Disposition of Oral Nalbuphine and Its Metabolites in Healthy Subjects and Subjects with Hepatic Impairment: Preliminary Modeling Results Using a Continuous Intestinal Absorption Model with Enterohepatic Recirculation
Abstract
1. Introduction
2. Materials and Methods
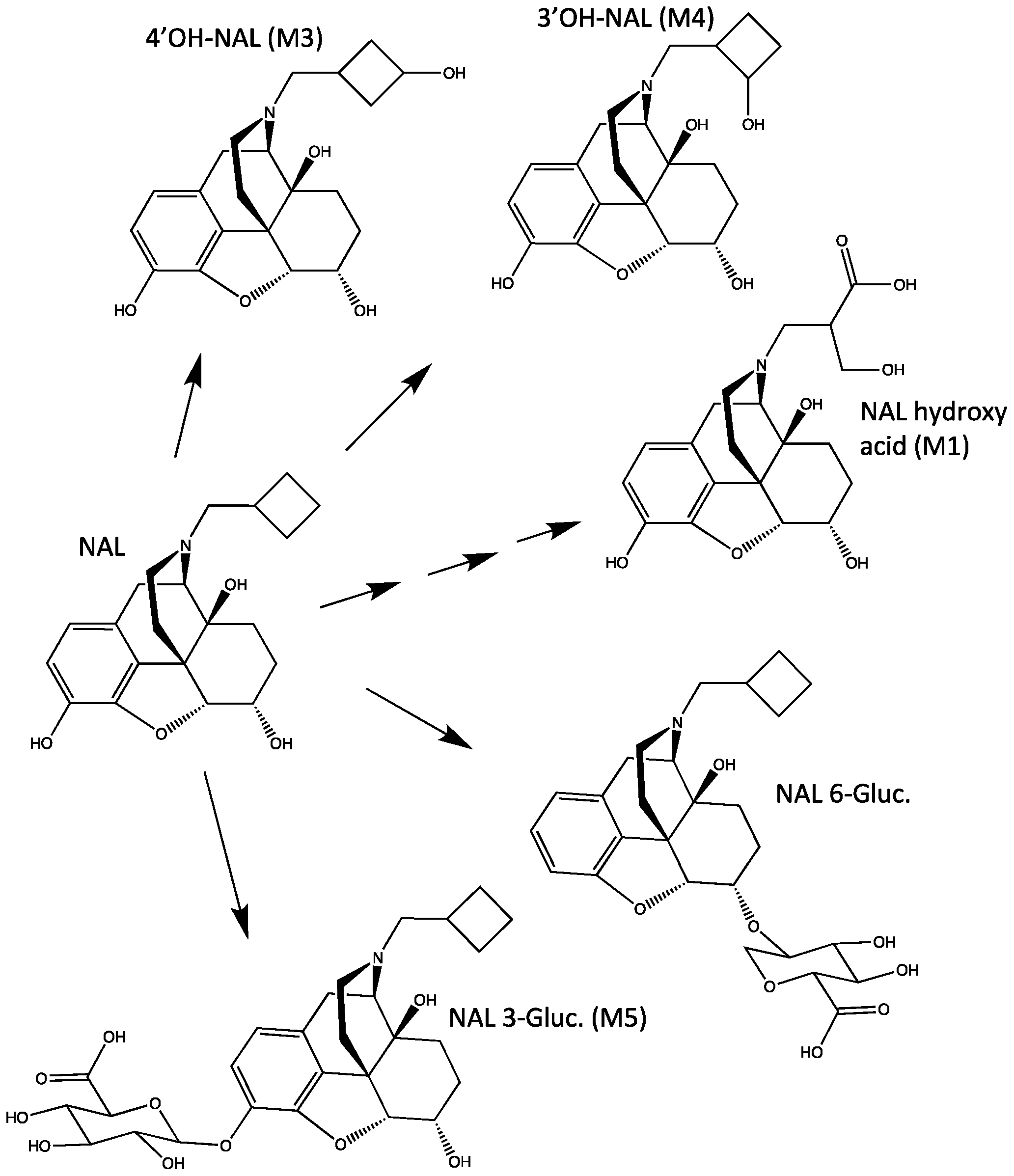
2.1. In Vitro Reaction Phenotyping Studies
2.2. Clinical Study Design
