Spatial Metabolomics Profiling Reveals Curcumin Induces Metabolic Reprogramming in Three-Dimensional Tumor Spheroids
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture
2.3. Cell Viability
2.4. Cell Migration
2.5. Cell Invasion
2.6. Caspase 3 Activity
2.7. MALDI-MSI Analysis
2.8. Analyte Identification
2.9. Quantitative Real-Time PCR (qRT-PCR) Assay
2.10. Data Processing
3. Results and Discussion
3.1. Curcumin Inhibits Cell Viability of MDA-MB-231 Cells
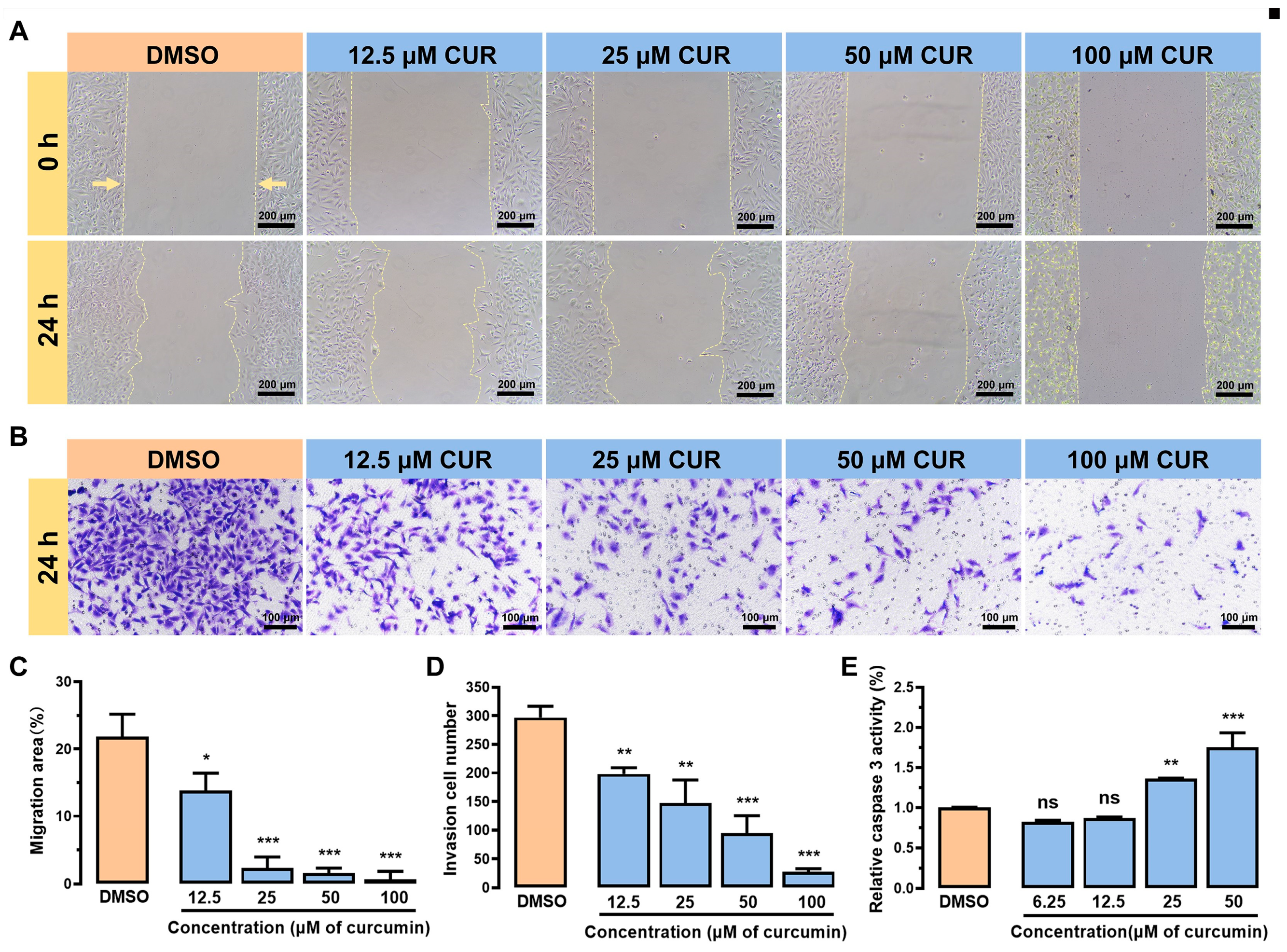
3.2. Curcumin Inhibits the Growth and Metastasis of MDA-MB-231 Cells
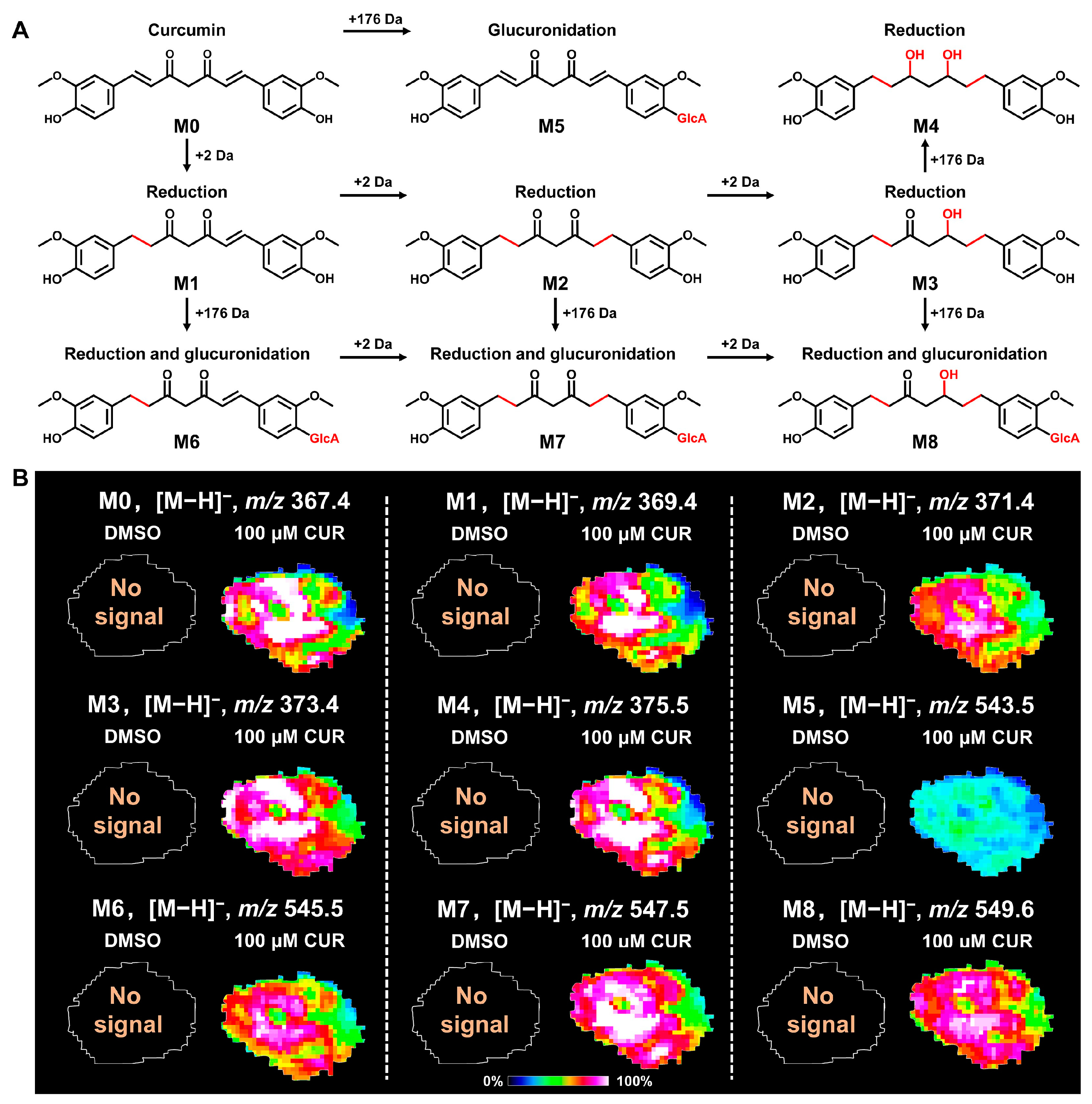
3.3. Spatial Distribution of Curcumin and Its Metabolites in 3D Tumor Spheroids
3.4. Spatial Metabolomics Emphasizes Metabolic Alterations in 3D Tumor Spheroids
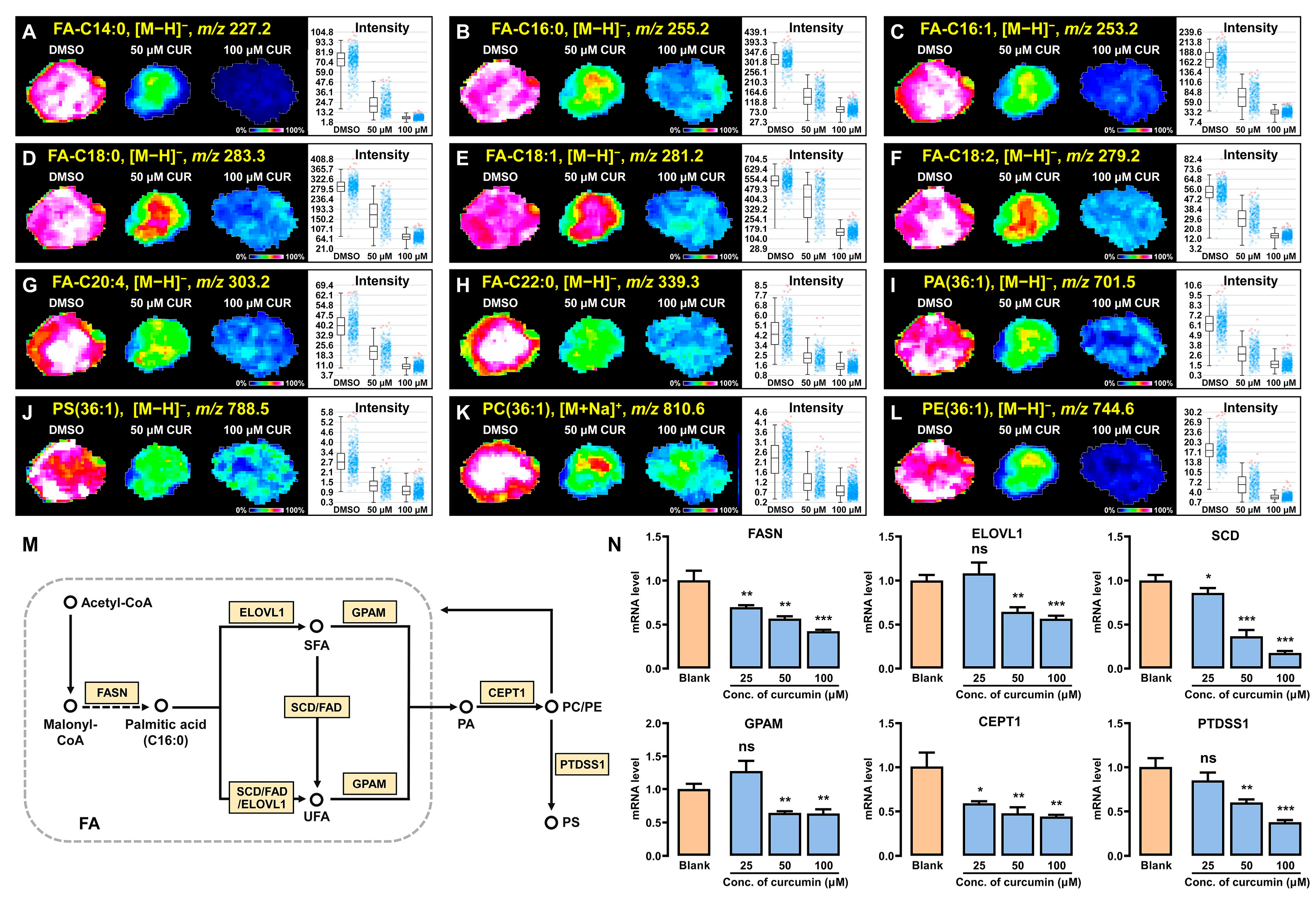
3.5. Curcumin Modulates Lipid Synthesis and Metabolism in 3D Breast Cancer Spheroids
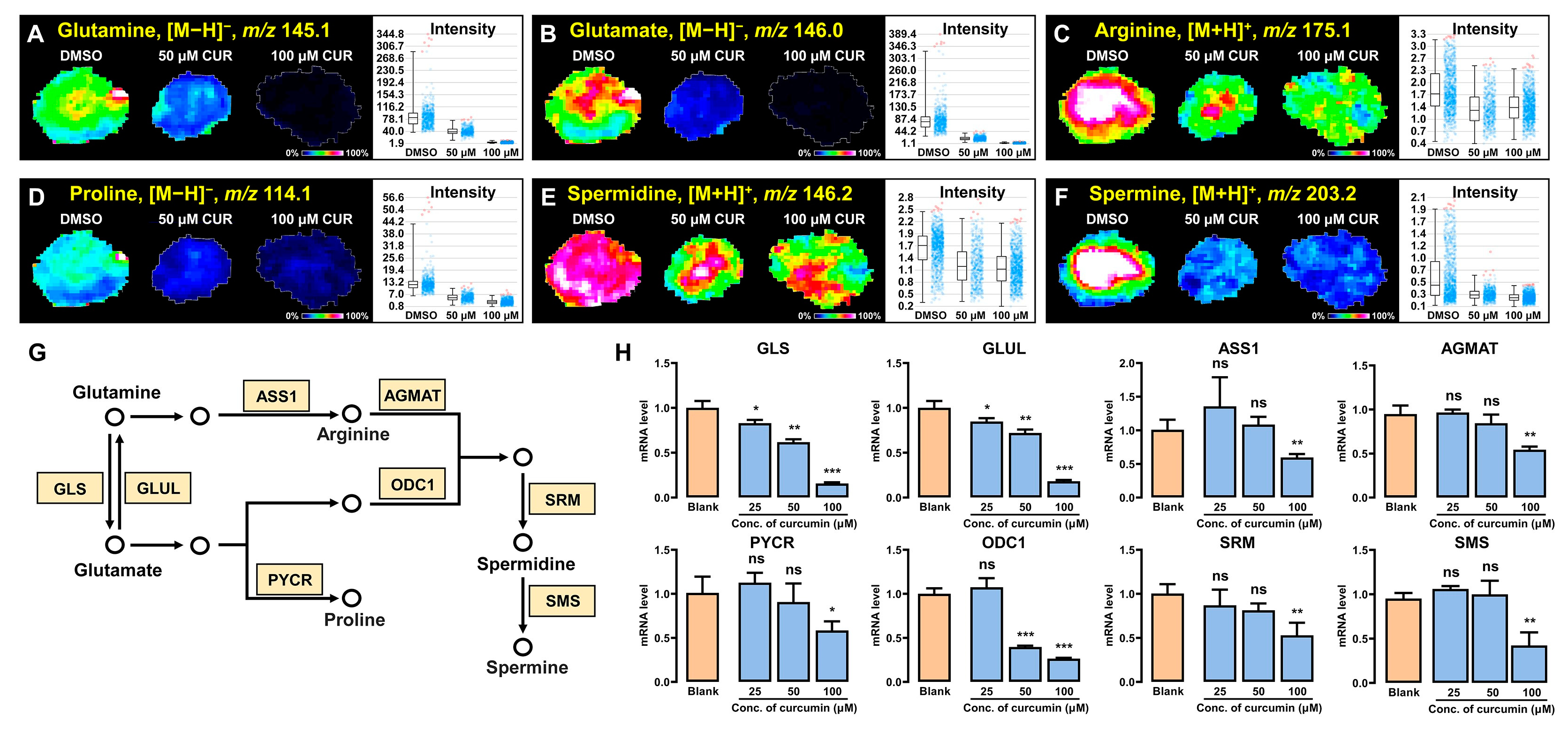
3.6. Curcumin Regulates Polyamine Synthesis and Metabolism in 3D Tumor Spheroids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Saha, T.; Solomon, J.; Samson, A.O.; Gil-Henn, H. Invasion and metastasis as a central hallmark of breast cancer. J. Clin. Med. 2021, 10, 3498. [Google Scholar] [CrossRef] [PubMed]
