Exploring Salivary Metabolic Alterations in Type 2 Diabetes: Implications for Dental Caries and Potential Influences of HbA1c and Vitamin D Levels
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Populations
2.1.1. Determination of Participant Whole-Blood Vitamin D (25-Hydroxyvitamin D) Concentrations for the ND and T2DM Groups
2.1.2. Sample Size Power Calculation for the Study
2.2. Sample Collection
2.3. Sample Preparation and 1H NMR Analysis
2.3.1. Sample Preparation
2.3.2. Acquisition of 1H NMR Spectra
2.3.3. Preprocessing of 1H NMR Profile Data
2.4. MV Metabolomics Analysis
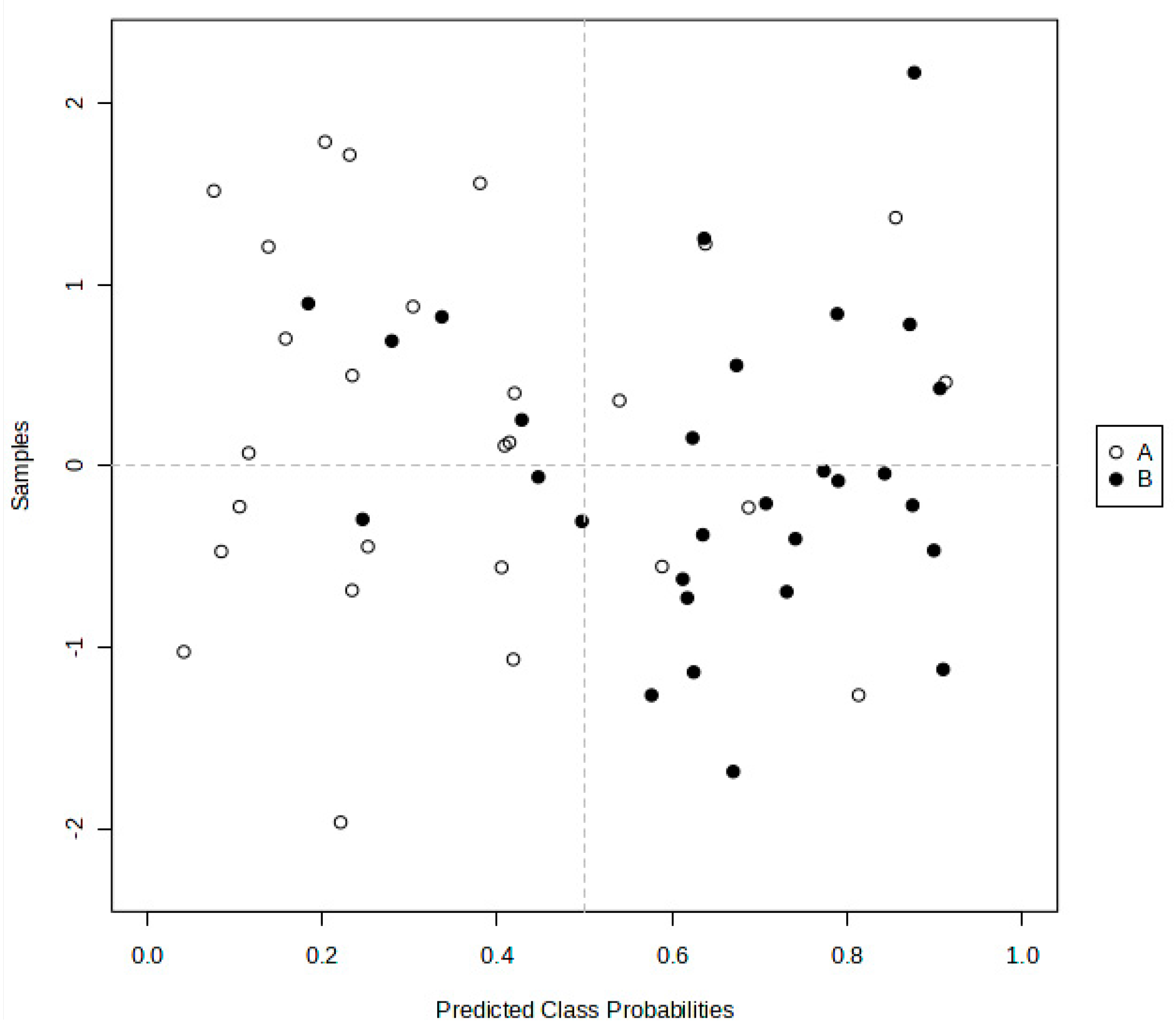
2.4.1. PLS-DA, OPLS-DA, and AUROC Biomarker Analyses
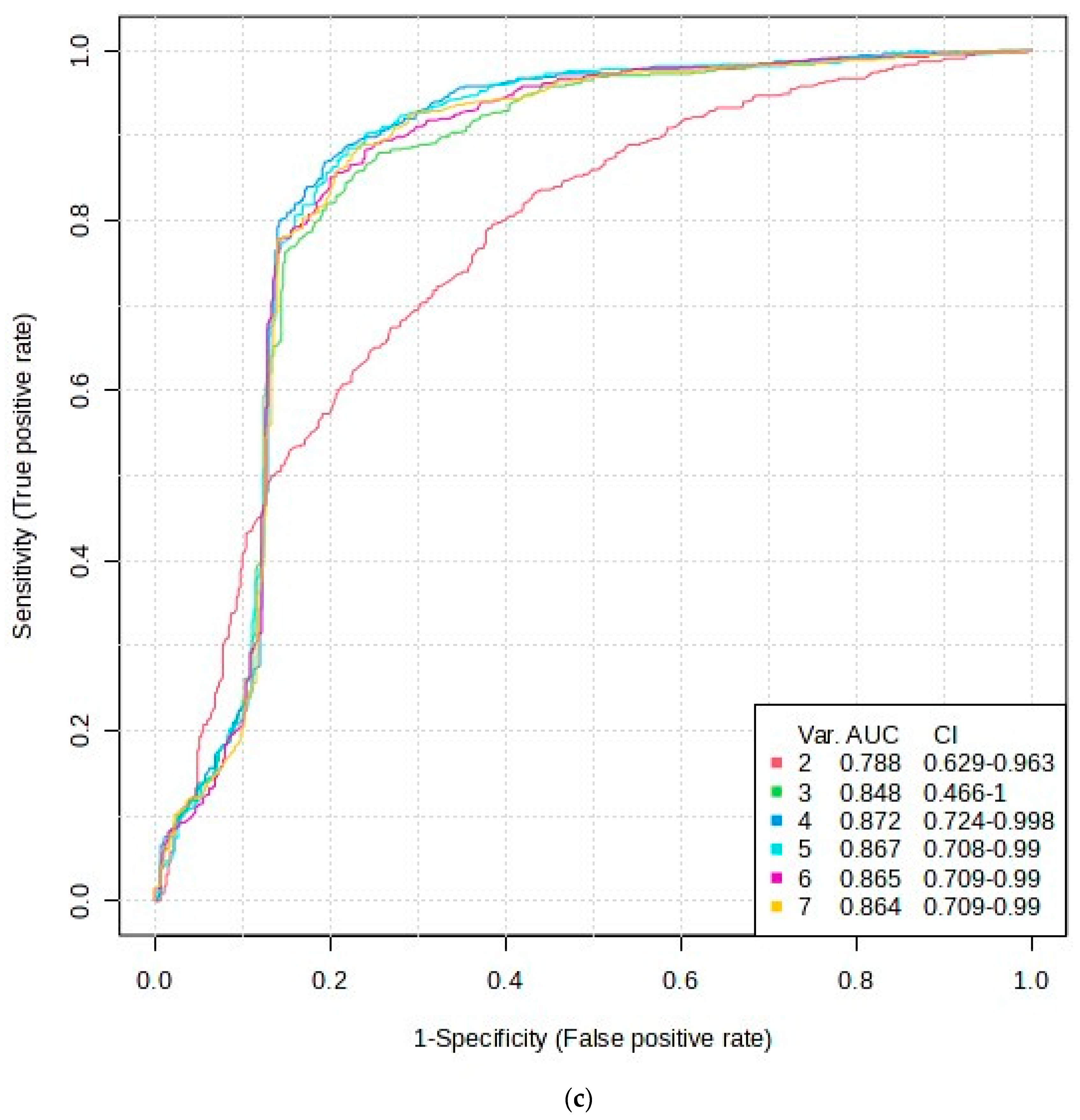
2.4.2. Further Assessment of AUROC Feature Selection Pathways
3. Results
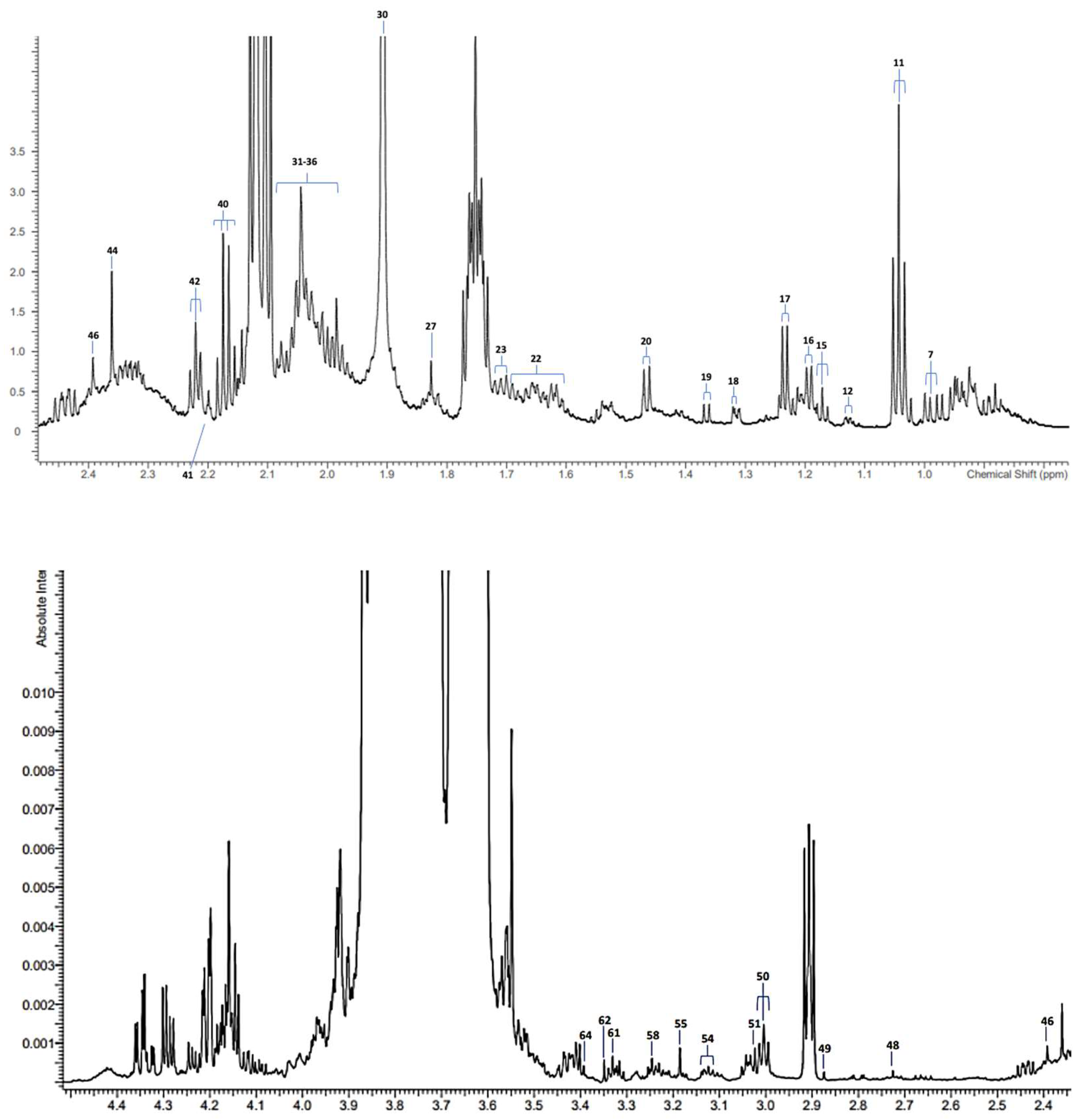
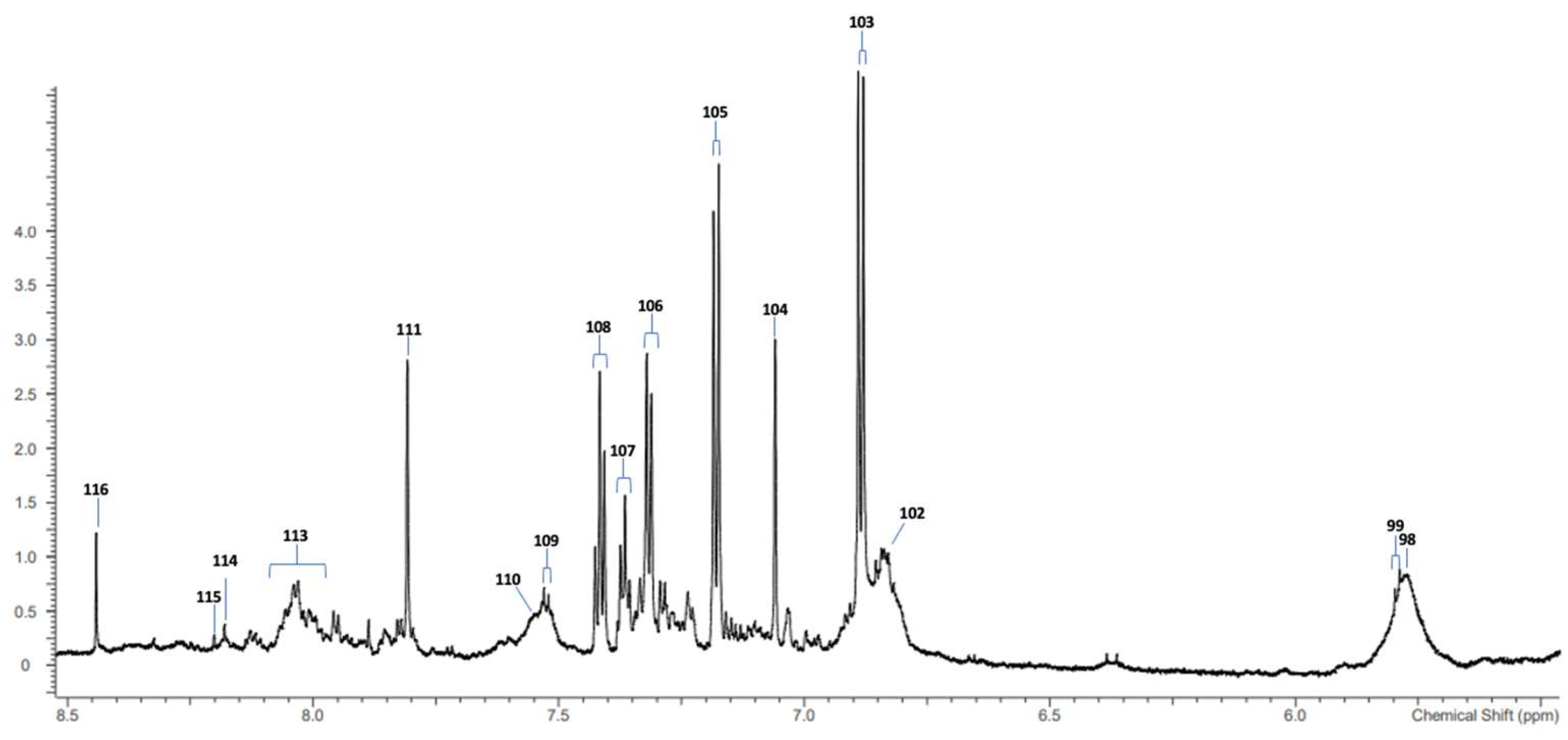
3.1. 1H NMR Analysis of SWS Supernatant Samples and Their Resonance Assignments
3.2. MV Metabolomics Analysis of 1H NMR Spectral Datasets
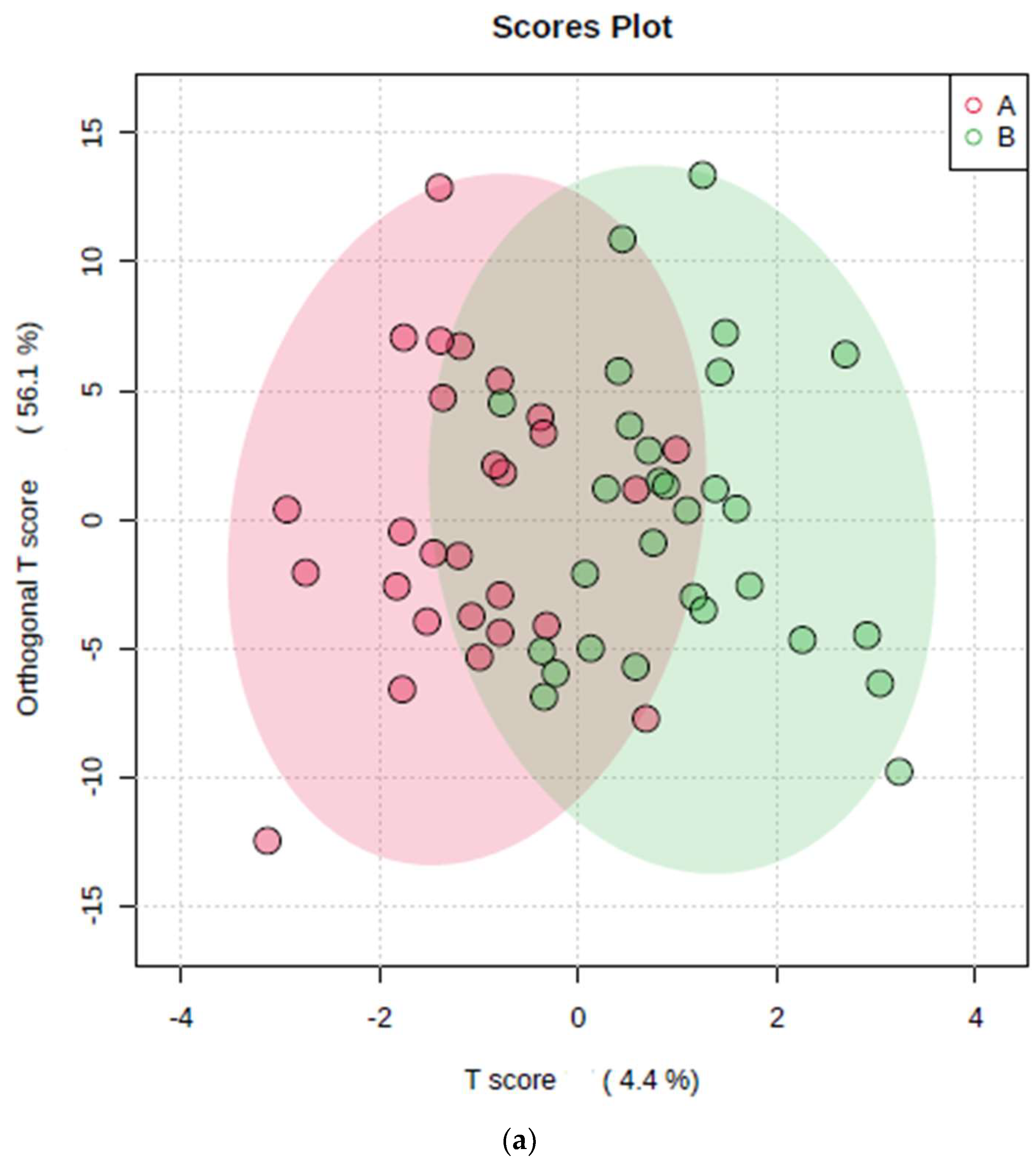
3.2.1. Application of PCA, PLS-DA, and OPLS-DA Techniques
3.2.2. AUROC Analysis of 1H NMR ISB Variables Alone
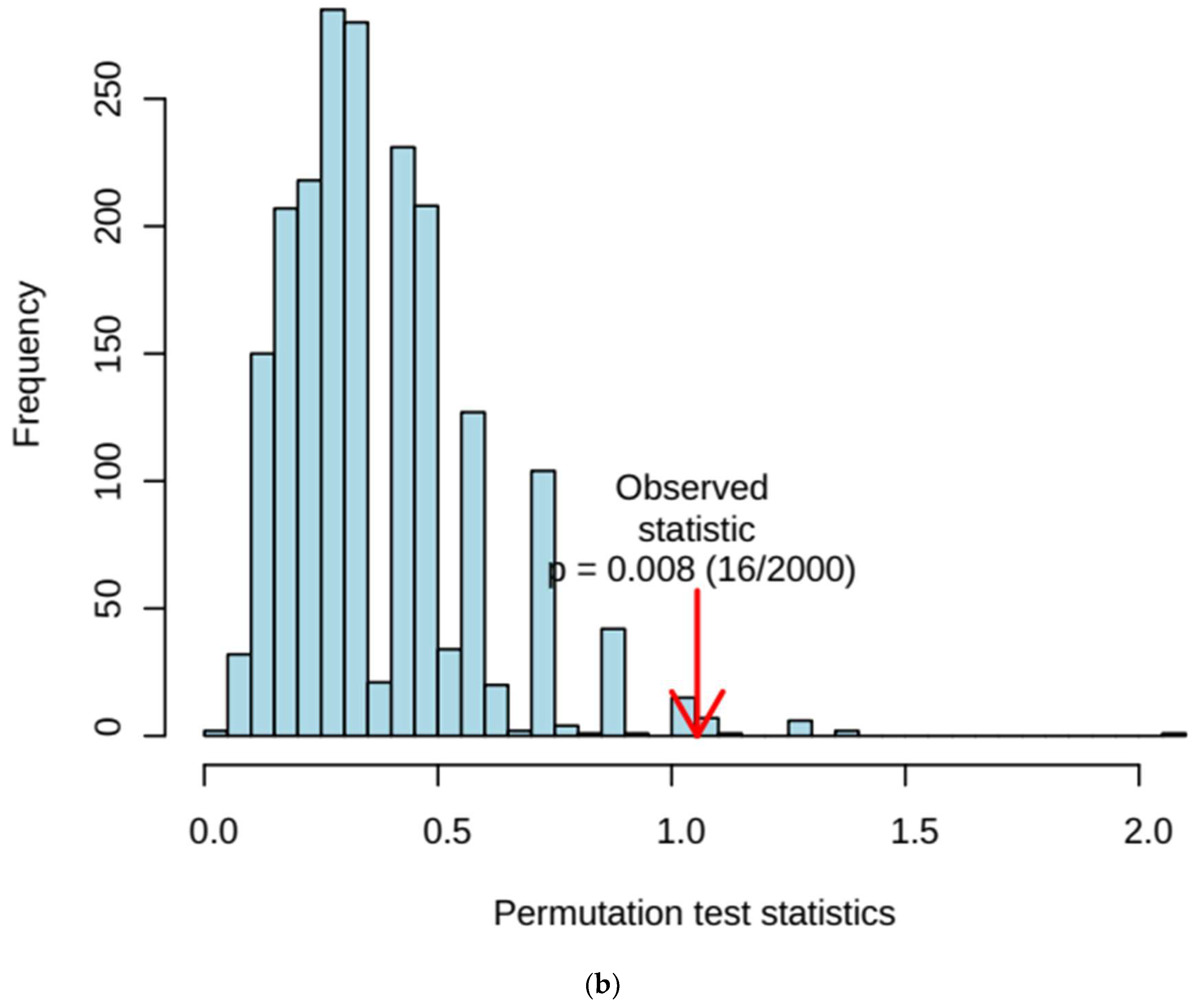
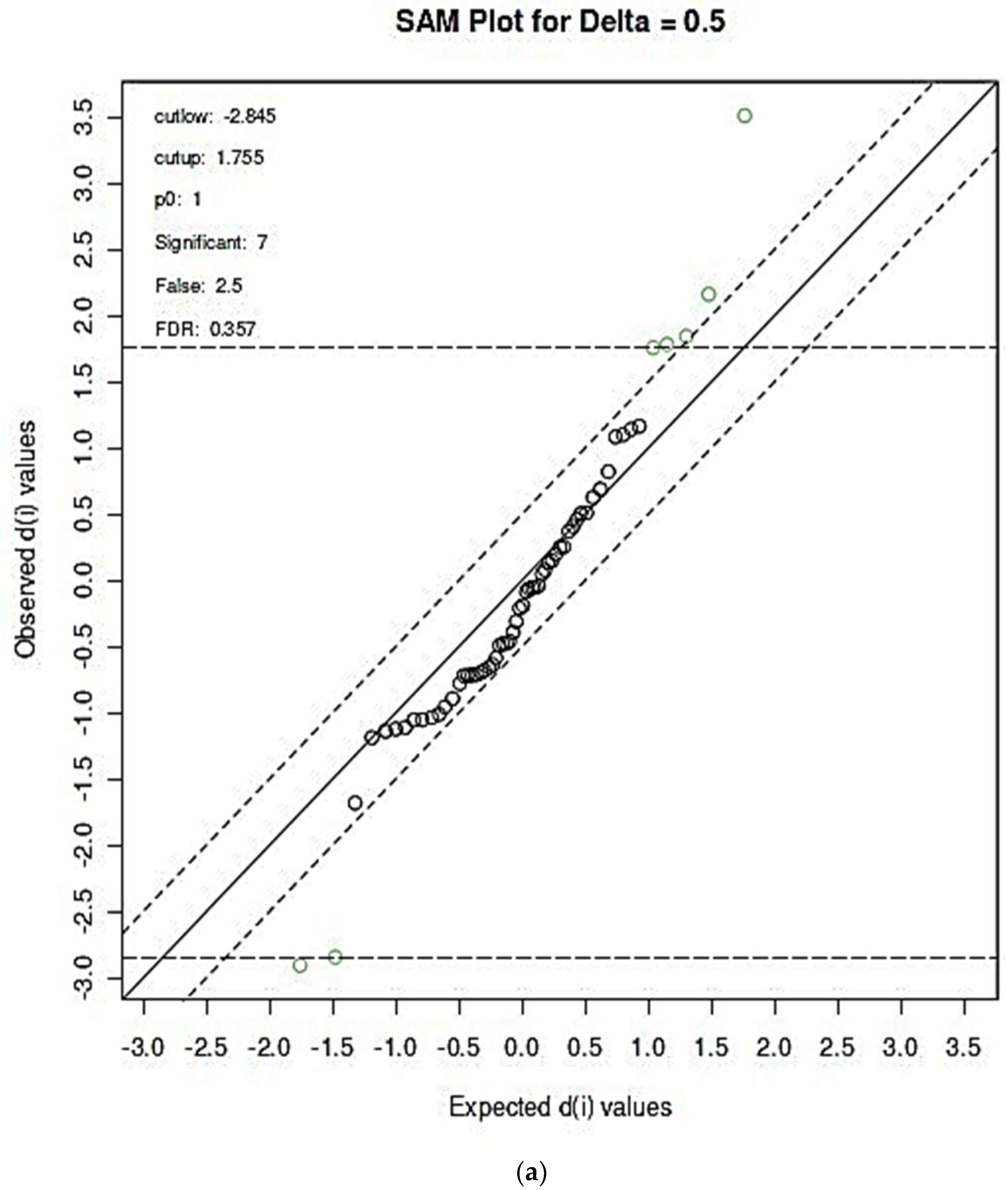
3.2.3. SAM- and EBAM-Supported AUROC Analysis: Significance Level Adjustments for FDR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.diabetes.org.uk/about-us/about-the-charity/our-strategy/statistics#:~:text=Our%20data%20shows%20that%20more,by%20148%2C591%20from%202020%2D2021 (accessed on 30 April 2024).
- Lamster, I.B.; Kunzel, C.; Lalla, E. Diabetes mellitus and oral health care: Time for the next step. J. Am. Dent. Assoc. 2012, 143, 208–210. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.J.; Ford, P.J.; Cullinan, M.P.; Leishman, S.; Yamazaki, K. Relationship between periodontal infections and systemic disease. Clin. Microbiol. Infect. 2007, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.A.; Aho, V.T.; Paulin, L.; Pekkonen, E.; Auvinen, P.; Scheperjans, F. Oral and nasal microbiota in Parkinson’s disease. Park. Relat. Disord. 2017, 38, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, J.D.; Calderaro, D.C.; Ferreira, G.A.; Mendonça, S.M.S.; Fernandes, G.R.; Xiao, E.; Teixeira, A.L.; Leys, E.J.; Graves, D.T.; Silva, T.A. Subgingival microbiota dysbiosis in systemic lupus erythematosus: Association with periodontal status. Microbiome 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Bracci, P.M. Oral Health and the oral microbiome in pancreatic cancer: An overview of epidemiological studies. Cancer J 2017, 23, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Segal, L.N.; Clemente, J.C.; Tsay, J.C.J.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A.; et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhao, Y.; Li, S.; Yang, L.; Wang, T.; Shi, B.; Gai, Z.; Heng, X.; Chunling, Z.; Yang, J.; et al. Variations in oral microbiome profiles in rheumatoid arthritis and osteoarthritis with potential biomarkers for arthritis screening. Sci. Rep. 2018, 8, 17126. [Google Scholar] [CrossRef] [PubMed]
- Casarin, R.C.; Barbagallo, A.; Meulman, T.; Santos, V.R.; Sallum, E.A.; Nociti, F.H.; Duarte, P.M.; Casati, M.Z.; Gonçalves, R.B. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J. Periodontal. Res. 2013, 48, 30–36. [Google Scholar] [CrossRef]
- Yamamoto, T.; Eguchi, T. Heat shock proteins and periodontitis—cross-reaction between bacterial and human HSP in periodontal infection linking with cardiovascular diseases. In Heat Shock Proteins in Inflammatory Diseases. Heat Shock Proteins; Asea, A.A.A., Kaur, P., Eds.; Springer: Cham, Switzerland, 2020; Volume 22. [Google Scholar] [CrossRef]
