Abstract
The grey tree frog, Dryophytes versicolor, survives whole-body freezing for weeks during cold winter months. Survival in a state devoid of available food, water, or oxygen forces a reliance on metabolic rate depression (MRD) and the reprioritization of bodily functions. This study utilizes next-generation sequencing (NGS) and bioinformatic analyses to characterize changes in the microRNAome of D. versicolor. When comparing control to frozen groups, five microRNAs (miRNA) were found to be differentially regulated (miR-143-3p, miR-30e-3p, miR-10a-5p, miR-140-3p, and miR-148a-3p), suggesting that they play key roles in freeze survival. The KEGG and GO analyses of these changes predicted a significant negative enrichment of terms associated with cell proliferation and active metabolism while simultaneously predicting the upregulation of cell signalling terms. These results suggest a fast-acting regulatory role for miRNA in contributing to the reorganization of gene expression and the limitation of energy-expensive processes during MRD in the hind leg skeletal muscle of the frog.
1. Introduction
Under extreme cold temperatures, poikilothermic animals are often required to employ one of two strategies to endure the subzero temperatures of winter. Some species use freeze avoidance, employing antifreeze proteins and cryoprotectants to enter a supercooled state [1]. Others employ freeze tolerance that allows ice formation in extracellular or extra-organ fluid spaces while protecting the internal organs from freezing. The grey tree frog, Dryophytes versicolor (formerly Hyla versicolor), is one of the few vertebrate species that have evolved freeze tolerance, surviving the conversion of up to about 60% of their total body water into extracellular ice. Notably, these frogs produce glycerol as a cryoprotectant that protects their intracellular environment from freezing while overwintering [2,3]. Although dissimilar to other well-studied frog species such as the wood frog, Rana sylvatica, that uses glucose as a cryoprotectant, glycerol is the cryoprotectant of choice for most invertebrates, some species of fish, and D. versicolor. Ice accumulating in extracellular spaces leads to ischemia and cessation of the heartbeat. In the frozen state, gas exchange and waste export from vital organs (lungs, kidneys, etc.) is also curtailed, leading to a reliance on intracellular fuel reserves and anaerobic metabolism [4,5]. The endogenous fuel reserves in the cells are often too little to sustain energy demand and, hence, extended periods of metabolic rate depression (MRD) are required for survival. Complex regulatory mechanisms elicit the MRD response, including miRNA-mediated gene suppression [6].
Studies in recent years have shown the importance of microRNA (miRNA) in the development, function, and regulation of cell functions and that these small molecules are involved in the regulation of numerous processes, including cell proliferation, differentiation, apoptosis, and angiogenesis. MicroRNAs (miRNAs) are small non-coding RNA molecules, typically 20–25 nucleotides in length, that play crucial roles in regulating gene expression in eukaryotic cells [7]. These small molecules are highly conserved throughout evolutionary history, and many miRNA families have been identified that are shared between distantly related species, indicating their important role in biological processes. The primary function of miRNAs is to regulate the expression of protein-coding gene transcripts by binding to target mRNA molecules, leading to their degradation or translational repression. This process is achieved through complementary base pairing between miRNA and mRNA targets, which results in the suppression of gene expression. Each miRNA type can regulate multiple target mRNAs, and a single mRNA type can also be targeted by multiple different miRNAs, allowing for the complex regulation of gene expression. Because miRNAs play a crucial role in the regulation of gene expression, they have emerged as important tools for understanding biological pathways and networks. By analyzing the miRNA expression profile of cells or tissues, it is possible to infer controls over metabolic pathways and identify potential biomarkers for various diseases [8,9,10]. Additionally, the analysis of miRNA–target interactions can reveal important information about the regulation of specific genes and the signalling pathways that they are involved in.
Since miRNAs have such widespread use in the regulation of pathways under normal conditions, there is a strong implication that they would also have key roles in navigating extreme stress situations. The grey tree frog (Dryophytes versicolor) can survive extended periods of time without food or normal levels of oxygen, and enduring subzero temperatures in a frozen state. Hence, it is an ideal organism to highlight the changes required to survive such ordeals. Previous studies of miRNA from D. versicolor highlighted changes in miRNA expression in the liver and miRNA action in downregulating energy-expensive pathways [11]. Since the liver is an internal organ with vital functions that support survival, it is likely that the liver also utilizes different survival mechanisms and pathways than does muscle tissue. As muscle is a superficial tissue that experiences no notable function when frozen for extended periods of time, its complete return to normal function upon thawing is even more profound. To build a better picture of how these frogs can survive for extended periods of time while frozen, and to understand the implications this could have on current biotechnology, an analysis of differentially expressed miRNAs is an important research target. Utilizing small RNA-seq and a comprehensive bioinformatics strategy, with elements of systems biology, the present study aimed to characterize the metabolic significance of differential miRNA expression in the muscle of frozen grey tree frogs in comparison to active controls. By integrating data gleaned through in silico analysis, this approach enables a discussion of a wide range of pathways and gauging overall trends. Identifying the regulatory changes within muscle required to transition to a hypometabolic, pro-survival frozen state could provide insight into the most relevant adaptations to extreme environmental stresses. This broad analysis of differential miRNA is designed to help future studies better target the downstream processes of importance to stress survival.
