Metabolite Profiling Analysis of the Tongmai Sini Decoction in Rats after Oral Administration through UHPLC-Q-Exactive-MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Plant Materials
2.2. Plant Extract Preparation
2.3. Animal and Drug Administration
2.4. Sample Collection and Pretreatment
2.5. Instrumentation and Conditions
3. Results and Discussion
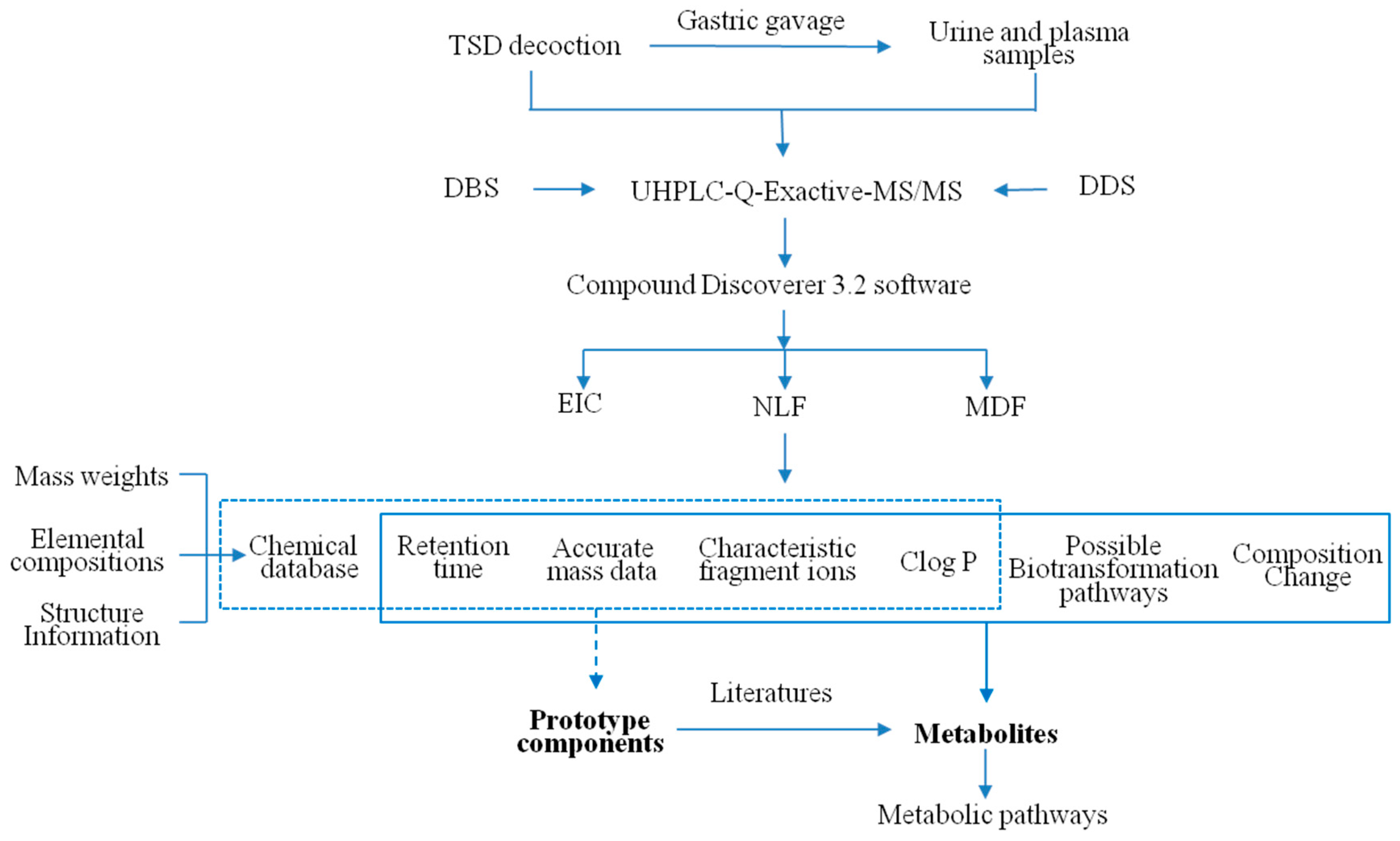
3.1. Systematic Analytical Strategy for Online Metabolite Analysis
3.2. Identification of Prototype Components
3.2.1. Identification of Alkaloid Components
3.2.2. Identification of Phenolic Compounds
3.2.3. Identification of Saponins
3.3. Identification of Metabolites
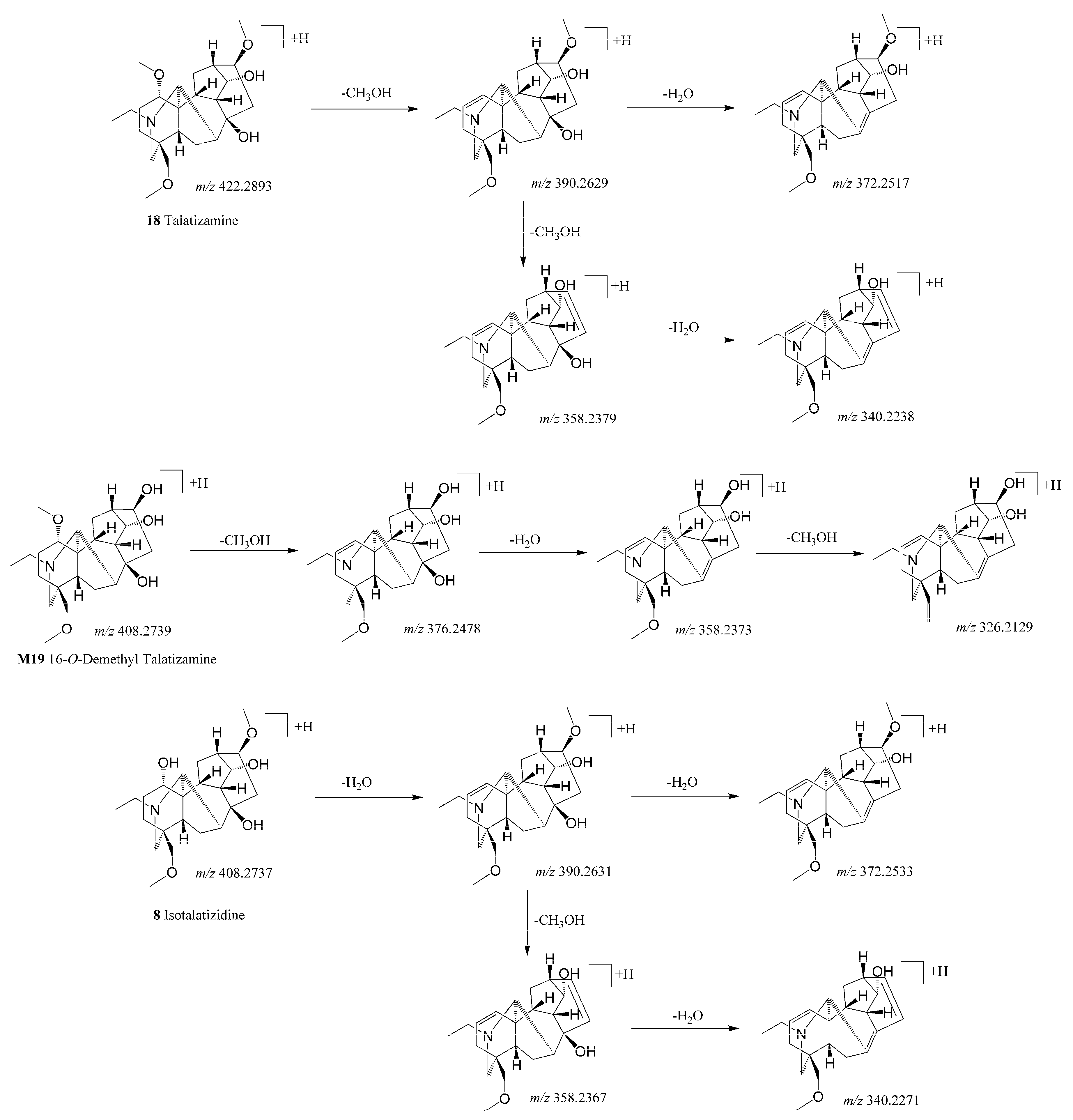
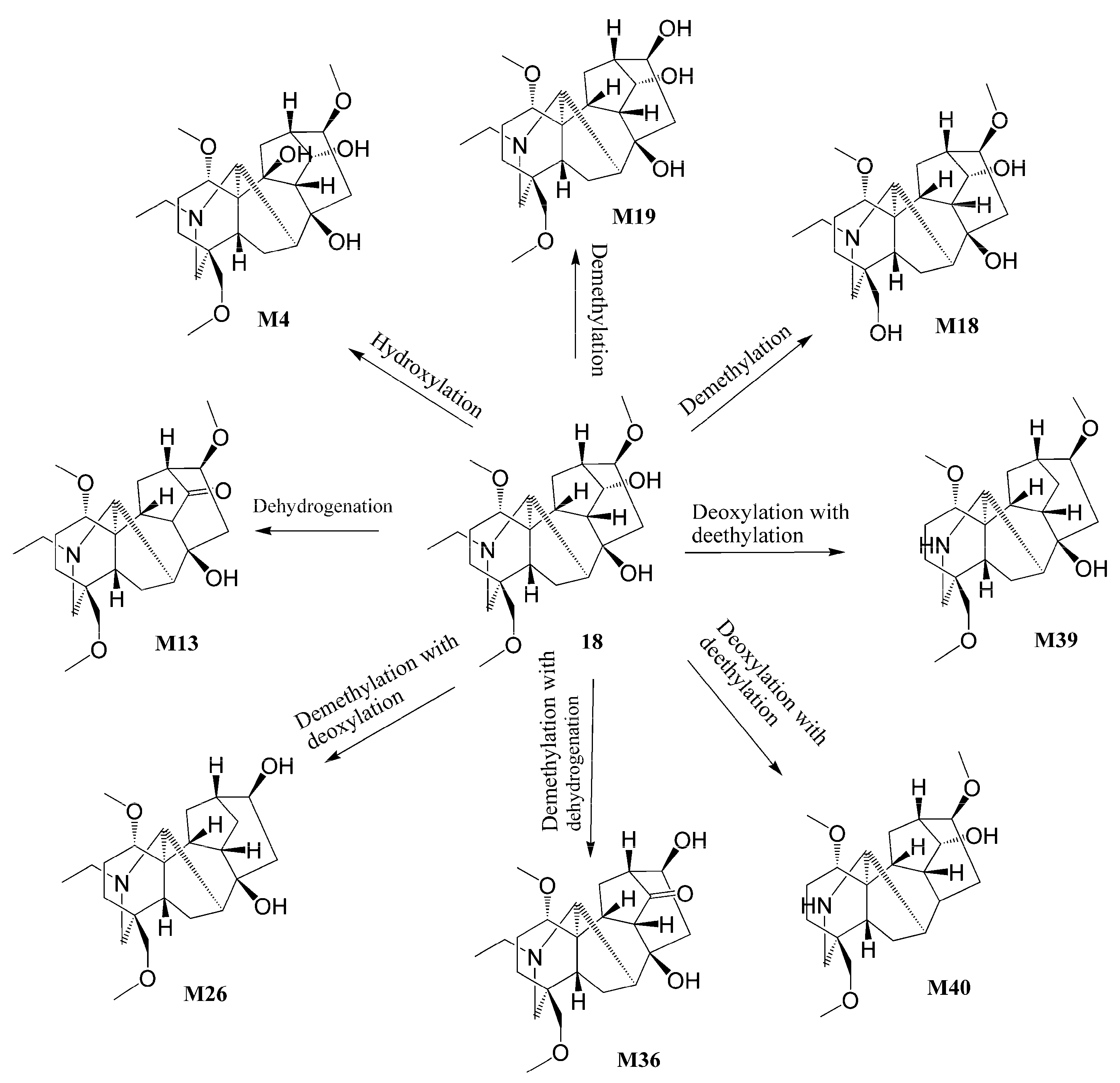
3.3.1. Identification of Alkaloid Metabolites
3.3.2. Identification of Phenolic Compound Metabolites
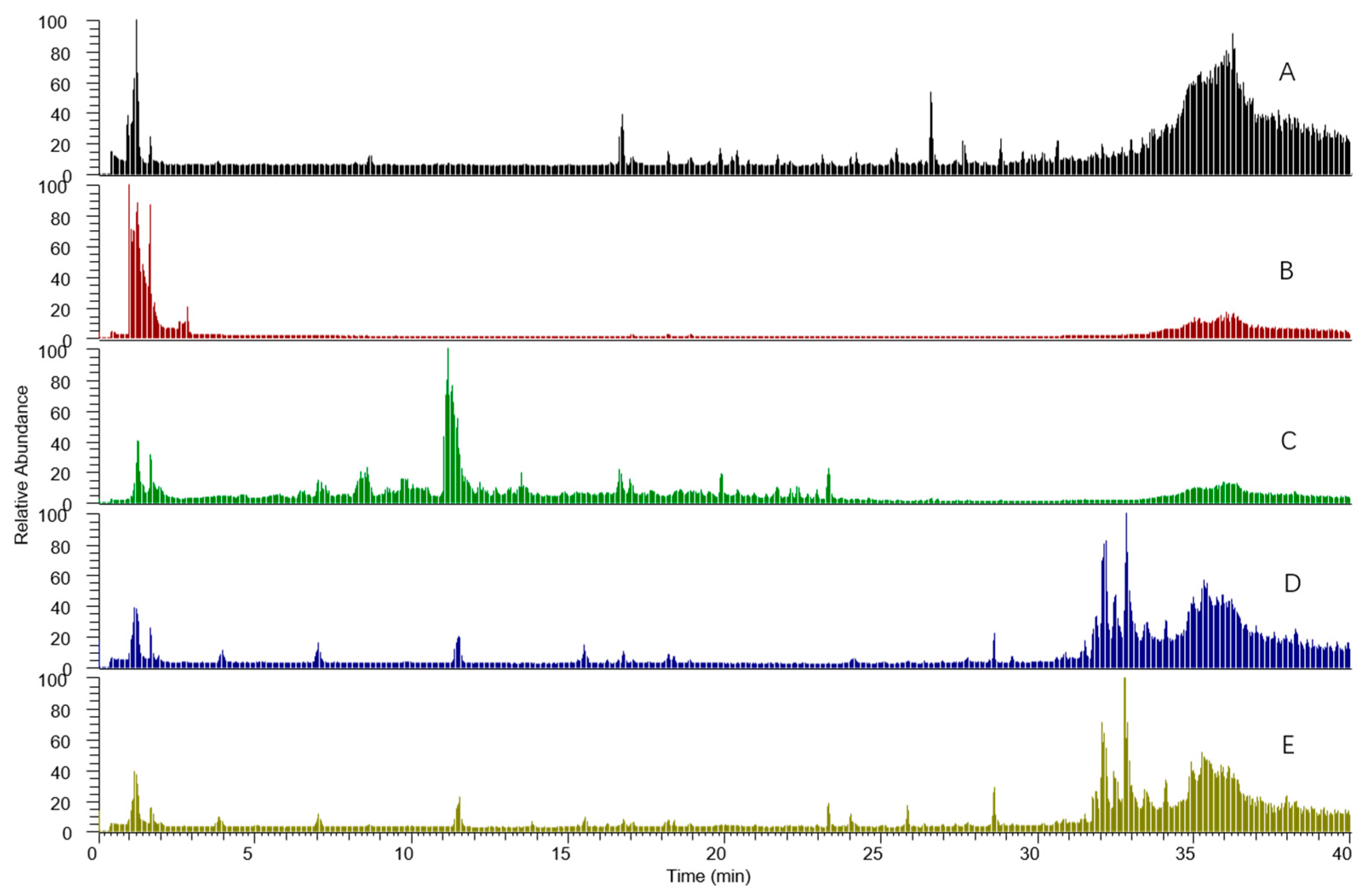
3.4. Difference between Urine and Plasma Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, L.Y.; Kang, Q.M.; Zhang, Y.; Chen, M.; Wang, Z.F.; Wu, Y.H.; Gao, H.T.; Zhong, Z.F.; Tan, W. Glycyrrhizae Radix et Rhizoma: The popular occurrence of herbal medicine applied in classical prescriptions. Phytother. Res. 2023, 37, 3135–3160. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.C.; Zhang, S.Y.; Lei, S.S.; Wang, D.N.; Peng, B.; Shi, R.P.; Chong, C.M.; Zhong, Z.F.; Wang, Y.T. A comprehensive review of the classical prescription Yiguan Jian: Phytochemistry, quality control, clinical applications, pharmacology, and safety profile. J. Ethnopharmacol. 2024, 319, 117230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Han, X.-X.; Mao, C.-Q.; Xie, H.; Chen, L.-H.; Mao, J.; Lu, T.-L.; Yan, G.-J. Opportunities and challenges in development of compound preparations of traditional Chinese medicine: Problems and countermeasures in research of ancient classical prescriptions. China J. Chin. Mater. Medica 2019, 44, 4300–4308. [Google Scholar]
- Chen, Z.K.; Wang, X.N.; Li, Y.Y.; Wang, Y.H.; Tang, K.L.; Wu, D.F.; Zhao, W.Y.; Ma, Y.M.; Liu, P.; Cao, Z.W. Comparative network pharmacology analysis of classical TCM prescriptions for chronic liver disease. Front. Pharmacol. 2019, 10, 1353. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.Y.; Wei, X.H.; Wu, X.F.; Chen, J.; Xia, H.; Xia, G.Y.; Lin, S.; Shang, H.C. Pharmacological research progress of five classical prescriptions in treatment of chronic heart failure. China J. Chin. Mater. Medica 2023, 48, 6324–6333. [Google Scholar]
- Zhou, Q.; Meng, P.; Zhang, Y.; Chen, P.; Wang, H.B.; Tan, G.G. The compatibility effects of sini decoction against doxorubicin-induced heart failure in rats revealed by mass spectrometry-based serum metabolite profiling and computational analysis. J. Ethnopharmacol. 2020, 252, 112618. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xiao, S.; Li, Z.H.; Ai, N.; Fan, X.H. Chemical and Metabolic Profiling of Si-Ni Decoction Analogous Formulae by High performance Liquid Chromatography-Mass Spectrometry. Sci. Rep. 2015, 5, 11638. [Google Scholar] [CrossRef]