- Li, L.; Messas, E.; Batista, E.L.; Levine, R.A.; Amar, S. Porphyromonas gingivalis infection accelerates the progression of atherosclerosis in a heterozygous apolipoprotein E-deficient murine model. Circulation 2002, 10, 861–867. [Google Scholar] [CrossRef]
- Pardo, A.; Signoriello, A.; Signoretto, C.; Messina, E.; Carelli, M.; Tessari, M.; De Manna, N.D.; Rossetti, C.; Albanese, M.; Lombardo, G.; et al. Detection of periodontal pathogens in oral samples and cardiac specimens in patients undergoing aortic valve replacement: A pilot study. J. Clin. Med. 2021, 10, 3874. [Google Scholar] [CrossRef]
- Mikkonen, J.J.W.; Singh, S.P.; Herrala, M.; Lappalainen, R.; Myllymaa, S.; Kullaa, A.M. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J. Periodont. Res. 2016, 51, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Grootveld, M.; Page, G.; Bhogadia, M.; Edgar, M. Updates and original case studies focused on the NMR-linked metabolomics analysis of human oral fluids. Part I: Emerging platforms and perspectives. Appl. Sci. 2022, 12, 1235. [Google Scholar] [CrossRef]
- Silwood, C.J.L.; Lynch, E.; Seddon, S.; Sheerin, A.; Claxson, A.W.D.; Grootveld, M.C. 1H NMR analysis of microbial-derived organic acids in primary root carious lesions and saliva. NMR Biomed. 1999, 12, 345–356. [Google Scholar] [CrossRef]
- Silwood, C.J.L.; Lynch, E.; Claxson, A.W.D.; Grootveld, M.C. 1H and 13C NMR spectroscopic analysis of human saliva. J. Dent. Res. 2002, 81, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Silwood, C.J.L.; Grootveld, M.; Lynch, E. 1H NMR Investigations of the molecular nature of low-molecular-mass calcium ions in biofluids. J. Biol. Inorg. Chem. 2002, 7, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Dame, Z.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.; Sajed, T.; Deng, L.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Alkahtani, A.; Anderson, P.; Baysan, A. The impact of sociodemographic determinants and diabetes type-2 on oral health outcomes: An analytical cross-sectional study. Clin. Exp. Dent. Res. 2024, 10, e846. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Song, Y.; Kim, S.; Kim, S.; Na, H.; Lee, S.; Chung, J.; Kim, S. Identification of a biomarker panel for diagnosis of early childhood caries using salivary metabolic profile. Metabolites 2023, 13, 356. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0—The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D6008–D6117. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; Tibshirani, R.; Storey, J.D.; Tusher, V. Empirical Bayes analysis of a microarray experiment. J. Amer. Statist. Assoc. 2001, 96, 1151–1160. [Google Scholar] [CrossRef]
- Pfaffe, T.; Cooper-White, J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic potential of saliva: Current state and future applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, E.; Lamster, I.B. The diagnostic applications of saliva—A review. Crit. Rev. Oral Biol. Med. 2002, 13, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Al-Tarawneh, S.K.; Border, M.B.; Dibble, C.F.; Bencharit, S. Defining salivary biomarkers using mass spectrometry-based proteomics: A systematic review. OMICS: J. Integr. Biol. 2001, 15, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Grootveld, M.; Page, G.; Bhogadia, M.; Hunwin, K.; Edgar, M. Updates and original case studies focused on the NMR-linked metabolomics analysis of human oral fluids. Part III: Implementations for the diagnosis of non-cancerous disorders, both oral and systemic. Metabolites 2023, 13, 66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Giannobile, W.V.; Beikler, T.; Kinney, J.S.; Ramseier, C.A.; Morelli, T.; Wong, D.T. Saliva as a diagnostic tool for periodontal disease: Current state and future directions. Periodontology 2009, 50, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, E.; Kashyap, B.; Kullaa, A.M. Oral sources of salivary metabolites. Metabolites 2023, 13, 498. [Google Scholar] [CrossRef] [PubMed]