2. Materials and Methods
2.1. Animal Treatments and Tissues
Adult male grey tree frogs, D. versicolor, weighing 6–11 g (mean = 7.883, sd = 1.197) were collected from breeding ponds in the Ottawa area (Ottawa, ON, Canada) as previously described [12,13]. The frogs were acclimated to temperatures of 5 °C for 2 weeks in plastic containers lined with damp sphagnum moss and without feeding. Control frogs were randomly selected from this group. Other groups of 4–5 frogs were placed in trays lined with damp paper towels and placed in an adjustable temperature incubator, first at −4 °C for 45 min to trigger ice nucleation when the frogs cooled below 0 °C. Subsequently, the temperature was raised to −2.5 °C and a 24 h freeze exposure was initiated. The euthanasia of both control and frozen frogs was performed via double-pithing, followed by the prompt dissection of selected tissues, including hind leg skeletal muscle. Tissues were flash-frozen in liquid nitrogen and stored at −80 °C until use. Animal care, experimentation, and euthanasia procedures adhered to the guidelines of the Canadian Council on Animal Care and had the prior approval of the Carleton University Animal Care Committee (protocol no. 13683).
2.2. Total RNA Extraction
Total RNA was isolated from the leg skeletal muscle of control (n = 4) and frozen (n = 3) frogs as previously described [14,15]. Approximately 50 mg of frozen muscle was crushed by a mortar and pestle under liquid nitrogen and then homogenized in 1 mL of TRIZOL reagent (Invitrogen; Cat. # 15596-018) using a Polytron PT1200 homogenizer (Kinematica, Werkstrasse, Switzerland). A 200 μL aliquot of chloroform was then added to each sample followed by centrifugation at 10,000× g at 4 °C for 15 min. Total RNA settled in the upper aqueous phase and was transferred to a new tube, and then the RNA was precipitated by adding isopropyl alcohol (500 μL) and incubating at room temperature for 10 min. Samples were then centrifuged at 10,000× g for 15 min at 4 °C followed by removal of the supernatant. RNA pellets were then washed twice with 70% ethanol and air-dried for 10 min. Next, RNA pellets were resuspended in 50 μL of RNAse-free water and then assessed for RNA concentration and purity using a Take3 micro-volume quantification plate and a PowerWave HT spectrophotometer. Following determination that the 260/280 nm ratio indicated RNA > 1.8, an aliquot of RNA sample was confirmed for integrity on a 1% agarose gel and stained with SYBR Green. The final RNA samples were then isolated and standardized to a concentration of 1 µg/µL followed by freezing and storage at −80 °C until use.
2.3. Small RNA Sequencing
RNA samples from the muscle of both control and 24 h frozen D. versicolor were sequenced by the Quebec Centre de Recherche (Laval, QC, Canada). Prior to microRNA library construction, RNA quality was assessed using a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA) and an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Subsequently, small RNA cDNA libraries were constructed, validated with the Bioanalyzer, and sequenced using an Illumina NovaSeq 6000 platform to produce single-end 100 base reads. All sequencing data are available on the Sequence Read Archive (SRA; BioProject ID: PRJNA1078871).
2.4. Data Processing
Raw read data were processed as outlined in previous papers using the RBioMIR pipeline [16]. CutAdapt was used to trim the adapters from all reads followed by the Phred quality scores being assessed by FastQC, which was used to validate adapter removal and remove reads with scores below 30 using fastq-mcf. A negative reference file compiled from the Rfam and piRNABank databases was used to filter out non-miRNA small RNA species (e.g., rRNA, tRNA, snRNA, snoRNA, and piRNA) and Bowtie was used to remove reads aligned to the negative reference file. The resulting miRNA reads were aligned to a positive reference file of mature miRNA sequences from miRbase using Bowtie. A seed sequence length parameter of 20 nucleotides was set with only perfect sequence matches being reported. Mature miRNA read counts with more than four reads were sorted and determined using SAMtools software and Unix command line tools for filtered reads. Finally, all filtered read counts were standardized using the Voom method as previously described [17,18].