- Chen, S.; Wu, S.; Li, W.H.; Chen, X.F.; Dong, X.; Tan, G.G.; Zhang, H.; Hong, Z.Y.; Zhu, Z.Y.; Chai, Y.F. Investigation of the therapeutic effectiveness of active components in Sini decoction by a comprehensive GC/LC-MS based metabolomics and network pharmacology approaches. Mol. Biosyst. 2014, 10, 3310–3321. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.G.; Wang, X.; Liu, K.; Dong, X.; Liao, W.T.; Wu, H. Correlation of drug-induced and drug-related ultra-high performance liquid chromatography-mass spectrometry serum metabolomic profiles yields discovery of effective constituents of Sini decoction against myocardial ischemia in rats. Food Funct. 2018, 9, 5528–5535. [Google Scholar] [CrossRef]
- Hu, Q.; Chen, M.; Yan, M.M.; Wang, P.L.; Lei, H.M.; Xue, H.Y.; Ma, Q. Comprehensive analysis of Sini decoction and investigation of acid-base self-assembled complexes using cold spray ionization mass spectrometry. Microchem. J. 2020, 173, 107008. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, M.; Zhang, W.; Chen, J.; Zhu, Z.Y.; Cao, H.; Chai, Y.F. Comparative pharmacokinetics of three monoester-diterpenoid alkaloids after oral administration of Acontium carmichaeli extract and its compatibility with other herbal medicines in Sini Decoction to rats. Biomed. Chromatogr. 2015, 29, 1076–1083. [Google Scholar] [CrossRef]
- Zhou, Q.; Meng, P.; Wang, H.B.; Dong, X.; Tan, G.G. Pharmacokinetics of monoester-diterpenoid alkaloids in myocardial infarction and normal rats after oral administration of Sini decoction by microdialysis combined with liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 2019, 33, e4406. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Chen, Q.S.; Ge, J.Y.; Liu, X.; Wang, X.X.; Zhan, Q.; Zhang, H.; Zhang, G.Q. Pharmacokinetic interaction of aconitine, liquiritin and 6-gingerol in a traditional Chinese herbal formula. Sini Decoction 2018, 48, 45–52. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, X.Q.; Shi, M.; Chen, C.W.; Sun, Y.; Li, J.J.; Xiong, Y.X.; Chen, J.J.; Li, F.Z. Serum metabolomics analysis reveals that obvious cardioprotective effects of low dose Sini decoction against isoproterenol-induced myocardial injury in rats. Phytomedicine 2017, 31, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.S.; Luo, D.W.; Zhong, G.Y.; Yang, S.L.; Ouyang, H.; Rao, X.Y.; Feng, Y.L. Exploration of plant metabolomics variation and absorption characteristics of water-extracted Rheum tanguticum and ethanol-extracted Rheum tanguticum by UHPLC-Q-TOF-MS/MS. Phytochem. Anal. 2024, 35, 288–307. [Google Scholar] [CrossRef]