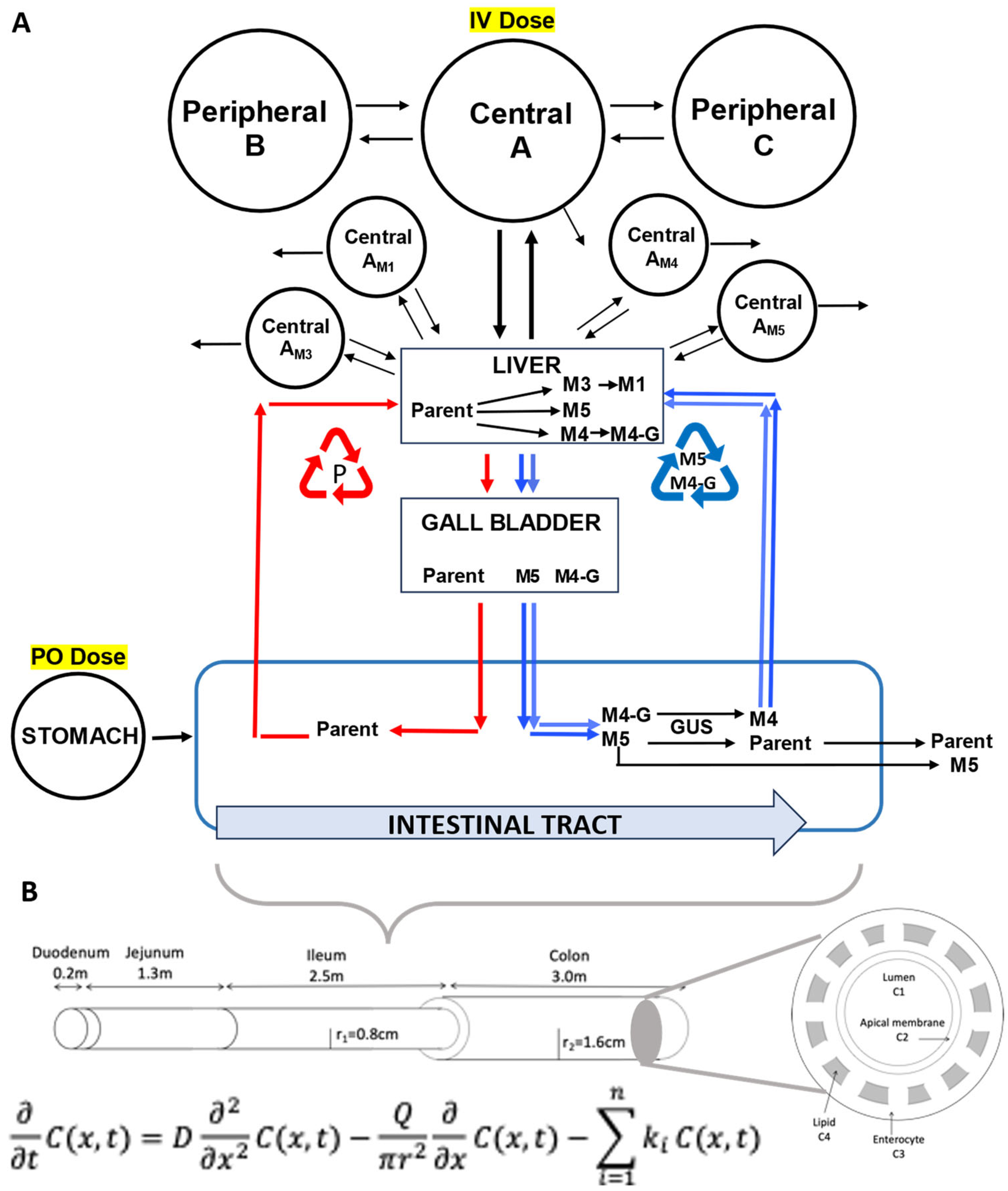
2.3. Modeling and Simulation
2.3.1. NAL Parent Drug Modeling
2.3.2. Metabolite M5 Modeling
2.3.3. Metabolite M3 Modeling
2.3.4. Metabolite M1 Modeling
2.3.5. Metabolite M4 Modeling
2.3.6. Other Parameters and Calculations
3. Results
3.1. In Vitro Reaction Phenotyping
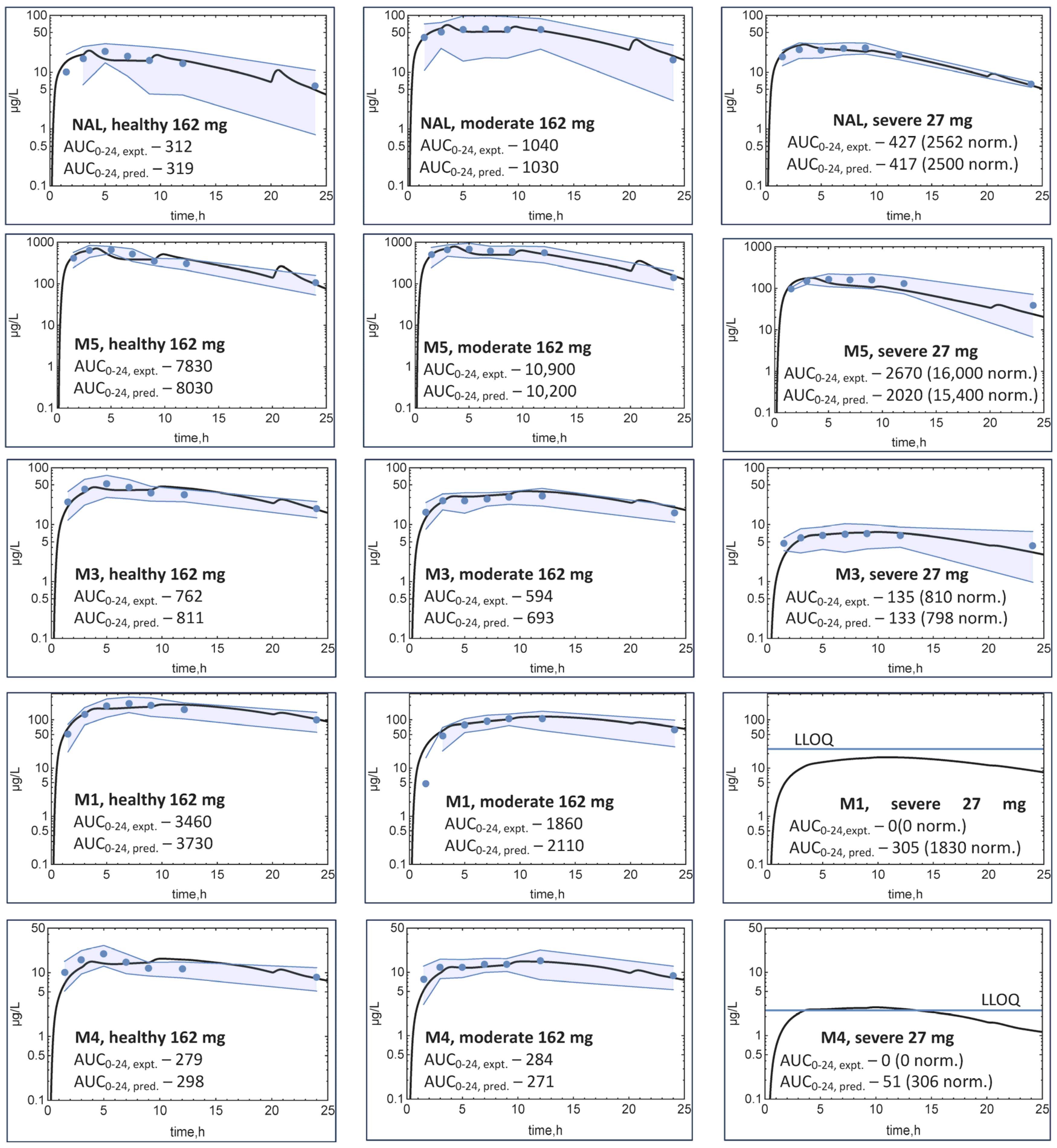
3.2. Clinical Results
3.3. Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maher, T.M.; Avram, C.; Bortey, E.; Hart, S.P.; Hirani, N.; Molyneux, P.L.; Porter, J.C.; Smith, J.A.; Sciascia, T. Nalbuphine Tablets for Cough in Patients with Idiopathic Pulmonary Fibrosis. NEJM Evid. 2023, 2, EVIDoa2300083. [Google Scholar] [CrossRef]
- Nubain Product Label. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/018024s042lbl.pdf (accessed on 24 July 2023).