2.5. Differential Expression Analysis
The differential expression of miRNA data from the control and frozen samples was analyzed using the linear models for microarray data (limma) R package with linear-model-fitting empirical Bayesian testing [19]. Differentially expressed miRNAs were considered significant if the false discovery rate (FDR)-adjusted p < 0.05 and a fold change was as follows:
|FC| ≥ 1.5
2.6. Gene Set Analysis
Gene set analysis was carried out using the RBiomirGS R package [16], implementing logistic regression-based analysis for significantly enriched gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [20]. Fully conserved human miRNA orthologs were used as substitutes for D. versicolor due to the lack of a sequenced genome for this species. RBiomirGS was used to calculate a miRNA score (SmiRNA) for each miRNA using the formula:
where p is the FDR-adjusted p-value and FC is the fold-change of the miRNA between the control and frozen frogs from the differential expression analysis. Additionally, a mRNA score (SmRNA) for each mRNA target was computed as:
where n is the number of miRNAs targeting a given mRNA, and is the SmiRNA of the ith targeting miRNA. Enriched gene sets with FDR-adjusted p-values ≥ 0.05 and corresponding model coefficients for each GO term and KEGG pathway were determined by RBiomirGS using the calculated SmRNA of mRNAs. Positive model coefficients indicated decreased negative regulation by microRNA, whereas negative coefficients indicated increased negative regulation by microRNA in the frozen group compared to the control group. The estimated model coefficients and standard error were calculated using logistic regression to determine statistical significance [16].
SmiRNA = −log10 p × signum(log2 FC)
3. Results
3.1. Analysis of Differentially Expressed miRNAs
From the total group of forty-nine mature miRNAs evaluated, five miRNAs were identified as differentially expressed (p < 0.05 and fold change >1.5) in the hind leg skeletal muscle of frozen D. versicolor as compared with control frogs. Of these, three miRNAs were downregulated whereas a significant upregulation of two miRNAs was observed in samples from frozen frogs. A volcano plot was used to visualize the differential expression of miRNAs from the muscle tissue of control and frozen frogs (Figure 1). The downregulated miRNAs were miR-143-3p, miR-30e-3p, and miR-10a-5p. The upregulated miRNAs were miR-140-3p and miR-148a-3p.
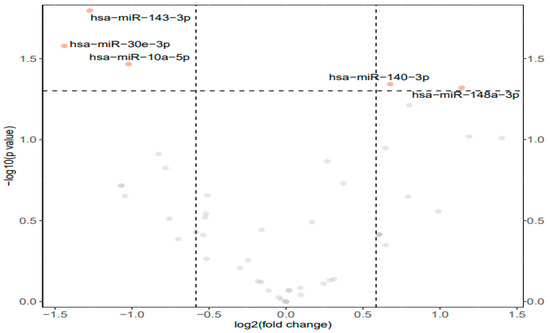
Figure 1.
Differential expression analysis of conserved miRNAs in the muscle tissue of D. versicolor. Volcano plot of the differential expression of conserved miRNAs showing −log10(p value) versus the log2(fold change) for individual miRNAs in muscle from frozen frogs versus the control group. Points in orange represent miRNAs that are significantly different from the control (>1.5-fold change and p < 0.01), grey points represent non-significant miRNAs.
3.2. Gene Set Analysis
Gene enrichment analyses of GO terms for molecular functions (GO MF) identified 33 significantly affected terms in the leg skeletal muscle of frozen frogs: 16 negative model and 17 positive model coefficients (Figure 2).
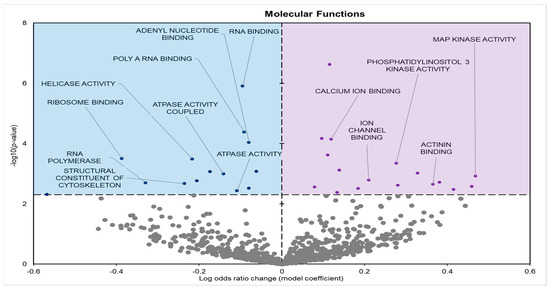
Figure 2.
Volcano plot demonstrating the enrichment of GO molecular function (GO MF) through a logistic regression-based gene set analysis of the mRNAs predicted to interact with differentially expressed (DE) miRNAs in frog muscle during freezing. Enriched terms are considered significantly altered if the FDR adjusted p-value is <0.05 and fold change >1.5. Negative model coefficients depicting decreased expression are shown as blue circles in the blue-shaded box (left), whereas coefficients depicting increased expression are shown by purple dots in the purple-shaded box (right).
Biological processes (GO BP) showed 141 significantly enriched terms when comparing controls to frozen groups; 69 had negative model coefficients whereas 72 had positive coefficients (Figure 3).
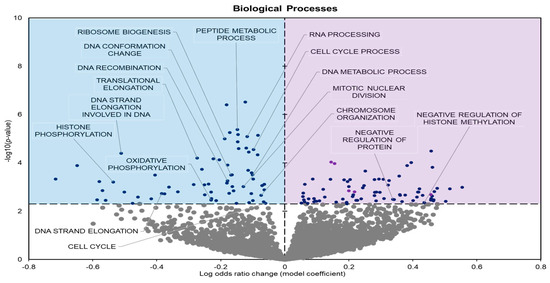
Figure 3.