- Ye, L.H.; He, X.X.; Yan, M.Z.; Chang, Q. Identification of in vivo components in rats after oral administration of lotus leaf flavonoids using ultra fast liquid chromatography with tandem mass spectrometry. Anal. Methods 2014, 6, 6088–6094. [Google Scholar] [CrossRef]
- Tao, J.H.; Zhao, M.; Jiang, S.; Zhang, W.; Xu, B.H.; Duan, J.A. UPLC-Q-TOF/MS-based metabolic profiling comparison of four major bioactive components in normal and CKD rat plasma, urine and feces following oral administration of Cornus officinalis Sieb and Rehmannia glutinosa Libosch herb couple extract. J. Pharm. Biomed. Anal. 2018, 161, 254–261. [Google Scholar] [CrossRef]
- Deng, F.; Li, X.M.; Gong, Q.Q.; Zheng, Z.X.; Zeng, L.; Zhang, M.J.; Duan, T.Y.; Liu, X.; Zhang, M.Z.; Guo, D.L. Identification of in vivo metabolites of Citri Sarcodactylis Fructus by UHPLC-Q/Orbitrap HRMS. Phytochem. Anal. 2023, 34, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Liu, S.; Xing, J.P.; Pi, Z.F.; Liu, Z.Q.; Song, F.R. Systematic study on metabolism and activity evaluation of Radix Scutellaria extract in rat plasma using UHPLC with quadrupole time-of-flight mass spectrometry and microdialysis intensity-fading mass spectrometry. J. Sep. Sci. 2018, 41, 1704–1710. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, J.P.; Bai, J.Q.; Wei, M.J.; Zhang, J.; Huang, Z.H.; Qu, G.H.; Xu, W.; Qiu, X.H. Chemical profiles and metabolite study of raw and processed Polygoni Multiflori Radix in rats by UPLC-LTQ-Orbitrap MSn spectrometry. Chin. J. Nat. Med. 2018, 16, 375–400. [Google Scholar]
- Su, C.Y.; Wang, J.H.; Chang, T.Y.; Shih, C.L. Mass defect filter technique combined with stable isotope tracing for drug metabolite identification using high-resolution mass spectrometry. Anal. Chim. Acta 2022, 1208, 339814. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.H.; Chen, X.; Yin, X.M.; Wang, M.Y.; Zhao, J.J.; Ren, Y. A strategy integrating parent ions list-modified mass defect filtering-diagnostic product ions for rapid screening and systematic characterization of flavonoids in Scutellaria barbata using hybrid quadrupole-orbitrap high-resolution mass spectrometry. J. Chromatogr. A 2022, 1674, 463149. [Google Scholar] [CrossRef]
- Wang, B.L.; Lu, Y.M.; Hu, X.L.; Feng, J.H.; Shen, W.; Wang, R.; Wang, H. Systematic Strategy for Metabolites of Amentoflavone In Vivo and In Vitro Based on UHPLC-Q-TOF-MS/MS Analysis. J. Agric. Food Chem. 2020, 68, 14808–14823. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, J.; Zhu, D.Y.; Huang, J.; Huang, Z.H.; Bai, J.Q.; Qiu, X.H. Rapid separation and characterization of diterpenoid alkaloids in processed roots of Aconitum carmichaeli using ultra high performance liquid chromatography coupled with hybrid linear ion trap-Orbitrap tandem mass spectrometry. J. Sep. Sci. 2014, 37, 2864–2873. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.F.; XU, Y.; Liu, H.P.; Shang, Q.; Qiu, J.Q.; Xu, W. Chemical analysis of classical prescription Qianghuo Shengshi standard decoction by UHPLC-Q Exactive Orbitrap MS. China J. Chin. Mater. Medica 2022, 47, 343–357. [Google Scholar]