- Yeeravalli, R.; Das, A. Molecular mediators of breast cancer metastasis. Hematol. Oncol. Stem Cell Ther. 2021, 14, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Nedeljković, M.; Damjanović, A. Mechanisms of chemotherapy resistance in triple-negative breast cancer—How we can rise to the challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lv, L.; Yang, S.; Zhu, Y.; Zhai, X.; Li, S.; Tao, X.; Dong, D. Relationship between metabolic reprogramming and drug resistance in breast cancer. Front. Oncol. 2022, 12, 942064. [Google Scholar] [CrossRef]
- Zheng, X.; Ma, H.; Wang, J.; Huang, M.; Fu, D.; Qin, L.; Yin, Q. Energy metabolism pathways in breast cancer progression: The reprogramming, crosstalk, and potential therapeutic targets. Transl. Oncol. 2022, 26, 101534. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Ferraro, G.B.; Ali, A.; Luengo, A.; Kodack, D.P.; Deik, A.; Abbott, K.L.; Bezwada, D.; Blanc, L.; Prideaux, B.; Jin, X. Fatty acid synthesis is required for breast cancer brain metastasis. Nat. Cancer 2021, 2, 414–428. [Google Scholar] [CrossRef]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef]
- Gupta, S.C.; Patchva, S.; Aggarwal, B.B. Therapeutic roles of curcumin: Lessons learned from clinical trials. AAPS J. 2013, 15, 195–218. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guo, T.; Lin, J.; Huang, X.; Ke, Q.; Wu, Y.; Fang, C.; Hu, C. Curcumin inhibits the invasion and metastasis of triple negative breast cancer via Hedgehog/Gli1 signaling pathway. J. Ethnopharmacol. 