Volcano plot demonstrating the enrichment of GO biological processes (GO BP) through a logistic regression-based gene set analysis of the mRNAs predicted to interact with DE miRNAs in frog muscle during freezing. All other information as in Figure 2.
The cellular components showed 34 significantly enriched terms with 29 negative model coefficients and 5 positive model coefficients (Figure 4).
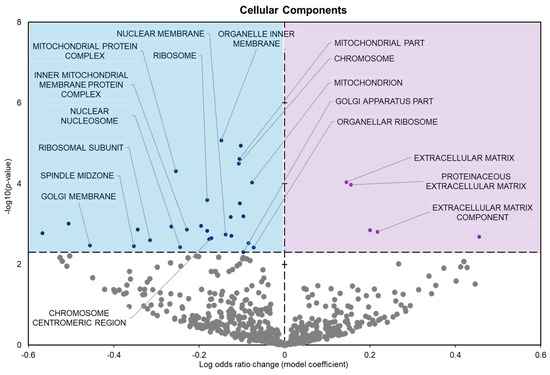
Figure 4.
Volcano plot demonstrating the enrichment of GO cellular components (GO CC) through a logistic regression-based gene set analysis of the mRNAs predicted to interact with DE miRNAs in frog muscle during freezing. All other information as in Figure 2.
KEGG enrichment analysis found five statistically enriched pathways in samples from frozen frogs; three with positive model coefficients and two with negative model coefficients (Figure 5). GO and KEGG term enrichment was predominantly focused on processes related to stimulus detection, metabolism, cell cycle, and ribosomal regulation.
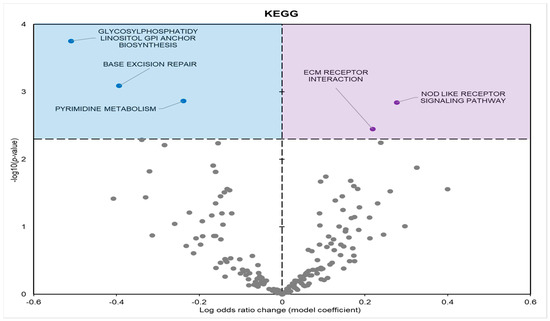
Figure 5.
Volcano plot demonstrating the enrichment of KEGG pathways through a logistic regression-based gene set analysis of the mRNAs predicted to interact with DE miRNAs in frog muscle during freezing. The KEGG pathway is considered significantly altered if the FDR adjusted p-value < 0.05. All other information as in Figure 2.
4. Discussion
An examination of the response to freezing in D. versicolor muscle tissue identified 49 miRNAs, with 5 of these displaying unique expression patterns under freezing stress (Figure 1). An increase in miRNA expression highlights the negative regulation of targeted pathways whereas a decrease in miRNA expression indicates a reduction/lack of inhibition on targets. It is important to note that this does not directly imply an increase in gene/protein expression, but just that a given pathway is not increasingly inhibited by miRNA expression. In this study, the upregulated miRNAs included miR-140-3p and miR148a-3p whereas the downregulated miRNAs included miR-10a-5p, miR-30e-3p, and miR-143-3p. As the downstream effects of miRNAs are the most important, the potential effects of these miRNAs were assessed via gene set analyses which covered GO terms and KEGG pathways. It was found that the miRNAs with significantly altered expression were largely linked to cell cycle regulation, gene expression, ribosomal regulation, histone modifications, and metabolism (Table A1). Since the miRNAs found to be over-expressed during freezing do not exclusively depict a strong inhibitory effect on these pathways, the elevated miRNA levels likely contribute to an overall regulation of pathways rather than just a single inhibitory one. As this is a novel study on miRNA expression in D. versicolor leg muscle, there are no comparisons that can be made to previous findings.
During freezing, terms heavily associated with the cell cycle and active cell division were significantly downregulated in D. versicolor muscle. The GO MF terms facing downregulation included helicase activity and cytoskeleton constituents, vital functions needed for DNA replication and cell proliferation (Figure 2) [21]. GO BP predicted significant reductions in processes including the cell cycle, DNA strand elongation, DNA recombination, and mitotic nuclear division among other related terms (Figure 3). GO CC also predicted significant reductions in terms related to the cell cycle with negative coefficients on cellular components such as the spindle midzone, chromosome centromeric region, and chromosome (Figure 4). GPI–anchor biosynthesis was one of the few KEGG terms that were significantly negatively enriched. However, its relevance to dynamic changes in cell proliferation through signal transduction is not dissimilar from the effects of other negatively enriched GO terms (Figure 5) [22]. These pathways and processes were predicted to be strongly suppressed due to miRNA activity and suggest a decrease in cell division and cell proliferation in the frozen state.