- Lu, F.Y.; Cai, H.; Li, S.M.; Xie, W.; Sun, R.J. The Chemical Signatures of Water Extract of Zingiber officinale Rosc. Molecules 2022, 27, 7818. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Y.; Li, H.L.; Song, F.R.; Liu, C.M.; Liu, Z.Q.; Liu, S.Y. Studies on Triterpenoids and Flavones in Glycyrrhiza uralensis Fisch by HPLC-ESI-MSn and FT-ICR-MSn. Chin. J. Chem. 2009, 27, 299–305. [Google Scholar] [CrossRef]
- Avula, B.; Bae, J.Y.; Chittiboyina, A.G.; Wang, Y.H.; Wang, M.; Zhao, J.P.; Ali, Z.; Brinckmann, J.A.; Li, J.; Wu, C. Chemometric analysis and chemical characterization for the botanical identification of Glycyrrhiza species (G. glabra, G. ura-lensis, G. inflata, G. echinata and G. lepidota) using liquid chromatography-quadrupole time of flight mass spectrometry (LC-QToF) Bharathi. J. Food Compos. Anal. 2022, 112, 104679. [Google Scholar]
- Zhang, J.; Huang, Z.H.; Qiu, X.H.; Yang, Y.M.; Zhu, D.Y.; Xu, W. Neutral fragment filtering for rapid identification of new diester-diterpenoid alkaloids in roots of Aconitum carmichaeli by ultra-high-pressure liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry. PLoS ONE 2012, 7, e52352. [Google Scholar] [CrossRef]
- He, G.N.; Wang, X.X.; Liu, W.R.; Li, Y.L.; Shao, Y.M.; Liu, W.D.; Liang, X.D.; Bao, X. Chemical constituents, pharmacological effects, toxicology, processing and compatibility of Fuzi (lateral root of Aconitum carmichaelii Debx): A review. J. Ethnopharmacol. 2023, 307, 116160. [Google Scholar] [CrossRef]
- Chen, X.; Tan, P.; He, R.; Liu, Y.G. Study on the fragmentation pathway of the aconitine-type alkaloids under electrospray ionization tandem mass spectrometry utilizing quantum chemistry. J. Pharm. Innov. 2013, 8, 83–89. [Google Scholar] [CrossRef]
- Hu, R.; Zhao, J.; Qi, L.W.; Li, P.; Jing, S.L.; Li, H.J. Structural characterization and identification of C19- and C20-diterpenoid alkaloids in roots of Aconitum carmichaeli by rapid-resolution liquid chromatography coupled with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 1619–1635. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.S.; Soroka, D.; Chen, X.X.; Leung, T.C.; Sang, S.M. 6-Gingerdiols as the major metabolites of 6-gingerol in cancer cells and in mice and their cytotoxic effects on human cancer cells. Agric Food Chem. 2012, 60, 11372–11377. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.N.; Tran, C.A.; Trinh, T.D.; Thi, N.L.N.; Phan, H.N.; Le, V.N.; Le, N.H.; Phung, V. UHPLC-Q-TOF-MS/MS Dereplication to identify chemical constituents of Hedera helix leaves in Vietnam. J. Anal. Methods Chem. 2022, 2022, 1167265. [Google Scholar] [CrossRef] [PubMed]