- Hawi, A.; Alcorn, H., Jr.; Berg, J.; Hines, C.; Hait, H.; Sciascia, T. Pharmacokinetics of nalbuphine hydrochloride extended release tablets in hemodialysis patients with exploratory effect on pruritus. BMC Nephrol. 2015, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Aitkenhead, A.; Lin, E.; Achola, K. The pharmacokinetics of oral and intravenous nalbuphine in healthy volunteers. Br. J. Clin. Pharmacol. 1988, 25, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.W.; Schary, W.L.; Whitney, C.C., Jr. The disposition and bioavailability of intravenous and oral nalbuphine in healthy volunteers. J. Clin. Pharmacol. 1987, 27, 866–873. [Google Scholar] [CrossRef]
- Wang, H.-J.; Hsiong, C.-H.; Pao, L.-H.; Chang, W.-L.; Zhang, L.-J.; Lin, M.-J.; Ho, S.-T.; Huang, P.-W.; Hu, O.Y.-P. New finding of nalbuphine metabolites in men: NMR spectroscopy and UPLC–MS/MS spectrometry assays in a pilot human study. Metabolomics 2013, 10, 709–718. [Google Scholar] [CrossRef]
- Liang, R.J.; Shih, Y.N.; Chen, Y.L.; Liu, W.Y.; Yang, W.L.; Lee, S.Y.; Wang, H.J. A dual system platform for drug metabolism: Nalbuphine as a model compound. Eur. J. Pharm. Sci. 2020, 141, 105093. [Google Scholar] [CrossRef]
- Daly, A.K.; Rettie, A.E.; Fowler, D.M.; Miners, J.O. Pharmacogenomics of CYP2C9: Functional and Clinical Considerations. J. Pers. Med. 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.E.; Min, B.J.; Heo, N.; Lee, K.H.; Kim, J.H. Comprehensive in vitro and in silico assessments of metabolic capabilities of 24 genomic variants of CYP2C19 using two different substrates. Front. Pharmacol. 2023, 14, 1055991. [Google Scholar] [CrossRef]
- Thornquest, J.; Powell, K.D.; Heasley, B.; LeBlanc, D.; Sciascia, T.; Hawi, A. Isolation, Characterization, and Synthesis of N-(4-Hydroxy-3-Methylene-Butanoic Acid) Nornalbuphine, a Previously Unidentified Nalbuphine Metabolite. Available online: https://www.trevitherapeutics.com/wp-content/uploads/2024/07/Nalubuphine-MetID-AAPS-2015-Poster.pdf (accessed on 24 July 2024).
- Pharmacokinetics in Subjects with Impaired Hepatic Function: Study Design, Data Analysis, and Impact on Dosing and Labeling. Available online: https://www.fda.gov/media/71311/download (accessed on 24 July 2024).
- Korzekwa, K.; Nagar, S.; Clark, D.; Sciascia, T.; Hawi, A. A Continuous Intestinal Absorption Model to Predict Drug Enterohepatic Recirculation in Healthy Humans: Nalbuphine as a Model Substrate. Mol. Pharm. 2024. [Google Scholar] [CrossRef] [PubMed]
- Nagar, S.; Korzekwa, R.C.; Korzekwa, K. Continuous Intestinal Absorption Model Based on the Convection-Diffusion Equation. Mol. Pharm. 2017, 14, 3069–3086. [Google Scholar] [CrossRef]
- Korzekwa, K.; Radice, C.; Nagar, S. A permeability- and perfusion-based PBPK model for improved prediction of concentration-time profiles. Clin. Transl. Sci. 2022, 15, 2035–2052. [Google Scholar] [CrossRef]
- Radice, C.; Korzekwa, K.; Nagar, S. Predicting Impact of Food and Feeding Time on Oral Absorption of Drugs with a Novel Rat Continuous Intestinal Absorption Model. Drug Metab. Dispos. 2022, 50, 750–761. [Google Scholar] [CrossRef]
- Rohatagi, S.; Marbury, C.; Wyatt, D.J.; Sciascia, T. Safety and Pharmacokinetics of Nalbuphine Following Administration of Nalbuphine ER Tablets in Subjects with Impaired Hepatic Function. Available online: https://www.morressier.com/o/event/61080a2c4517da001280e092/article/6143051f87a68d83cb5d42be (accessed on 24 July 2024).