Two main reasons for processes related the cell cycle being downregulated in the frozen state can be postulated: (a) preventing disease during prolonged winter freezing, and (b) conserving energy. It is well documented that under extreme stress conditions, organisms conserve as much energy as possible via metabolic rate depression (MRD) [23,24]. In D. versicolor, and animals that undergo comparable stresses like Rana sylvatica, a steady decrease in ATP production often warrants an equally significant suppression of ATP-expensive processes. This prioritizes the use of low energy stores in crucial pro-survival pathways. As cell division has little to contribute to freezing survival and can also strip vital resources from cells already reliant on low fuel/energy stores, the process of downregulation is an effective means of maintaining MRD [25]. Previous studies corroborate evidence of cell cycle arrest/suppression under stress conditions that similarly expose cells to nutrient, oxygen, and/or hydration limitations [26,27,28]. By limiting cell division via reductions in cell cycle-promoting pathways and processes, a two-pronged result of saving energy and avoiding a diseased state while frozen can more easily be achieved.
The expression of miRNAs in D. versicolor muscle during freezing was largely associated with gene transcription and translation terms. GO analyses consistently revealed the downregulation of terms associated with active transcription and translation. GO MF analyses predicted the downregulation of RNA polymerase, poly-A RNA binding, RNA-binding functions, and ribosome binding, terms strongly associated with gene expression (Figure 2) [29,30,31]. GO BP terms relevant to gene expression were also strongly negatively affected, including ribosome biogenesis and protein maturation (Figure 3). Related GO cell cycle terms that were downregulated included components of the ribosome and ribosome subunit (Figure 4).
Many terms were found to be associated with the downregulation of gene transcription and protein translation. RNA polymerases are enzymes required for gene transcription and a decrease in their function would suggest a limitation on actively transcribed genes. Similarly, RNA binding plays major roles in transcription factor (TF) function, with its downregulation implying an added regulatory form of negative control on active transcription by miRNAs [30]. Viewing gene expression from the scope of translation shows a similar scenario. GO terms related to ribosomes, which are largely responsible for protein synthesis, were negatively enriched across biological processes and cellular components. Given the energetic cost of both ribosome biosynthesis and protein synthesis, the negative regulation of these processes is an effective means of conserving energy while in a state of hypometabolism in response to stress [5,24,32,33]. The predicted downregulation of terms associated with active gene expression support findings that miRNA may play important roles in regulating the MRD response in freezing.
Interestingly, studies of diverse tissues in similar organisms found dissimilar results. Studies of miRNA expression in Scaphiopus couchii and Xenopus laevis heart in response to a significant loss of body water under dry conditions (these species undergo summer estivation) revealed the upregulation of both protein translation and transcription terms [14,15]. Since the heart is a core organ that is still active and vital to survival across a variety of stresses, a more superficial tissue like muscle understandably has a more complete downregulation of energy-expensive processes. These notable differences in expression profile increase evidence of the tissue-specific regulation of gene expression during MRD, in this case caused by miRNA action.
GO BP predicted terms associated with histones, namely histone methylation and phosphorylation, to be negatively enriched during freezing (Figure 3). Post-translational modifications (PTMs) of histones are regulatory mechanisms that can control gene expression. At a transcriptional level, the modification of specific residues influences chromatin structure. This can play an important role in metabolic control and cell survival when facing environmental stresses.
Histone phosphorylation is often associated with the relaxation of chromatin via various means. The phosphorylation of H3S10ph influences H3 acetylation, a transcription activator. H3S28ph in combination with H3K27ac plays roles in gene activation [34]. The phosphorylation of T41 on H3 has also been stated to lead to transcriptional activation via inhibiting chromatin binding to heterochromatin protein 1 α (HP1α). Adenosine monophosphate-activated protein kinase (AMPK) also activates transcription through association with H2Bph at S36 [35,36]. Although histone phosphorylation on four residues of H3 (T3, S10, T11, S28) is associated with chromatin compaction, its effects are largely involved in chromatin relaxation alongside its regulation of overall gene expression.
The functional outcome of histone modifications often depends on the residue being modified. In the case of methylation, the number of methyl groups attached (mono-, di-, tri-methylated) is also key to the modification on gene expression. H3K4me3 is associated with active transcription and H3K4me1 is associated with an enhancer function, whereas H3K27me3 is linked to a repressed chromatin state [37]. The predicted negative regulation imposed by miRNA on histone modifications is complex, as histones play a large role in gene regulation as well. Although an overarching conclusion of decreased gene expression can be drawn from the present predictive results, possibilities such as the negative regulation of modifications that downregulate gene expression require further in-depth study.