2022, 283, 114689. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Wang, D.; Zou, D.; Wang, C.; Lopes-Bastos, B.; Jiang, W.G.; Chester, J.; Zhou, Q.; Cai, J. Re-purposing of curcumin as an anti-metastatic agent for the treatment of epithelial ovarian cancer: In vitro model using cancer stem cell enriched ovarian cancer spheroids. Oncotarget 2016, 7, 86374. [Google Scholar] [CrossRef] [PubMed]
- Hossain, D.M.S.; Bhattacharyya, S.; Das, T.; Sa, G. Curcumin: The multi-targeted therapy for cancer regression. Front. Biosci. -Sch. 2012, 4, 335–355. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef]
- Huang, F.; Ni, M.; Chalishazar, M.D.; Huffman, K.E.; Kim, J.; Cai, L.; Shi, X.; Cai, F.; Zacharias, L.G.; Ireland, A.S. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab. 2018, 28, 369–382.e5. [Google Scholar] [CrossRef]
- Huang, J.; Liu, D.; Wang, Y.; Liu, L.; Li, J.; Yuan, J.; Jiang, Z.; Jiang, Z.; Hsiao, W.W.; Liu, H. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 2022, 71, 734–745. [Google Scholar] [CrossRef]
- Liu, D.; Huang, J.; Gao, S.; Jin, H.; He, J. A temporo-spatial pharmacometabolomics method to characterize pharmacokinetics and pharmacodynamics in the brain microregions by using ambient mass spectrometry imaging. Acta Pharm. Sin. B 2022, 12, 3341–3353. [Google Scholar] [CrossRef]