- Lanza, I.R.; Zhang, S.; Ward, L.E.; Karakelides, H.; Raftery, D.; Nair, K.S. Quantitative metabolomics by 1H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS ONE 2010, 5, e10538. [Google Scholar] [CrossRef]
- Grootveld, M.; Percival, B.C.; Page, G.; Hunwin, K.; Bhogadia, M.; Chan, W.; Edgar, M. Updates and original case studies focused on the NMR-linked metabolomics analysis of human oral fluids. Part II: Applications to the diagnosis and prognostic monitoring of oral and systemic cancers. Metabolites 2022, 12, 778. [Google Scholar] [CrossRef]
- Soininen, P.; Haarala, J.; Vepsäläinen, J.; Niemitz, M.; Laatikainen, R. Strategies for organic impurity quantification by 1H NMR spectroscopy: Constrained total-line-shape fitting. Anal. Chim. Acta 2005, 542, 178–185. [Google Scholar] [CrossRef]
- Gardner, A.; Parkes, H.G.; Carpenter, G.H.; So, P.W. Developing and standardizing a protocol for quantitative proton nuclear magnetic resonance (1H NMR) spectroscopy of saliva. J. Proteome Res. 2018, 17, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.; Ceriello, A.; Buysschaert, M.; Chapple, I.; Demmer, R.T.; Graziani, F.; Vegh, D. Scientific evidence on the links between periodontal diseases and diabetes: Consensus report and guidelines of the joint workshop on periodontal diseases and diabetes by the International Diabetes Federation and the European Federation of Periodontology. Diabetes Res. Clin. Prac. 2018, 137, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Laouali, N.; El Fatouhi, D.; Aguayo, G.; Balkau, B.; Boutron-Ruault, M.C.; Bonnet, F.; Fagherazzi, G. Type 2 diabetes and its characteristics are associated with poor oral health: Findings from 60,590 senior women from the E3N study. BMC Oral Health 2021, 21, 315. [Google Scholar] [CrossRef] [PubMed]
- Saremi, A.; Nelson, R.G.; Tulloch-Reid, M.; Hanson, R.L.; Sievers, M.L.; Taylor, G.W.; Shlossman, M.; Bennett, P.H.; Genco, R.; Knowler, W.C. Periodontal disease and mortality in type 2 diabetes. Diabetes Care 2005, 28, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Santos, V.R.; Lima, J.A.; Miranda, T.S.; Feres, M.; Zimmermann, G.S.; Nogueira-Filho, G.R.; Duarte, P.M. Relationship between glycemic subsets and generalized chronic periodontitis in type 2 diabetic Brazilian subjects. Arch. Oral Biol. 2012, 57, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Fidalgo, T.K.; Freitas-Fernandes, L.B.; Angeli, R.; Muniz, A.M.; Gonsalves, E.; Santos, R.; Nadal, J.; Almeida, F.C.; Valente, A.P.; Souza, I.P. Salivary metabolite signatures of children with and without dental caries lesions. Metabolomics 2013, 9, 657–666. [Google Scholar] [CrossRef]
- Lamster, I.B.; Lalla, E.; Borgnakke, W.S.; Taylor, G.W. The relationship between oral health and diabetes mellitus. J. Am. Dent. Assoc. 2008, 139, 19S–24S. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.; Beighton, D.; Curtis, M.A.; Cury, J.; Dige, I.; Dommisch, H.; Ellwood, R.; Giacaman, R.A.; Herrera, D.; Herzberg, M.C.; et al. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J. Clin. Periodontol. 2017, 44 (Suppl. 18), S5–S11. [Google Scholar] [CrossRef]
- Moussa, D.G.; Paras, A.; Mansour, T.A.; Siqueira, W.L. Current state and challenges of the global outcomes of dental caries research in the meta-omics era. Front. Cell. Infect. Microbiol. 2022, 12, 887907. Available online: https://www.frontiersin.org/articles/10.3389/fcimb.2022.887907 (accessed on 24 April 2024). [CrossRef]