A crucial target of miRNA regulation is cell signalling pathways. Predicted upregulated GO MF terms included phosphatidylinositol kinase (PI3K) and mitogen-activated protein kinase (MAPK) activities (Figure 2). Positive enrichment of the KEGG term “Nod-like receptor (NLR) signalling pathway” is also of importance, since the stimulation of the NLR signalling pathway has been found to activate MAPKs and other signal-transducing enzymes [38,39].
PI3K and MAPK coordinate extracellular cues to cells, leading to a large variety of downstream effects, and play an important role in cell fate [40,41,42]. As crucial elements in signal transduction pathways, these two kinases play major roles in controlling cell survival, division, and metabolism [43]. These signalling pathways can also have varying effects depending on the stimulus. For example, depending on the stimulus and the cell type under stress, MAPK pathways can be either pro-survival or pro-apoptotic [44].
Past studies have made it clear that there are countless stress factors associated with freezing survival that can induce apoptosis in cryopreserved cells, including vascular damage by ice formation, energy limitations, and osmotic imbalance. The involvement of apoptosis in cryopreservation failure has also been widely reported [45]. It is plausible that a lack of miRNA inhibition on MAPK and PI3K function during freezing is an important means by which to promote cell survival. One study found that the activation of a MAPK subfamily, p38MAPK, was protective against oxidative stress-induced apoptosis, which is experienced in a cryoprotective state [46]. JNK (a member of the MAPK subfamily) can also exhibit anti-apoptotic functions via the protein phosphorylation of Bcl-2-associated death promoter (BAD) and preventing BAD-induced apoptosis [44]. PI3K activation is linked to the phosphorylation of AKT and is highly conserved [47]. The effect of AKT is based largely on its ability to phosphorylate transcription factors that promote cell survival genes, such as nuclear factor κB (NFκB), and its negative regulation of transcription factors that promote cell death genes (e.g., forkhead box proteins) [47]. Like the JNK/MAPK subfamily, AKT mediated phosphorylation of BAD also inhibits its apoptotic effects [48]. NLR signalling pathways, aside from the activation of MAPK, similarly activate downstream molecules that could be involved in promoting cell survival, such as NFκB [49].
Despite the overarching downregulation of processes in D. versicolor muscle, the lack of inhibition on cell signalling pathways implies active cell regulation in frozen muscle tissue cells. The suppression of apoptosis via cell signalling pathways is an effective means of aiding freezing survival and maintaining cell viability over long periods of time. This could be especially important in hind leg muscle tissue as it is largely inactive throughout freezing episodes and susceptible to dysfunction when thawed. This study thereby highlights the genes in regulatory pathways prominently affected by miRNA for future targeted downstream analyses.
5. Conclusions
The results herein demonstrate a subset of five miRNA molecules (miR-140-3p, miR148a-3p, miR-10a-5p, miR-30e-3p, and miR-143-3p) that are predicted to be the most important to the freeze response in skeletal muscle. Bioinformatic analysis predicted a significant downregulation of pathways associated with the cell cycle, active protein translation, and targets early in gene expression pathways. The recurring theme of miRNA suppression of energy-expensive processes is likely an effective means of conserving vital fuel/energy stores until a return to normal muscle function is permitted by environmental conditions. Pathways associated with cell signalling and stimulus detection were predicted to be upregulated even during a frozen state in muscle tissue. The lack of miRNA inhibition of the MAPK and PI3K cell signalling processes highlights the role of cell fate determination in freezing, with a focus on anti-apoptosis to prolong cell survival. An overall downregulation of histone modifications by miRNA interference warrants further study given the complexity of the current findings. This study was able to identify key miRNAs and their targets that are involved in muscle tissue responses to aid freeze survival. These findings highlight new avenues for the freeze tolerance studies of tissues from varying stress-tolerant organisms and indicate the importance of miRNA on global MRD during the freezing of freeze-tolerant species.
Author Contributions
Conceptualization, S.R. and K.B.S.; methodology, K.B.S.; software, K.B.S.; validation, S.R.; formal analysis, S.R.; investigation, S.R.; resources, K.B.S.; data curation, S.R.; writing—original draft preparation, S.R.; writing—review and editing, S.R. and K.B.S.; visualization, S.R.; supervision, K.B.S.; project administration, S.R.; funding acquisition, K.B.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Sciences and Engineering Council (NSERC) of Canada, grant number RGPIN-2020-04733.
Institutional Review Board Statement
All animal care, experimentation, and euthanasia procedures were previously approved by the Carleton University Animal Care Committee (animal protocol no. 13683) in accordance with the guidelines of the Canadian Council on Animal Care.