- Savarino, P.; Demeyer, M.; Decroo, C.; Colson, E.; Gerbaux, P. Mass spectrometry analysis of saponins. Mass Spectrom. Rev. 2023, 42, 954–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jiang, M.Y.; Wei, X.Y.; Shi, J.F.; Geng, Z.; Yang, S.S.; Fu, C.M.; Guo, L. Rapid discovery of chemical constituents and absorbed components in rat serum after oral administration of Fuzi-Lizhong pill based on high-throughput HPLC-Q-TOF/MS analysis. Chin. Med. 2019, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Chen, X.F.; Lu, D.Y.; Dong, X.; Zhang, G.Q.; Chai, Y.F. Using cell membrane chromatography and HPLC-TOF/MS method for in vivo study of active components from roots of Aconitum carmichaeli. J. Pharm. Anal. 2011, 1, 125–134. [Google Scholar] [PubMed]
- Zhang, M.; Peng, C.; Li, X.B. In vivo and in vitro metabolites from the main diester and monoester diterpenoid alkaloids in a traditional Chinese herb, the Aconitum species. Evid. Based Complement. Altern. Med. 2015, 2015, 252434. [Google Scholar]
- Zhang, L.; Wang, C.X.; Wu, J.; Wang, T.Y.; Zhong, Q.Q.; Du, Y.; Ji, S.; Wang, L.; Guo, M.Z.; Xu, S.Q. Metabolic profiling of mice plasma, bile, urine and feces after oral administration of two licorice flavonones. J. Ethnopharmacol. 2020, 257, 112892. [Google Scholar] [CrossRef]
- Li, Y.Y.; Yang, L.; Chai, X.; Yang, J.J.; Wang, Y.F.; Zhu, Y. Four major urinary metabolites of liquiritigenin in rats and their anti-platelet aggregation activity. Chem. Nat. Compd. 2018, 54, 443–446. [Google Scholar] [CrossRef]
- Xiong, L.; Peng, C.; Xie, X.F.; Guo, L.; He, C.J.; Geng, Z.; Wan, F.; Dai, O.; Zhou, Q.M. Alkaloids Isolated from the Lateral Root of Aconitum carmichaelii. Molecules 2012, 17, 9939–9946. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.X.; Jian, X.X.; Cai, X.F.; Chao, R.B.; Chen, Q.H.; Chen, D.L.; Wang, X.L.; Wang, F.P. Cardioactive C19-diterpenoid alkaloids from the lateral roots of Aconitum carmichaeli “Fu Zi”. Chem. Pharm. Bull. 2012, 60, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Q.; Qi, J.Y.; He, Q.Q.; Ma, D.L.; Li, J.; Chu, X.; Zuo, S.J.; Zhang, Y.X.; Li, L.; Chu, L. Liquiritigenin protects against myocardial ischemic by inhibiting oxidative stress, apoptosis, and L-type Ca2+ channels. Phytother. Res. 2022, 36, 3619–3631. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, J.J.; Guo, H.L.; Sun, S.N.; Wang, S.F.; Zhang, Y.L.; Li, S.Y.; Qiao, Y.J. [6]-Gingerol: A Novel AT1 Antagonist for the Treatment of Cardiovascular Disease. Planta Medica 2013, 79, 322–326. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Liu, X.Y.; Jiang, Y.P.; Wang, N.N.; Li, F.; Xin, H.L. 6-Gingerol attenuates ischemia-reperfusion-induced cell apoptosis in human AC16 cardiomyocytes through HMGB2-JNK1/2-NF-κB pathway. Evid. Based Complement. Altern. 2019, 2019, 8798653. [Google Scholar] [CrossRef] [PubMed]