- Moussa, D.G.; Sharma, A.K.; Mansour, T.A.; Witthuhn, B.; Perdigão, J.; Rudney, J.D.; Aparicio, C.; Gomez, A. Functional signatures of ex-vivo dental caries onset. J. Oral Microbiol. 2022, 14, 2123624. [Google Scholar] [CrossRef] [PubMed]
- Vajargah, K.F.; Mehdizadeh, R.; Sadeghi-Bazargani, H. Applications of OPLS statistical method in medicine. J. Math. Comput. Sci. 2014, 8, 411–422. [Google Scholar] [CrossRef]
- Xu, W.; Xue, W.; Zhou, Z.; Wang, J.; Qi, H.; Sun, S.; Jin, T.; Yao, P.; Zhao, J.Y.; Lin, F. Formate might be a novel potential serum metabolic biomarker for type 2 diabetic peripheral neuropathy. Diabetes Metab. Synd. Obes. 2023, 16, 3147–3160. [Google Scholar] [CrossRef] [PubMed]
- Adeva-Andany, M.; López-Ojén, M.; Funcasta-Calderón, R.; Ameneiros-Rodríguez, E.; Donapetry-García, C.; Vila-Altesor, M.; Rodríguez-Seijas, J. Comprehensive review on lactate metabolism in human health. Mitochondrion 2014, 17, 76–100. [Google Scholar] [CrossRef] [PubMed]
- Juraschek, S.P.; Shantha, G.P.S.; Chu, A.Y.; Miller, E.R., 3rd; Guallar, E.; Hoogeveen, R.C.; Ballantyne, C.M.; Brancati, F.L.; Schmidt, M.I.; Pankow, J.S.; et al. Lactate and risk of incident diabetes in a case-cohort of the atherosclerosis risk in communities (ARIC) study. PLoS ONE 2013, 8, e55113. [Google Scholar] [CrossRef] [PubMed]
- van Houte, J.; Lopman, J.; Kent, R. The predominant cultivable flora of sound and carious human root surfaces. J. Dent. Res. 1994, 73, 1727–1734. [Google Scholar] [CrossRef]
- Koo, H.; Duarte, S.; Murata, R.M.; Scott-Anne, K.; Gregoire, S.; Watson, G.E.; Singh, A.P.; Vorsa, N. Influence of cranberry proanthocyanidins on formation of biofilms by Streptococcus mutans on saliva-coated apatitic surface and on dental caries development in vivo. Caries Res. 2010, 44, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lee, S.Y. Identification of non-streptococcal organisms from human dental plaque grown on the Streptococcus-selective medium mitis-salivarius agar. Arch. Oral Biol. 2015, 60, 267–271. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, T.; Zhang, J.; Zhou, X. Characterization of the Actinomyces naeslundii ureolysis and its role in bacterial aciduricity and capacity to modulate pH homeostasis. Microbiol. Res. 2006, 161, 304–310, Erratum in Microbiol. Res. 2011, 4, 336. [Google Scholar]
- Nascimento, M.M.; Gordan, V.V.; Garvan, C.W.; Browngardt, C.M.; Burne, R.A. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol. Immunol. 2009, 24, 89–95. [Google Scholar] [CrossRef]
- Takahashi, N.; Yamada, T. Glucose and lactate metabolism by Actinomyces naeslundii. Crit. Rev. Oral Biol. Med. 1999, 10, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nyvad, B. Caries ecology revisited: Microbial dynamics and the caries process. Caries Res. 2008, 42, 409–418. [Google Scholar] [CrossRef]
- Brighenti, F.L.; Salvador, M.J.; Delbem, A.C.B.; Delbem, Á.C.B.; Oliveira, M.A.C.; Soares, C.P.; Freitas, L.S.; Koga-Ito, C.Y. Systematic screening of plant extracts from the Brazilian Pantanal with antimicrobial activity against bacteria with cariogenic relevance. Caries Res. 2014, 48, 353–360. [Google Scholar] [CrossRef]
- Shu, M.; Wong, L.; Miller, J.H.; Sissons, C.H. Development of multi-species consortia biofilms of oral bacteria as an enamel and root caries model system. Arch. Oral Biol. 2000, 45, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, J. Bacterial metabolism in dental biofilms. Adv. Dent. Res. 1997, 11, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Dame-Teixeira, N.; Parolo, C.C.F.; Maltz, M.; Tugnait, A.; Devine, D.; Do, T. Actinomyces spp. gene expression in root caries lesions. J. Oral Microbiol. 2016, 8, 32383. [Google Scholar] [CrossRef] [PubMed]