- Panayotis, N.; Freund, P.A.; Marvaldi, L.; Shalit, T.; Brandis, A.; Mehlman, T.; Tsoory, M.M.; Fainzilber, M. β-sitosterol reduces anxiety and synergizes with established anxiolytic drugs in mice. Cell Rep. Med. 2021, 2, 100281. [Google Scholar] [CrossRef]
- Liu, R.; Bao, Z.; Zhao, P.; Li, G. Advances in the study of metabolomics and metabolites in some species interactions. Molecules 2021, 26, 3311. [Google Scholar] [CrossRef] [PubMed]
- Kirla, K.T.; Groh, K.J.; Steuer, A.E.; Poetzsch, M.; Banote, R.K.; Stadnicka-Michalak, J.; Eggen, R.I.; Schirmer, K.; Kraemer, T. From the cover: Zebrafish larvae are insensitive to stimulation by cocaine: Importance of exposure route and toxicokinetics. Toxicol. Sci. 2016, 154, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, W.; Mu, Y.; Wang, X. 1, 1′-binaphthyl-2, 2′-diamine as a novel MALDI matrix to enhance the in situ imaging of metabolic heterogeneity in lung cancer. Talanta 2020, 209, 120557. [Google Scholar] [CrossRef]
- Huang, H.; Liu, H.; Ma, W.; Qin, L.; Chen, L.; Guo, H.; Xu, H.; Li, J.; Yang, C.; Hu, H. High-throughput MALDI-MSI metabolite analysis of plant tissue microarrays. Plant Biotechnol. J. 2023, 21, 2574–2584. [Google Scholar] [CrossRef]