| Alkaloids (from RALP) | Phenolic and Saponin Compounds (from RGP and RZ) | |
|---|---|---|
| Karakolidine | Liquiritigenin | Formononetin |
| Fuziline | Isoliquiritigenin | Ononin |
| Neoline | Liquiritin | Glycyrrhizic Acid |
| Songorine | Licochalcone B | Glycyrrhetinic Acid |
| 14-Benzoylhypaconine | Licochalcone C | Uralsaponin C |
| Talatizamine | Licochalcone D | Licoricesaponin G2 |
| Karakoline | Licoflavone C | Glycycoum-Arim |
| 14-Benzoylmesaconine | Licoflavone A | Glycyrol |
| Mesaconitine | Licoricidin | Glycyrin |
| Hypaconitine | Licoleafol | 6-Gingerol |
| Aconitine | Gancaonin M | |
| ID | [M+H]+(m/z) | Formula | tR (min) | Error (ppm) | ms/ms | Identification | ClogP | Area | |
|---|---|---|---|---|---|---|---|---|---|
| Urine Plasma | |||||||||
| Alkaloids | |||||||||
| 1. | 394.25839 | C22H35NO5 | 3.51 | −1.58 | 376.2474, 358.2376, 344.2229, 326.2116, 243.2516 | Karakolidine | +++ | ||
| 2. | 394.25820 | C22H35NO5 | 4.02 | −1.42 | 376.2476, 358.2367, 340.2268, 328.2260, 307.4473, 218.6333 | Chuanfumine | +++ | ||
| 3. | 439.25229[M-H]− | C23H37NO7 | 4.25 | 0.56 | 392.2438, 344.2226, 295.8235, 193.8604, 146.9375 | 9-Hydroxysenbusine A | + | ||
| 4. | 424.26871 | C23H37NO6 | 6.69 | −1.55 | 406.2584, 388.2478, 356.2207, 154.1227 | Senbusine A | −2.70 | +++ | |
| 5. | 486.26941 | C24H39NO9 | 7.01 | −0.72 | 454.2438, 436.2322, 404.2069, 378.1887, 372.1793, 319.9836 | Mesaconine | ++ | ||
| 6. | 424.26871 | C23H37NO6 | 7.74 | −1.55 | 406.2581, 388.2472, 356.2210, 154.1231 | Senbusine B | 0.16 | ++ | |
| 7. | 378.26306 | C22H35NO4 | 8.20 | −2.18 | 360.2524, 342.2431, 328.2268, 242.3140 | Karakoline | ++ | ||
| 8. | 408.27371 | C23H37NO5 | 8.26 | −1.81 | 390.2631, 372.2533, 358.2367, 340.2271 | Isotalatizidine | ++ | ||
| 9. | 358.23691 | C22H31NO3 | 9.17 | −2.13 | 340.2265 | Songorine | + | ||
| 10. | 360.25293 | C22H33NO3 | 9.18 | −1.09 | - | Napelline | ++ | ||
| 11. | 330.20569 | C20H27NO3 | 9.70 | −2.07 | 236.8785, 170.7432, 152.4712 | Hetisine | ++ | ||
| 12. | 470.27435 | C24H39NO8 | 10.51 | −1.05 | - | Hypaconine | ++ | ||
| 13. | 454.27933 | C24H39NO7 | 11.33 | −1.26 | 436.2685, 418.2609, 404.2422, 154.1227 | Fuziline | +++ | ||
| 14. | 438.28445 | C24H39NO6 | 11.58 | −1.28 | 420.2736, 402.2617, 388.2472, 356.2214, 278.6899 | Neoline | +++ | ||
| 15. | 420.27390 | C24H37NO5 | 12.31 | −1.32 | 402.2632, 384.2512, 370.2359, 342.2414, 324.2322, 251.1396 | 14-Acetylkarakoline | + | ||
| 16. | 484.28937 | C25H41NO8 | 12.41 | −2.33 | - | Deoxyaconine | + | ||
| 17. | 342.16931 | C20H23NO4 | 12.89 | −1.96 | 297.1120, 282.0887, 237.0910, 219.0804, 191.0860 | N-Methyl-laurotetanine | +++ | ||