- Hinderling, P.H. Red blood cells: A neglected compartment in pharmacokinetics and pharmacodynamics. Pharmacol. Rev. 1997, 49, 279–295. [Google Scholar]
- Korzekwa, K.; Nagar, S. On the Nature of Physiologically-Based Pharmacokinetic Models -A Priori or A Posteriori? Mechanistic or Empirical? Pharm. Res. 2017, 34, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.; Ye, M.; Nagar, S.; Korzekwa, K. Prediction of tissue-plasma partition coefficients using microsomal partitioning: Incorporation into physiologically based pharmacokinetic models and steady-state volume of distribution predictions. Drug Metab. Dispos. 2019, 47, 1050–1060. [Google Scholar] [CrossRef]
- Korzekwa, K.; Nagar, S. Drug distribution part 2. Predicting volume of distribution from plasma protein binding and membrane partitioning. Pharm. Res. 2017, 34, 544–551. [Google Scholar] [CrossRef]
- El-Khateeb, E.; Achour, B.; Al-Majdoub, Z.M.; Barber, J.; Rostami-Hodjegan, A. Non-uniformity of Changes in Drug-Metabolizing Enzymes and Transporters in Liver Cirrhosis: Implications for Drug Dosage Adjustment. Mol. Pharm. 2021, 18, 3563–3577. [Google Scholar] [CrossRef] [PubMed]
- Drozdzik, M.; Lapczuk-Romanska, J.; Wenzel, C.; Skalski, L.; Szeląg-Pieniek, S.; Post, M.; Parus, A.; Syczewska, M.; Kurzawski, M.; Oswald, S. Protein Abundance of Drug Metabolizing Enzymes in Human Hepatitis C Livers. Int. J. Mol. Sci. 2023, 24, 4543. [Google Scholar] [CrossRef]
- Elmeliegy, M.; Yang, D.Z.; Salama, E.; Parivar, K.; Wang, D.D. Discordance Between Child-Pugh and National Cancer Institute Classifications for Hepatic Dysfunction: Implications on Dosing Recommendations for Oncology Compounds. J. Clin. Pharmacol. 2020, 61, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Lewinsky, R.H.; Gardner-Stephen, D.A.; Mackenzie, P.I. Regulation of UDP glucuronosyltransferases in the gastrointestinal tract. Toxicol. Appl. Pharmacol. 2004, 199, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Kawana, K.; Nakajin, S. Contribution of UDP-glucuronosyltransferase 1A1 and 1A8 to morphine-6-glucuronidation and its kinetic properties. Drug Metab. Dispos. 2008, 36, 688–694. [Google Scholar] [CrossRef]
- Li, X.; Bratton, S.; Radominska-Pandya, A. Human UGT1A8 and UGT1A10 mRNA are expressed in primary human Hepatocytes. Drug Metab. Pharmacokinet. 2007, 22, 152–161. [Google Scholar] [CrossRef]
- Takayama, K.; Ito, K.; Matsui, A.; Yamashita, T.; Kawakami, K.; Hirayama, D.; Kishimoto, W.; Nakase, H.; Mizuguchi, H. In Vivo Gene Expression Profile of Human Intestinal Epithelial Cells: From the Viewpoint of Drug Metabolism and Pharmacokinetics. Drug Metab. Dispos. 2021, 49, 221–232. [Google Scholar] [CrossRef]
- Ladumor, M.K.; Storelli, F.; Liang, X.; Lai, Y.; Enogieru, O.J.; Chothe, P.P.; Evers, R.; Unadkat, J.D. Predicting changes in the pharmacokinetics of CYP3A-metabolized drugs in hepatic impairment and insights into factors driving these changes. CPT Pharmacomet. Syst. Pharmacol. 2023, 12, 261–273. [Google Scholar] [CrossRef]