- Mattos-Graner, R.O.; Klein, M.I.; Smith, D.J. Lessons learned from clinical studies: Roles of mutans streptococci in the pathogenesis of dental caries. Curr. Oral Health Rep. 2014, 1, 70–78. [Google Scholar] [CrossRef]
- Mozaffari, M.S.; Borke, J.L. Taurine in submandibular gland of the rat: Effect of muscarinic drugs. J. Histochem. Cytochem. 2022, 50, 527–532. [Google Scholar] [CrossRef]
- Hansen, S.H. The role of taurine in diabetes and the development of diabetic complications. Diabetes/Metab. Res. Rev. 2001, 17, 330–346. [Google Scholar] [CrossRef]
- Tao, X.; Zhang, Z.; Yang, Z.; Rao, B. The effects of taurine supplementation on diabetes mellitus in humans: A systematic review and meta-analysis. Food Chem. Molecular Sci. 2022, 4, 100106. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Roshan, V.D.; Aslani, E.; Stannard, S.R. Taurine supplementation has anti-atherogenic and anti-inflammatory effects before and after incremental exercise in heart failure. Ther. Advan. Cardiovasc. Dis. 2017, 11, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Qaradakhi, T.; Gadanec, L.K.; McSweeney, K.R.; Abraham, J.R.; Apostolopoulos, V.; Zulli, A. The anti-inflammatory effect of taurine on cardiovascular disease. Nutrients 2020, 12, 2847. [Google Scholar] [CrossRef]
- Jakaria, M.; Azam, S.; Haque, M.E.; Jo, S.H.; Uddin, M.S.; Kim, I.S.; Choi, D.K. Taurine and its analogs in neurological disorders: Focus on therapeutic potential and molecular mechanisms. Redox Biol. 2019, 24, 101223. [Google Scholar] [CrossRef]
- Stacy, A.; Andrade-Oliveira, V.; McCulloch, J.A.; Hild, B.; Oh, J.H.; Perez-Chaparro, P.J.; Sim, C.K.; Lim, A.I.; Link, V.M.; Enamorado, M.; et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 2021, 184, 615–627.e17. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Song, X.; Dong, X.; Yang, Y.; Zhang, Z.; Wen, J.; Li, Y.; Zhou, L.; Zhao, N.; Zhu, X.; et al. High prevalence of chronic kidney disease in population-based patients diagnosed with type 2 diabetes in downtown Shanghai. J. Diabetes Complicat. 2008, 22, 96–103. [Google Scholar] [CrossRef]
- Dabla, P.K. Renal function in diabetic nephropathy. World J. Diabetes 2010, 1, 48–56. [Google Scholar] [CrossRef]
- Liebsch, C.; Pitchika, V.; Pink, C.; Samietz, S.; Kastenmüller, G.; Artati, A.; Suhre, K.; Adamski, J.; Nauck, M.; Völzke, H.; et al. The saliva metabolome in association to oral health status. J. Dent. Res. 2019, 98, 642–651. [Google Scholar] [CrossRef]
- Romano, F.; Meoni, G.; Manavella, V.; Baima, G.; Tenori, L.; Cacciatore, S.; Aimetti, M. Analysis of salivary phenotypes of generalized aggressive and chronic periodontitis through nuclear magnetic resonance-based metabolomics. J. Periodontol. 2018, 89, 1452–1460. [Google Scholar] [CrossRef]
- Vangipurapu, J.; Stancáková, A.; Smith, U.; Kuusisto, J.; Laakso, M. Nine amino acids are associated with decreased insulin secretion and elevated glucose levels in a 7.4-year follow-up study of 5181 Finnish men. Diabetes 2019, 68, 1353–1358. [Google Scholar] [CrossRef]
- Rzeznik, M.; Triba, M.N.; Lévy, P.; Jungo, S.; Botosoa, E.; Duchemann, B.; Le Moyec, L.; Bernaudin, J.F.; Savarin, P.; Guez, D. Identification of a discriminative metabolomic fingerprint of potential clinical relevance in saliva of patients with periodontitis using 1H nuclear magnetic resonance (NMR) spectroscopy. PLoS ONE 2017, 12, e0182767. [Google Scholar] [CrossRef] [PubMed]
- Minh, T.D.; Oliver, S.R.; Ngo, J.; Flores, R.; Midyett, J.; Meinardi, S.; Carlson, M.K.; Rowland, F.S.; Blake, D.R.; Galassetti, P.R. Noninvasive measurement of plasma glucose from exhaled breath in healthy and type 1 diabetic subjects. Am. J. Physiol.-Endocrinol. Metab. 2011, 300, E1166–E1175. [Google Scholar] [CrossRef]