| 18. | 422.28931 | C24H39NO5 | 13.64 | −1.88 | 390.2629, 372.2517, 358.2379, 340.2238, 98.0970 | Talatizamine | ++++ | ++ | |
| 19. | 420.23825 | C23H33NO6 | 15.48 | −0.86 | 402.2268, 370.1989, 293.7002, 154.1224 | Giraldine F | ++ | ||
| 20. | 452.29996 | C25H41NO6 | 15.54 | −1.57 | 420.2740, 388.2465, 356.2219, 209.1644, 154.1228, 114.0916 | Chasmanine | ++++ | ||
| 21. | 464.30038 | C26H41NO6 | 16.933 | −0.60 | 432.2740, 414.2626, 400.2474, 372.2535, 265.1608, 235.1487, 154.1225 | 14-Acetyltalatizamine | ++++ | ++ | |
| 22. | 606.28992 | C31H43NO11 | 17.26 | −1.60 | 574.2627, 556.2545, 524.2269, 506.2188, 492.1945, 261.0641, 173.0955, 105.0341 | 14-Benzoyl-10-OH-mesaconine | ++ | ||
| 23. | 544.28955 | C30H41NO8 | 19.47 | −0.95 | 512.2635, 494.2548, 480.2364, 462.2258, 390.2286, 270.0846, 105.0340 | Gadenine | + | ||
| 24. | 590.29490 | C31H43NO10 | 19.51 | −1.78 | 558.2616, 540.2575, 419.7593, 307.8019, 246.8854, 105.0339 | 14-Benzoylmesaconine | ++ | ||
| 25. | 540.29486 | C31H41NO7 | 20.01 | −1.33 | 504.2730, 462.2614, 382.2463, 340.2256, 322.2149, 304.2042 | Aconicarchamine B | + | ||
| 26. | 604.31060 | C32H45NO10 | 20.53 | −1.68 | 572.2811, 554.2750, 522.2495, 490.2176, 340.3151, 105.0341 | 14-Benzoylaconine | ++ | ||
| 27. | 574.30010 | C31H43NO9 | 21.20 | −1.65 | 542.2744, 510.2461, 304.5384, 198.1281, 105.0339 | 14-Benzoylhypaconine | +++ | ||
| 28. | 618.29210[M-H]− | C32H45NO11 | 21.21 | 0.31 | 384.9167, 351.8983, 270.7405, 190.9267 | 14-Benzoyl-10-OH-aconine | ++ | ||
| 29. | 648.30023 | C33H45NO12 | 21.93 | −1.98 | 588.2775, 556.2513, 455.3509, 370.1645, 105.0340 | 10-OH-mesaconitine | ++ | ||
| 30. | 558.30530 | C31H43NO8 | 22.07 | −1.52 | 526.2800, 508.2674, 232.0710182.0626, 105.0341 | 14-Benzoyl-doxyhypaconine | ++ | ||
| 31. | 588.31561 | C32H45NO9 | 22.32 | −1.24 | 556.2905, 524.2639, 506.2443, 346.4250, 253.7027, 154.1226, 105.0341 | 14-Benzoyldeoxyaconine | + | ||
| 32. | 542.31061 | C31H43NO7 | 23.18 | −1.15 | 510.2846, 492.2735, 482.2483, 460.2504, 154.1231 | 14-Benzoylneoline | + | ||
| 33. | 632.30591 | C33H45NO11 | 23.18 | −0.63 | 572.2844, 540.2551, 522.2487, 508.2299, 354.1694, 105.0341 | Mesaconitine * | +++ | ||
| 34. | 662.31683 | C34H47NO12 | 23.39 | −0.27 | - | Aconifine | ++ | ||
| 35. | 614.29553 | C33H43NO10 | 24.17 | −0.72 | 554.2743, 494.2534, 372.2162, 344.21622, 203.5583, 105.0341 | 2,3-didehydrohypaconitine | + | ||
| 36. | 646.32135 | C34H47NO11 | 24.58 | −0.84 | 586.3002, 554.2727, 526.2797, 494.2520, 368.1843, 105.0340 | Aconitine * | ++ | ||
| 37. | 616.31079 | C33H45NO10 | 24.61 | −0.83 | 556.2903, 524.2634, 496.2750, 464.2434, 338.1741, 310.1812, 105.0341 | Hypaconitine * | ++++ | ||
| 38. | 600.31592 | C33H45NO9 | 24.96 | −1.32 | 540.2948, 508.2683, 480.2747, 476.2424, 448.2475, 354.2031, 254.4337, 105.0339 | Secoyunaconitine | + | ||