- Al-Qassabi, J.; Tan, S.P.F.; Phonboon, P.; Galetin, A.; Rostami-Hodjegan, A.; Scotcher, D. Facing the Facts of Altered Plasma Protein Binding: Do Current Models Correctly Predict Changes in Fraction Unbound in Special Populations? J. Pharm. Sci. 2024, 113, 1664–1673. [Google Scholar] [CrossRef]
- Jamei, M.; Marciniak, S.; Feng, K.; Barnett, A.; Tucker, G.; Rostami-Hodjegan, A. The Simcyp population-based ADME simulator. Expert. Opin. Drug Metab. Toxicol. 2009, 5, 211–223. [Google Scholar] [CrossRef]
- Gagnon, R.; Grogan, G.; Levitt, M.S.; Roberts, S.M.; Wan, P.W.H.; Willetts, A.J. Biological Baeyer-Villiger Oxidation of Some Monocyclic and Bicyclic Ketones Using Monooxygenases from Acinetobacter-Calcoaceticus Ncimb-9871 and Pseudomonas-Putida Ncimb-10007. J. Chem. Soc. Perkin Trans. 1994, 1, 2537–2543. [Google Scholar] [CrossRef]
- Swinney, D.C.; Mak, A.Y. Androgen formation by cytochrome P450 CYP17. Solvent isotope effect and pL studies suggest a role for protons in the regulation of oxene versus peroxide chemistry. Biochemistry 1994, 33, 2185–2190. [Google Scholar] [CrossRef]
- Vaz, A.D.N.; Kessell, K.J.; Coon, M.J. Aromatization of a Bicyclic Steroid Analog, 3-Oxodecalin-4-ene-10-carboxaldehyde, by Liver Microsomal Cytochrome P450 2B4. Biochemistry 2002, 33, 13651–13661. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, Y.; McCarty, K.D.; Martin, M.V.; Guengerich, F.P. Oxygen-18 Labeling Defines a Ferric Peroxide (Compound 0) Mechanism in the Oxidative Deformylation of Aldehydes by Cytochrome P450 2B4. ACS Catal. 2024, 14, 2388–2394. [Google Scholar] [CrossRef]
- Matsumoto, K.; Hasegawa, T.; Ohara, K.; Takei, C.; Kamei, T.; Koyanagi, J.; Takahashi, T.; Akimoto, M. A metabolic pathway for the prodrug nabumetone to the pharmacologically active metabolite, 6-methoxy-2-naphthylacetic acid (6-MNA) by non-cytochrome P450 enzymes. Xenobiotica 2020, 50, 783–792. [Google Scholar] [CrossRef]
- Partani, P.; Verma, S.M.; Gurule, S.; Khuroo, A.; Monif, T. Simultaneous quantitation of atorvastatin and its two active metabolites in human plasma by liquid chromatography/(–) electrospray tandem mass spectrometry. J. Pharm. Anal. 2014, 4, 26–36. [Google Scholar] [CrossRef][Green Version]
- Nagar, S.; Radice, C.; Tuohy, R.; Stevens, R.; Bennyhoff, D.; Korzekwa, K. The Rat Continuous Intestine Model Predicts the Impact of Particle Size and Transporters on the Oral Absorption of Glyburide. Mol. Pharm. 2023, 20, 219–231. [Google Scholar] [CrossRef]


| Test System | NAL Loss (%) | M1 (pmol) | M3 (pmol) | M4 (pmol) |
|---|---|---|---|---|
| Control Bactosomes® | No loss | ND 1 | ND | ND |
| Reductase Control | 3.3 | ND | ND | ND |
| CYP1A2 | 1.6 | ND | ND | ND |
| CYP2B6 + b5 | No loss | ND | ND | ND |
| CYP2C8 + b5 | 3.6 | ND | 16.6 | ND |
| CYP2C9 + b5 | 4.0 | ND | 305 | 51.0 |
| CYP2C19 + b5 | 27 | 1.81 2 | 1390 | 93.2 |
| CYP2D6 | No loss | ND | 5.95 | 31.5 |
| CYP3A4 + b5 | 10.3 | ND | 6.01 | ND |
| % Inhibition (SD 1) | ||||
|---|---|---|---|---|
| Targeted CYP | Inhibitor (μM) | NAL | M3 | M4 |
| CYP1A2 | Furafylline (10) | None | 14.6 (0.4) | 7.5 (0.4) |
| CYP2B6 | Phencyclidine (30) | None | None | 18.3 (0.4) |
| CYP2C8 | Gemfibrozil glucuronide (100) | None | 0.6 (0.03) | 20.3 (0.6) |