- Tang, W.; Shi, J.J.; Liu, W.; Lu, X.; Li, B. MALDI Imaging Assisted Discovery of a Di-O-glycosyltransferase from Platycodon grandiflorum Root. Angew. Chem. Int. Ed. Engl. 2023, 62, e202301309. [Google Scholar] [CrossRef]
- Chen, P.; Han, Y.; Wang, L.; Zheng, Y.; Zhu, Z.; Zhao, Y.; Zhang, M.; Chen, X.; Wang, X.; Sun, C. Spatially Resolved Metabolomics Combined with the 3D Tumor-Immune Cell Coculture Spheroid Highlights Metabolic Alterations during Antitumor Immune Response. Anal. Chem. 2023, 95, 15153–15161. [Google Scholar] [CrossRef]
- Dilillo, M.; Ait-Belkacem, R.; Esteve, C.; Pellegrini, D.; Nicolardi, S.; Costa, M.; Vannini, E.; Graaf, E.d.; Caleo, M.; McDonnell, L. Ultra-high mass resolution MALDI imaging mass spectrometry of proteins and metabolites in a mouse model of glioblastoma. Sci. Rep. 2017, 7, 603. [Google Scholar] [CrossRef]
- Chen, P.; Zhu, Z.; Geng, H.; Cui, X.; Han, Y.; Wang, L.; Zhang, Y.; Lu, H.; Wang, X.; Zhang, Y. Integrated Spatial Metabolomics and Transcriptomics Decipher the Hepatoprotection Mechanisms of Wedelolactone and Demethylwedelolactone on Non-alcoholic Fatty Liver Disease. J. Pharm. Anal. 2024, 14, 100910. [Google Scholar] [CrossRef]
- Sun, C.; Wang, A.; Zhou, Y.; Chen, P.; Wang, X.; Huang, J.; Gao, J.; Wang, X.; Shu, L.; Lu, J. Spatially resolved multi-omics highlights cell-specific metabolic remodeling and interactions in gastric cancer. Nat. Commun. 2023, 14, 2692. [Google Scholar] [CrossRef]