Data Availability Statement
All sequencing data are available on Sequence Read Archive (SRA; BioProject ID: PRJNA1078871). The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank J. M. Storey for editorial review of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Significantly enriched GO term statistical values.
Table A1.
Significantly enriched GO term statistical values.
| Category | GO Term | Coefficient | p-Value |
|---|---|---|---|
| Molecular Functions | Helicase activity | −0.2165 | 0.000327 |
| RNA polymerase | −0.32913 | 0.00201 | |
| MAPK activity | 0.468236 | 0.00120 | |
| Biological Processes | Ribosome biogenesis | −0.18786 | 0.0000102 |
| Cell cycle | −0.05995 | 0.004741 | |
| DNA recombination | −0.17865 | 0.000559 | |
| Cellular Components | Chromosome | −0.10697 | 0.0000320 |
| Ribosome | −0.18159 | 0.000256 | |
| Spindle midzone | −0.4559 | 0.00355 |
References
- Block, W. To Freeze or Not to Freeze? Invertebrate Survival of Sub-Zero Temperatures. Funct. Ecol. 1991, 5, 284. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Molecular Physiology of Freeze Tolerance in Vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Molecular Biology of Freezing Tolerance. Compr. Physiol. 2013, 3, 1283–1308. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Freeze Tolerance: Constraining Forces, Adaptive Mechanisms. Can. J. Zool. 1988, 66, 1122–1127. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic Rate Depression and Biochemical Adaptation in Anaerobiosis, Hibernation and Estivation. Q. Rev. Biol. 1990, 65, 145–174. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Storey, K.B. The Emerging Roles of MicroRNAs in the Molecular Responses of Metabolic Rate Depression. J. Mol. Cell Biol. 2011, 3, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Salerno, M.; Bertozzi, G.; Cipolloni, L.; Messina, G.; Aromatario, M.; Polo, L.; Turillazzi, E.; Pomara, C. MiRNAs as Novel Biomarkers of Chronic Kidney Injury in Anabolic-Androgenic Steroid Users: An Experimental Study. Front. Pharmacol. 2020, 11, 1454. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. MicroRNA Functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161. [Google Scholar]
- Ingelson-Filpula, W.A.; Hadj-Moussa, H.; Storey, K.B. MicroRNA Transcriptomics in Liver of the Freeze-Tolerant Gray Tree Frog (Dryophytes versicolor) Indicates Suppression of Energy-Expensive Pathways. Cell Biochem. Funct. 2023, 41, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.M.; Storey, K.B. Adaptations of Metabolism for Freeze Tolerance in the Gray Tree Frog, Hyla Versicolor. Can. J. Zool. 1985, 63, 49–54. [Google Scholar] [CrossRef]
- Rehman, S.; Storey, K.B. Dynamics of Epigenetic Regulation in Dryophytes versicolor Skeletal Muscle: Lysine Methylation and Acetylation Involvement in Metabolic Rate Depression. J. Therm. Biol. 2024, 122, 103865. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, M.; Breedon, S.A.; Storey, K.B. Cardiac MicroRNA Expression Profile in Response to Estivation. Biochimie 2023, 210, 22–34. [Google Scholar] [CrossRef]
- Hawkins, L.J.; Storey, K.B. MicroRNA Expression in the Heart of Xenopus Laevis Facilitates Metabolic Adaptation to Dehydration. Genomics 2020, 112, 3525–3536. [Google Scholar] [CrossRef]
- Zhang, J.; Storey, K.B. RBiomirGS: An All-in-One MiRNA Gene Set Analysis Solution Featuring Target MRNA Mapping and Expression Profile Integration. PeerJ 2018, 6, e4262. [Google Scholar] [CrossRef]
- Zhang, J.; Storey, K.B. RBioplot: An Easy-to-Use R Pipeline for Automated Statistical Analysis and Data Visualization in Molecular Biology and Biochemistry. PeerJ 2016, 4, e2436. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X. DEAD-Box RNA Helicases in Cell Cycle Control and Clinical Therapy. Cells 2021, 10, 1540. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T. Glycosylphosphatidylinositol (GPI) Anchors: Biochemistry and Cell Biology: Introduction to a Thematic Review Series. J. Lipid Res. 2016, 57, 4. [Google Scholar] [CrossRef] [PubMed]