- Gupta, S.; Nayak, M.T.; Sunitha, J.D.; Dawar, G.; Sinha, N.; Rallan, N.S. Correlation of salivary glucose level with blood glucose level in diabetes mellitus. J. Oral Maxillofac. Pathol. 2017, 21, 334–339. [Google Scholar] [PubMed]
- Gupta, V.; Kaur, A. Salivary glucose levels in diabetes mellitus patients: A case-control study. J. Oral Maxillofac. Pathol. 2020, 24, 187. [Google Scholar]
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; Devizio, W.; Trivedi, H.M.; Jönsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef]
- Vibhakar, P.; Patankar, S.R.; Yadav, M.; Vibhakar, P. Correlation of salivary glucose levels with dental caries: A biochemical study. Int. J. Oral Maxillofac. Pathol. 2014, 5, 17–21. [Google Scholar]
- Otvos, J.D.; Shalaurova, I.; Wolak-Dinsmore, J.; Connelly, M.A.; Mackey, R.H.; Stein, J.H.; Tracy, R.P. GlycA: A composite nuclear magnetic resonance biomarker of systemic inflammation. Clin. Chem. 2015, 61, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Connelly, M.A.; Otvos, J.D.; Shalaurova, I.; Playford, M.P.; Mehta, N.N. GlycA, a novel biomarker of systemic inflammation and cardiovascular disease risk. J. Translatl. Med. 2017, 15, 219. [Google Scholar] [CrossRef] [PubMed]
- Biri, S.R.K.; Ch, S.S.; Gundu, R.; Vadlakonda, A. A study on evaluating blood urea and serum creatinine in diabetes mellitus patients. Int. J. Clin. Biochem. Res. 2023, 8, 285–288. [Google Scholar]
- Ivanovski, K.; Naumovski, V.; Kostadinova, M.; Pesevska, S.; Drijanska, K.; Filipce, V. Xerostomia and salivary levels of glucose and urea in patients with diabetes. Contrib. Macedon. Acad. Sci. Arts 2012, 33, 219–229. [Google Scholar]
- Liu, X.; Gao, J.; Chen, J.; Wang, Z.; Shi, Q.; Man, H.; Guo, S.; Wang, Y.; Li, Z.; Wang, W. Identification of metabolic biomarkers in patients with type 2 diabetic coronary heart diseases based on metabolomic approach. Sci. Rep. 2016, 6, 30785. [Google Scholar] [CrossRef] [PubMed]
- Del Coco, L.; Vergara, D.; De Matteis, S.; Mensà, E.; Sabbatinelli, J.; Prattichizzo, F.; Bonfigli, A.R.; Storci, G.; Bravaccini, S.; Pirini, F.; et al. NMR-based metabolomic approach tracks potential serum biomarkers of disease progression in patients with type 2 diabetes mellitus. J. Clin. Med. 2019, 8, 720. [Google Scholar] [CrossRef] [PubMed]
- Van Wuyckhuyse, B.C.; Perinpanayagam, H.E.R.; Bevacqua, D.; Raubertas, R.F.; Billings, R.J.; Bowen, W.H.; Tabak, L.A. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J. Dent. Res. 1995, 74, 686–690. [Google Scholar] [CrossRef] [PubMed]
- McClure, F.J.; Folk, J.E. Lysine and the cariogenicity of two experimental rat diets. Science 1955, 122, 557. [Google Scholar] [CrossRef] [PubMed]
- Najjar, B.; Kashtanov, M.; Chikindas, M.L. Natural antimicrobials ε-poly-l-lysine and Nisin A for control of oral microflora. Probiotics Antimicrob. Proteins 2009, 1, 143–147. [Google Scholar] [CrossRef] [PubMed]
- McClure, F.J. Effect of lysine provided by different routes on cariogenicity of lysine deficient diet. Proc. Soc. Exp. Biol. Med. 1957, 96, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Englander, H.R.; Keyes, P.H.; Fitzgerald, R.J. Effect of amino acid supplements on dental caries in the syrian hamster. Arch. Oral Biol. 1965, 10, 599–604. [Google Scholar] [CrossRef]
- Levine, M.; Lohinai, Z.M. Resolving the contradictory functions of lysine decarboxylase and butyrate in periodontal and intestinal diseases. J. Clin. Med. 2021, 10, 2360. [Google Scholar] [CrossRef]
- Gavião, M.B.D.; Van der Bilt, A.J. Salivary secretion and chewing: Stimulatory effects from artificial and natural foods. Appl. Oral Sci. 2004, 12, 159–163. [Google Scholar] [CrossRef]