- Zang, Q.; Sun, C.; Chu, X.; Li, L.; Gan, W.; Zhao, Z.; Song, Y.; He, J.; Zhang, R.; Abliz, Z. Spatially resolved metabolomics combined with multicellular tumor spheroids to discover cancer tissue relevant metabolic signatures. Anal. Chim. Acta. 2021, 1155, 338342. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Jamil, Q.U.A.; Jaerapong, N.; Zehl, M.; Jarukamjorn, K.; Jäger, W. Metabolism of curcumin in human breast cancer cells: Impact of sulfation on cytotoxicity. Planta Med. 2017, 83, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Fhu, C.W.; Ali, A. Fatty acid synthase: An emerging target in cancer. Molecules 2020, 25, 3935. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Cheng, M.; Bhujwalla, Z.M.; Glunde, K. Targeting phospholipid metabolism in cancer. Front. Oncol. 2016, 6, 266. [Google Scholar] [CrossRef]
- Stoica, C.; Ferreira, A.K.; Hannan, K.; Bakovic, M. Bilayer forming phospholipids as targets for cancer therapy. Int. J. Mol. Sci. 2022, 23, 5266. [Google Scholar] [CrossRef]
- Holbert, C.E.; Cullen, M.T.; Casero, R.A., Jr.; Stewart, T.M. Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 2022, 22, 467–480. [Google Scholar] [CrossRef]