| CYP2C9 | Tienilic acid (20) | 34.6 (1.6) | 60.8 (4.9) | 84.1 (1.6) |
| CYP2C19 | Esomeprazole (10) | 28.7 (1.4) | 22.5 (1.7) | 24.6 (1.6) |
| CYP2D6 | Paroxetine (5) | None | 7.9 (1.6) | 22.9 (0.5) |
| CYP3A4/5 | Troleandomycin (50) | 17.0 (0.5) | 4.2 (ND 2) | 19.0 (1.2) |
| Test System | NAL Loss (%) | M5 (pmol) |
|---|---|---|
| Control Supersomes | 3.9 | ND 1 |
| UGT1A1 | 3.4 | ND |
| UGT1A3 | 22.5 | 162 |
| UGT1A4 | 3.7 | ND |
| UGT1A6 | 2.9 | ND |
| UGT1A8 | 69.9 | 621 |
| UGT1A9 | 15.0 | 84.8 |
| UGT2B4 | 2.4 | ND |
| UGT2B7 | 26.0 | 169 |
| UGT2B15 | 1.7 | ND |
| Parameter (Units) | Healthy | Moderate | Severe |
|---|---|---|---|
| CLgi,p = Clgi,m | 0.25 CLint,H | 0.25 CLint,H | 0.1 CLint,H |
| Observed CL/F (L/h) | 465 | 142 | 58 |
| VD (L) simulated | 267 | 267 | 267 |
| Initial factor “a” for change in CL and F 1 | NA 2 | 1.8 | 2.5 |
| Optimized “a” | NA 2 | 1.4 | 1.7 |
| CLH (L/h) | 78.8 | 51.5 | 32.4 |
| CLnH (L/h) | 4.14 | 4.14 | 4.14 |
| CLint,H(L/h) | 322 | 92 | 42 |
| Fraction of NAL CLint forming M3 | 0.5 | 0.35 | 0.3 |
| Fraction of NAL CLint forming M4 | 0.1 | 0.1 | 0.1 |
| Fraction of NAL CLint forming M5 | 0.4 | 0.55 | 0.6 |
| Formation CLint for M1 (L/h) | 60 | 40 | 30 |
| Formation CLint for M3 (L/h) | 161 | 32.3 | 12.5 |
| Formation CLint for M4 (L/h) | 32.2 | 9.2 | 4.2 |
| Formation CLint for M4-G (L/h) | 5 | 5 | 5 |
| Formation CLint for M5 (L/h) | 129 | 51 | 25 |
| M1 VD (L) | 53 | 53 | 53 |
| M3 VD (L) | 214 | 214 | 214 |
| M4 VD (L) | 214 | 214 | 214 |
| M5 VD (L) | 15 | 15 | 15 |
| Elimination CL for M1 (L/h) | 20 | 20 | 20 |
| Elimination CL for M3 (L/h) | 4 | 4 | 4 |
| Elimination CL for M4 (L/h) | 40 | 40 | 40 |
| Elimination CL for M5 (L/h) | 17 | 17 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagar, S.; Hawi, A.; Sciascia, T.; Korzekwa, K. Disposition of Oral Nalbuphine and Its Metabolites in Healthy Subjects and Subjects with Hepatic Impairment: Preliminary Modeling Results Using a Continuous Intestinal Absorption Model with Enterohepatic Recirculation. Metabolites 2024, 14, 471. https://doi.org/10.3390/metabo14090471
Nagar S, Hawi A, Sciascia T, Korzekwa K. Disposition of Oral Nalbuphine and Its Metabolites in Healthy Subjects and Subjects with Hepatic Impairment: Preliminary Modeling Results Using a Continuous Intestinal Absorption Model with Enterohepatic Recirculation. Metabolites. 2024; 14(9):471. https://doi.org/10.3390/metabo14090471
Chicago/Turabian StyleNagar, Swati, Amale Hawi, Thomas Sciascia, and Ken Korzekwa. 2024. "Disposition of Oral Nalbuphine and Its Metabolites in Healthy Subjects and Subjects with Hepatic Impairment: Preliminary Modeling Results Using a Continuous Intestinal Absorption Model with Enterohepatic Recirculation" Metabolites 14, no. 9: 471. https://doi.org/10.3390/metabo14090471
APA StyleNagar, S., Hawi, A., Sciascia, T., & Korzekwa, K. (2024). Disposition of Oral Nalbuphine and Its Metabolites in Healthy Subjects and Subjects with Hepatic Impairment: Preliminary Modeling Results Using a Continuous Intestinal Absorption Model with Enterohepatic Recirculation. Metabolites, 14(9), 471. https://doi.org/10.3390/metabo14090471