- Neyraud, E.; Tremblay-franco, M.; Gregoire, S.; Berdeaux, O.; Canlet, C. Relationships between the metabolome and the fatty acid composition of human saliva; effects of stimulation. Metabolomics 2013, 9, 213–222. [Google Scholar] [CrossRef]
- Dlugash, G.; Schultheiss, O.C. Suitability of saliva stimulants for valid assessment of steroid hormones via radioimmunoassay. Psychoneuroendocrinol 2021, 127, 105175. [Google Scholar] [CrossRef] [PubMed]




| Clinical Characteristic | ND | T2DM | p Value |
|---|---|---|---|
| Blood HbA1c concentration (mean ± SD) | 35.70 ± 5.94 mmol/mol | 54.15 ± 13.03 mmol/mol | 1.11 × 10−8 |
| Severity of dental caries: mean of ICDAS ± SD | 0.57 ± 0.24 | 0.67 ± 0.32 | 0.014 |
| Distribution of root carious lesions (%) | 21% | 76.9% | na |
| Extreme risk of dental caries (%) | 48.9% | 16.48% | na |
| Assignments | Assignment Code | Chemical Shift (δ/ppm) | Coupling Pattern |
|---|---|---|---|
| Leucine-CH3 | 7 | 0.962 | t |
| Propioniate-CH3 | 11 | 1.058 | t |
| Iso-Butyrate-CH3 | 12 | 1.125 | t |
| Ethanol-CH3 | 15 | 1.183 | d |
| Methylmalonate-CH3/α-L-Fucose-CH3 | 16 | 1.211 | d/d |
| 3-D-hydroxybutyrate-CH3/β-Fucose-CH3 | 17 | 1.242 | d |
| Lactate-CH3 | 18 | 1.330 | d |
| Acetoin-CH3 | 19 | 1.371 | d |
| Alanine-CH3 | 20 | 1.486 | d |
| 5-Aminovalerate-β,γ-CH2′ s | 22 | 1.641 | m |
| Leucine-CH2 | 23 | 1.685 | m |
| Senicioate-CH3 | 27 | 1.823 | s |
| Thymine-CH3 | Uncoded | 1.860 | s |
| Acetate-CH3 | 30 | 1.920 | s |
| 2-Hydroxyglutarate-γ-CH2 | 31 | 1.954 | m |
| Proline-γ-CH2/N-Acetylneuraminate-C3H | 32 | 2.005 | m |
| Glycoprotein carbohydrate side-chain N-acetylsugar-NHCOCH3 groups (Glyc A sugnal) | 33 | 2.020–2.080 (3 partially resolved signals) | broad |
| N-Acetylglutamate-/N-Acetylaspartate–NHCOCH3 (2 signals) | 34 | 2.025/2.030 | s |
| N-Acetylglucosamine-/N-Acetylneuraminate-NHCOCH3 | 35 | 2.040/2.060 | s |
| Unassigned N-Acetylated metabolite-NHCOCH3 (2 signals), including N-Acetylneuraminate (2.060 ppm) | 36 | 2.053/2.060 | 2 × s |
| Propioniate-CH2 | 40 | 2.193 | q |
| Acetone-CO-CH3 | 41 | 2.215 | s |
| 5-Aminovalerate-α-CH2 | 42 | 2.235 | t |
| Pyruvate-CH3 | 44 | 2.377 | s |
| Succinate-CH2 | 46 | 2.415 | s |
| Dimethylamine-N(CH3)2 | 48 | 2.723 | s |
| Trimethylamine-N(CH3)3 | 49 | 2.872 | s |
| 5-Aminovalerate-δ-CH2/Lysine-ε-CH2 | 50 | 3.004 | t/t |
| Creatine-N(CH3) | 51 | 3.022 | s |
| Dimethylsulphone-CH3 (3.10 ppm)/1/2 His- and Phe-β-CH2 (3.14 ppm) | 54 | 3.10–3.15 | s/m |
| 1,9-Dimethylurate-N1(CH3) | 55 | 3.183 | s |
| Betaine-N(CH3)3+/Taurine-CH2NH3+ | 58 | 3.242 | s/t |
| Paraxanthine-N(CH3) | 61 | 3.328 | s |
| 1,3,7-Trimethylurate-N7(CH3) | 62 | 3.348 | s |
| Methanol-CH3/1,3-Dimethyluracil-N1(CH3) | 64 | 3.386 | s/s |
| Urea-CO-NH2 | 98 | 5.790 | broad signals |
| Uracil-C2H | 99 | 5.800 | d |
| Protein aromatic amino acid residue(s) | 102 | 6.850 | broad |
| Tyrosine aromatic ring-C2H/C6H | 103 | 6.880 | d |
| Histidine imidazole ring-C5H | 104 | 7.071 | s |
| Hydroxyphenylacetate-aromatic ring-C2H/C6H | Uncoded | 7.17 | d |
| Tyrosine aromatic ring-C3H/C5H | 105 | 7.237 | d |
| Phenylalanine aromatic ring-C2H/C6H | 106 | 7.320 | m |
| Phenylalanine aromatic ring-C4H | 107 | 7.375 | m |
| Phenylalanine aromatic ring-C3H/C5H | 108 | 7.430 | m |
| Uracil-C1H | 109 | 7.533 | d |
| Protein aromatic amino acid residue(s) | 110 | 7.552 | m |
| Histidine imidazole ring-C2H | 111 | 7.812 | s |
| Protein aromatic amino acid residue(s) | 113 | 8.050 | 2 × broad signals |
| Hypoxanthine-C8H | 114 | 8.175 | s |
| Hypoxanthine-C3H | 115 | 8.219 | s |
| Formate-H | 116 | 8.456 | s |