- Mandal, S.; Mandal, A.; Johansson, H.E.; Orjalo, A.V.; Park, M.H. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2169–2174. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Liu, Y. New insights into the roles and mechanisms of spermidine in aging and age-related diseases. Aging Dis. 2021, 12, 1948. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Z.; Zhang, Y.; Wang, L.; Geng, H.; Li, M.; Chen, S.; Wang, X.; Chen, P.; Sun, C.; Zhang, C. Spatial Metabolomics Profiling Reveals Curcumin Induces Metabolic Reprogramming in Three-Dimensional Tumor Spheroids. Metabolites 2024, 14, 482. https://doi.org/10.3390/metabo14090482
Zhu Z, Zhang Y, Wang L, Geng H, Li M, Chen S, Wang X, Chen P, Sun C, Zhang C. Spatial Metabolomics Profiling Reveals Curcumin Induces Metabolic Reprogramming in Three-Dimensional Tumor Spheroids. Metabolites. 2024; 14(9):482. https://doi.org/10.3390/metabo14090482
Chicago/Turabian StyleZhu, Zihan, Yaqi Zhang, Lei Wang, Haoyuan Geng, Min Li, Shiping Chen, Xiao Wang, Panpan Chen, Chenglong Sun, and Chao Zhang. 2024. "Spatial Metabolomics Profiling Reveals Curcumin Induces Metabolic Reprogramming in Three-Dimensional Tumor Spheroids" Metabolites 14, no. 9: 482. https://doi.org/10.3390/metabo14090482
APA StyleZhu, Z., Zhang, Y., Wang, L., Geng, H., Li, M., Chen, S., Wang, X., Chen, P., Sun, C., & Zhang, C. (2024). Spatial Metabolomics Profiling Reveals Curcumin Induces Metabolic Reprogramming in Three-Dimensional Tumor Spheroids. Metabolites, 14(9), 482. https://doi.org/10.3390/metabo14090482






