair, A.; Pandit, A.; Shivaprasad, P.V. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 2019, 70, 4775–4792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dai, Z.; Ferrier, T.; Orduña, L.; Santiago, A.; Peris, A.; Wong, D.C.J.; Kappel, C.; Savoi, S.; Matus, J.T.; et al. MYB24 orchestrates terpene and flavonol metabolism as light responses to anthocyanin depletion in variegated grape berries. Plant Cell 2023, 35, 4238–4265. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, S.; Chen, B.; Bai, X.; Li, Y.; Yang, J.; Wang, W.; Li, C.; Li, Y.; Li, Z. The MADS-box transcription factor GlMADS1 regulates secondary metabolism in Ganoderma lucidum. Mycologia 2021, 113, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, K.; Tang, B.; Su, D.; He, X.; Deng, H.; Wu, M.; Bouzayen, M.; Grierson, D.; Liu, M. The MADS-box protein SlTAGL1 regulates a ripening-associated SlDQD/SDH2 involved in flavonoid biosynthesis and resistance against Botrytis cinerea in post-harvest tomato fruit. Plant J. 2023, 115, 1746–1757. [Google Scholar] [CrossRef] [PubMed]

| ID | Metabolite (MS2) | Superclass(MS2) | A2_vs_A1 | J2_vs_J1 | M2_vs_M1 | VIP (Max) |
|---|---|---|---|---|---|---|
| neg-M327T262 | (9R,10S,12Z)-9,10-Dihydroxy -8-oxo-12-octadecenoic acid | Lipids and lipid-like molecules | down | up | up | 2.278544 |
| neg-M329T274 | 9,10,13-TriHOME | Lipids and lipid-like molecules | down | up | up | 1.814051 |
| pos-M131T92 | Citraconic acid | Lipids and lipid-like molecules | up | down | down | 1.416736 |
| neg-M347T197 | Foeniculoside VII | Lipids and lipid-like molecules | down | down | up | 2.343692 |
| pos-M520T375 | LysoPC 18:2 | Lipids and lipid-like molecules | up | down | down | 3.387475 |
| pos-M518T352 | LysoPC 18:3 | Lipids and lipid-like molecules | up | down | down | 3.638893 |
| pos-M478T366 | LysoPE 18:2 | Lipids and lipid-like molecules | up | down | down | 2.335447 |
| neg-M474T349 | LysoPE 18:3 | Lipids and lipid-like molecules | up | down | down | 2.492346 |
| pos-M275T262 | Nandrolone | Lipids and lipid-like molecules | down | up | up | 1.599161 |
| neg-M206T142 | 1H-Indole-3-carboxylic acid | Organoheterocyclic compounds | down | down | up | 1.887678 |
| neg-M111T112 | 2-Furoic acid | Organoheterocyclic compounds | up | down | down | 1.542474 |
| neg-M129T93 | 4-Hydroxy-2-butenoic acid gamma-lactone | Organoheterocyclic compounds | up | down | down | 3.307597 |
| pos-M233T170 | Alantolactone | Organoheterocyclic compounds | down | up | up | 1.549641 |
| pos-M353T274 | Anileridine | Organoheterocyclic compounds | down | up | up | 1.585172 |
| neg-M179T144 | Dihydroxyacetone (dimer) | Organoheterocyclic compounds | down | up | up | 1.376759 |
| pos-M86T80 | Piperidine | Organoheterocyclic compounds | up | up | up | 1.961697 |
| pos-M72T54 | Pyrrolidine | Organoheterocyclic compounds | up | up | up | 1.652866 |
| neg-M133T55_2 | D-(+)-Malic acid | Organic acids and derivatives | up | down | down | 2.619117 |
| pos-M130T71 | D-Pyroglutamic acid | Organic acids and derivatives | up | up | up | 1.936217 |
| neg-M115T55_2 | Maleic acid | Organic acids and derivatives | up | down | down | 2.572134 |
| pos-M172T54 | Tetrahydrodipicolinate | Organic acids and derivatives | up | down | up | 1.995907 |
| neg-M85T93 | Diacetyl | Organic oxygen compounds | up | down | down | 2.641908 |
| neg-M509T169 | Linalool 3,6-oxide primeveroside | Organic oxygen compounds | up | up | up | 3.128103 |
| neg-M71T55 | Malondialdehyde | Organic oxygen compounds | up | down | down | 2.262774 |
| neg-M87T93 | Malonic semialdehyde | Organic oxygen compounds | up | down | down | 2.00654 |
| pos-M268T72 | Adenosine | Nucleosides, nucleotides | down | down | down | 2.409461 |
| pos-M664T64 | beta-Nicotinamide adenine dinucleotide | Nucleosides, nucleotides | down | up | up | 1.850993 |
| neg-M262T188 | Gemcitabine | Nucleosides, nucleotides | up | up | up | 2.460595 |
| pos-M318T324 | Phytosphingosine | Organic nitrogen compounds | up | down | down | 1.991469 |
| pos-M203T41 | Spermine | Organic nitrogen compounds | up | up | down | 1.932743 |
| pos-M625T140 | Peonidin-3,5-O-di-beta-glucopyranoside | Phenylpropanoids and polyketides | down | up | up | 2.273243 |
| pos-M167T186 | 4-Ethyl-1,2-dimethoxybenzene | Benzenoids | down | down | up | 2.080155 |
| pos-M600T165 | (7′R)-(+)-Lyoniresinol 9′-glucoside | Lignans, neolignans | down | up | up | 2.050689 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Zhang, L.; Wang, M.; Zhu, Y.; Xun, Z.; Dai, X.; Zhao, Q. Integrated Transcriptomic and Metabolomic Insights into Flavor-Related Metabolism in Grape Berries Across Cultivars and Developmental Stages. Metabolites 2025, 15, 648. https://doi.org/10.3390/metabo15100648
Huang L, Zhang L, Wang M, Zhu Y, Xun Z, Dai X, Zhao Q. Integrated Transcriptomic and Metabolomic Insights into Flavor-Related Metabolism in Grape Berries Across Cultivars and Developmental Stages. Metabolites. 2025; 15(10):648. https://doi.org/10.3390/metabo15100648
Chicago/Turabian StyleHuang, Liping, Linan Zhang, Min Wang, Yue Zhu, Zhili Xun, Xi Dai, and Qifeng Zhao. 2025. "Integrated Transcriptomic and Metabolomic Insights into Flavor-Related Metabolism in Grape Berries Across Cultivars and Developmental Stages" Metabolites 15, no. 10: 648. https://doi.org/10.3390/metabo15100648
APA StyleHuang, L., Zhang, L., Wang, M., Zhu, Y., Xun, Z., Dai, X., & Zhao, Q. (2025). Integrated Transcriptomic and Metabolomic Insights into Flavor-Related Metabolism in Grape Berries Across Cultivars and Developmental Stages. Metabolites, 15(10), 648. https://doi.org/10.3390/metabo15100648