| Chemical Shift (ppm) | Corresponding Assignment | VIP Value | Regulatory Status * |
|---|---|---|---|
| 3.40–3.46 | Taurine-CH2SO3− | 3.14 | ↑ |
| 3.03–3.08 | Creatinine-NCH3 | 2.92 | ↑ |
| 3.10–3.15 | ½ Histidine-/Phenylalanine-β-CH2/Dimethylsulphone-SO2(CH3)2 | 2.91 | ↑ |
| 5.24–5.28 | α-Glucose-C1H | 1.88 | ↓ |
| HbA1c | n/a | 1.78 | ↑ |
| 1.30–1.33 ppm | Lactate-CH3 | 1.75 | ↑ |
| Vitamin D | n/a | 1.49 | ↓ |
| 3.27–3.30 ppm | Unassigned | 1.39 | ↑ |
| 2.00–2.06 ppm | Glyc A glycoprotein-NHCOCH3 | 1.17 | ↓ |
| 5.74–5.80 | Urea-CONH2 | 1.17 | ↓ |
| 7.39–7.43 | Phenylalanine aromatic ring-C3H/C5H | 1.02 | ↑ |
| 4.13–4.19 | Lactate-CH | 1.02 | ↑ |
| 3.30–3.36 | Methanol-CH3 | 1.01 | ↑ |
| 8.41–8.47 | Formate-H | 0.90 | ↑ |
| Chemical Shifts (ppm) | Corresponding Assignment | VIP Value | Regulatory Status * |
|---|---|---|---|
| HbA1c | n/a | 3.44 | ↑ |
| Vitamin D | n/a | 3.38 | ↓ |
| 1.30–1.33 ppm | Lactate-CH3 | 2.06 | ↑ |
| 2.00–2.06 ppm | Glyc A glycoprotein-NHCOCH3 | 1.69 | ↓ |
| 4.13–4.19 ppm | Lactate-CH | 1.43 | ↑ |
| 1.95–1.97 ppm | Proline-γ-CH2 | 1.32 | ↓ |
| 1.66–1.72 ppm | Lysine-CH2 | 1.29 | ↓ |
| 3.40–3.46 ppm | Taurine-CH2SO3− | 1.28 | ↑ |
| 1.97–2.00 ppm | Proline-β-CH2 | 1.27 | ↓ |
| 7.77–7.82 ppm | Histidine imidazole ring-CH | 1.19 | ↓ |
| 3.10–3.15 ppm | ½ Histidine-/Phenylalanine-β-CH2/Dimethylsulphone-SO2(CH3)2 | 1.16 | ↑ |
| 3.03–3.08 ppm | Creatinine-NCH3 | 1.11 | ↑ |
| 6.87–6.93 ppm | Tyrosine aromatic ring-C3/C5H | 1.01 | ↓ |
| Variable or 1H NMR Bucket | 1H NMR Assignment | Regulatory Status * | Univariate AUROC Value |
|---|---|---|---|
| HbA1c | n/a | ↑ | 0.736 |
| Vitamin D | n/a | ↓ | 0.727 |
| 5.24–5.28 ppm | α-Glucose-C1H | ↓ | 0.700 |
| 1.30–1.34 ppm | Lactate-CH3 | ↑ | 0.683 |
| 5.72–5.74 ppm | Urea-CONH2 | ↓ | 0.625 |
| 5.74–5.76 ppm | Urea-CONH2 | ↓ | 0.617 |
| 7.39–7.43 ppm | Phenylalanine-C3H/C5H | ↑ | 0.611 |
| 5.08–5.14 ppm | Unassigned | ↑ | 0.567 |
| 8.41–8.47 ppm | Formate-H | ↑ | 0.582 |
| 4.13–4.19 ppm | Lactate-CH | ↑ | 0.571 |
| 7.04–7.09 ppm | Histidine imidazole ring-C5H | ↓ | 0.531 |
| 7.99–8.01 ppm | Unassigned | ↑ | 0.531 |
| 7.35–7.39 ppm | Phenylalanine aromatic ring-C3H/C5H | ↑ | 0.610 |
| 4.11–4.13 ppm | Unassigned | ↓ | 0.521 |
| 7.12–7.17 ppm | 4-Hydroxyphenylacetate aromatic ring-C2H/C6H | ↑ | 0.554 |
| 5.19–5.24 ppm | Unassigned (apparent triplet) | ↓ | 0.548 |
| 1.01–1.07 ppm | Valine-CH3 | ↑ | 0.525 |
| 3.30–3.36 ppm | Methanol-CH3 | ↑ | 0.581 |
| 7.30–7.35 ppm | Phenylalanine aromatic ring-C4H | ↑ | 0.521 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkahtani, A.; Grootveld, M.; Bhogadia, M.; Baysan, A. Exploring Salivary Metabolic Alterations in Type 2 Diabetes: Implications for Dental Caries and Potential Influences of HbA1c and Vitamin D Levels. Metabolites 2024, 14, 372. https://doi.org/10.3390/metabo14070372
Alkahtani A, Grootveld M, Bhogadia M, Baysan A. Exploring Salivary Metabolic Alterations in Type 2 Diabetes: Implications for Dental Caries and Potential Influences of HbA1c and Vitamin D Levels. Metabolites. 2024; 14(7):372. https://doi.org/10.3390/metabo14070372
Chicago/Turabian StyleAlkahtani, Ashwaq, Martin Grootveld, Mohammed Bhogadia, and Aylin Baysan. 2024. "Exploring Salivary Metabolic Alterations in Type 2 Diabetes: Implications for Dental Caries and Potential Influences of HbA1c and Vitamin D Levels" Metabolites 14, no. 7: 372. https://doi.org/10.3390/metabo14070372
APA StyleAlkahtani, A., Grootveld, M., Bhogadia, M., & Baysan, A. (2024). Exploring Salivary Metabolic Alterations in Type 2 Diabetes: Implications for Dental Caries and Potential Influences of HbA1c and Vitamin D Levels. Metabolites, 14(7), 372. https://doi.org/10.3390/metabo14070372