- Al-attar, R.; Wijenayake, S.; Storey, K.B. Metabolic Reorganization in Winter: Regulation of Pyruvate Dehydrogenase (PDH) during Long-Term Freezing and Anoxia. Cryobiology 2019, 86, 10–18. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic Rate Depression in Animals: Transcriptional and Translational Controls. Biol. Rev. Camb. Philos. Soc. 2004, 79, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Storey, K.B. Evidence for Cell Cycle Suppression and MicroRNA Regulation of Cyclin D1 during Anoxia Exposure in Turtles. Cell Cycle 2012, 11, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Pietenpol, J.A.; Stewart, Z.A. Cell Cycle Checkpoint Signaling: Cell Cycle Arrest versus Apoptosis. Toxicology 2002, 181–182, 475–481. [Google Scholar] [CrossRef]
- Imadome, K.; Iwakawa, M.; Nojiri, K.; Tamaki, T.; Sakai, M.; Nakawatari, M.; Moritake, T.; Yanagisawa, M.; Nakamura, E.; Tsujii, H.; et al. Upregulation of Stress-Response Genes with Cell Cycle Arrest Induced by Carbon Ion Irradiation in Multiple Murine Tumors Models. Cancer Biol. Ther. 2008, 7, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Moxnes, J.F.; Haux, J.; Hausken, K. The Dynamics of Cell Proliferation. Med. Hypotheses 2004, 62, 556–563. [Google Scholar] [CrossRef]
- Bradrick, S.S.; Dobrikova, E.Y.; Kaiser, C.; Shveygert, M.; Gromeier, M. Poly(A)-Binding Protein Is Differentially Required for Translation Mediated by Viral Internal Ribosome Entry Sites. RNA 2007, 13, 1582. [Google Scholar] [CrossRef]
- Oksuz, O.; Henninger, J.E.; Warneford-Thomson, R.; Zheng, M.M.; Erb, H.; Vancura, A.; Overholt, K.J.; Hawken, S.W.; Banani, S.F.; Lauman, R.; et al. Transcription Factors Interact with RNA to Regulate Genes. Mol. Cell 2023, 83, 2449–2463.e13. [Google Scholar] [CrossRef]
- Breedon, S.A.; Storey, K.B. Lost in Translation: Exploring MicroRNA Biogenesis and Messenger RNA Fate in Anoxia-Tolerant Turtles. Oxygen 2022, 2, 227–245. [Google Scholar] [CrossRef]
- Fuery, C.J.; Withers, P.C.; Hobbs, A.A.; Guppy, M. The Role of Protein Synthesis during Metabolic Depression in the Australian Desert Frog Neobatrachus Centralis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 1998, 119, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Pakay, J.L.; Withers, P.C.; Hobbs, A.A.; Guppy, M. In Vivo Downregulation of Protein Synthesis in the Snail Helix Apersa during Estivation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.N.I.; Cheung, P. Histone Code Pathway Involving H3 S28 Phosphorylation and K27 Acetylation Activates Transcription and Antagonizes Polycomb Silencing. Proc. Natl. Acad. Sci. USA 2011, 108, 2801–2806. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone Phosphorylation: A Chromatin Modification Involved in Diverse Nuclear Events. Epigenetics 2012, 7, 1098. [Google Scholar] [CrossRef] [PubMed]
- Bungard, D.; Fuerth, B.J.; Zeng, P.Y.; Faubert, B.; Maas, N.L.; Viollet, B.; Carling, D.; Thompson, C.B.; Jones, R.G.; Berger, S.L. Signaling Kinase AMPK Activates Stress-Promoted Transcription via Histone H2B Phosphorylation. Science 2010, 329, 1201. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like Receptors in Microbial Recognition and Host Defense. Immunol. Rev. 2009, 227, 106. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Signaling to NF-KappaB. Genes Dev. 2004, 18, 2195–2224. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605. [Google Scholar] [CrossRef]
- Morrison, D.K. MAP Kinase Pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef] [PubMed]
- Ingelson-Filpula, W.A.; Storey, K.B. Hibernation-Induced MicroRNA Expression Promotes Signaling Pathways and Cell Cycle Dysregulation in Ictidomys Tridecemlineatus Cardiac Tissue. Metabolites 2023, 13, 1096. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-MTOR Pathways: Cross-Talk and Compensation. Trends Biochem. Sci. 2011, 36, 320. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; López, J.M. Understanding MAPK Signaling Pathways in Apoptosis. Int. J. Mol. Sci. 2020, 21, 2346. [Google Scholar] [CrossRef] [PubMed]
- Baust, J.G.; Gao, D.; Baust, J.M. Cryopreservation. Organogenesis 2009, 5, 90–96. [Google Scholar] [CrossRef]
- Shao, Q.; Han, F.; Peng, S.; He, B. Nur77 Inhibits OxLDL Induced Apoptosis of Macrophages via the P38 MAPK Signaling Pathway. Biochem. Biophys. Res. Commun. 2016, 471, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Ouyang, G.; Bao, S. The Activation of Akt/PKB Signaling Pathway and Cell Survival. J. Cell Mol. Med. 2005, 9, 59–71. [Google Scholar] [CrossRef]
- Saxena, M.; Yeretssian, G. NOD-like Receptors: Master Regulators of Inflammation and Cancer. Front. Immunol. 2014, 5, 98103. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).