| 39. | 572.32117 | C32H45NO8 | 24.95 | −1.10 | 484.2688, 456.2745, 382.2002, 322.1798, 294.1857, 158.0964 | 14-O-Anisoylneoline | + | ||
| 40. | 630.32635 | C34H47NO10 | 26.07 | −1.36 | 570.3046, 538.2788, 510.2882, 506.2528, 478.2571, 352.1898, 314.5361, 105.0341 | 3-Deoxyaconitine | +++ | ||
| 41. | 614.33173 | C34H47NO9 | 27.60 | −0.53 | - | Chasmaconitine | ++ | ||
| Phenolic compounds | |||||||||
| 42. | 209.04474[M-H]− | C10H10O5 | 8.56 | −3.85 | 165.0545, 121.0281, 103.9187, 87.9238, 59.0123 | Hydroxyferulic acid | +++ | ++ | |
| 43. | 433.13394[M-H]− | C18H24O12 | 9.88 | 0.08 | 161.0442, 125.0230, 99.0436 | Asperulosidic acid | ++ | ||
| 44. | 433.11407[M-H]− | C21H22O10 | 13.87 | −0.19 | 271.0615, 151.0024 | 5-Hydroxyliquiritin | ++ | ||
| 45. | 593.15137[M-H]− | C27H30O15 | 15.51 | 0.29 | 473.1098, 383.9785, 353.0774 | Vitexin II | +++ | ||
| 46. | 563.14055[M-H]− | C26H28O14 | 15.83 | −0.51 | 473.1089, 443.0985, 383.0769, 253.0502, 146.9367 | Vitexin I | + | ||
| 47. | 417.11890[M-H]− | C21H22O9 | 16.74 | −0.32 | 255.0662, 153.0182, 135.0074, 119.0488 | Liquiritin * | ++++ | ||
| 48. | 505.13339 | C24H24O12 | 18.72 | −0.89 | 257.0809, 137.0234 | Malonyl liquiritin | + | ||
| 49. | 505.13358 | C24H24O12 | 18.99 | −0.07 | 257.0810, 137.0234 | Malonyl liquiritin | + | ||
| 50. | 431.13280 | C22H22O9 | 20.26 | −1.97 | 269.0809 | Ononin | ++++ | ||
| 51. | 417.11908[M-H]− | C21H22O9 | 20.39 | −1.01 | 255.0662, 153.0180, 135.0072, 119.0481 | Neoliquiritin | 0.75 | +++ | |
| 52. | 417.11900[M-H]− | C21H22O9 | 20.74 | −2.47 | 255.0662, 153.01816, 135.0074, 119.0488 | Isoliquiritin | 1.28 | ++ | |
| 53. | 285.07670[M-H]− | C16H14O5 | 21.23 | −0.31 | 270.0536, 253.0505, 177.0182, 150.0310, 108.0203 | Licochalcone B | ++ | ||
| 54. | 255.06560 | C15H10O4 | 21.30 | 0.10 | 227.0704, 199.0754, 145.0286, 137.0234 | Dihydroxyflavone | ++++ | ||
| 55. | 255.06580[M-H]− | C15H12O4 | 21.70 | −0.36 | 153.0180, 135.0073, 119.0487, 91.0173 | Liquiritigenin * | ++++ | +++ | |
| 56. | 295.19040 | C17H26O4 | 23.34 | −0.22 | 177.0914, 163.0755, 137.0598, 131.0493, 99.0809 | 6-Gingerol | ++++ | ++ | |
| 57. | 269.04530[M-H]− | C15H10O5 | 24.34 | 0.38 | 233.1537, 181.0644 | Genistein | +++ | ||
| 58. | 255.06586[M-H]− | C15H12O4 | 26.29 | −0.49 | 153.0179, 135.0073, 119.0487, 91.0174 | Isoliquiritigenin * | ++++ | +++ | |
| 59. | 269.08170 | C16H12O4 | 26.58 | 1.04 | 253.0497, 237.0554, 213.0911, 118.0418, 107.0497 | Formononetin * | ++++ | +++ | |
| 60. | 367.11790[M-H]− | C21H20O6 | 27.75 | −1.99 | 352.0944, 309.0400, 298.0476, 283.0247 | Glycycoum-arim/Licocoumarione | +++ | ||
| 61. | 271.09565 | C16H14O4 | 28.56 | 1.19 | 254.2579, 161.0599, 137.0598, 123.04440, 100.0763 | Echinatin | ++ | ||
| 62. | 355.11835[M-H]− | C20H20O6 | 28.60 | −1.07 | 328.1265, 269.11820, 269.11820, 178.9975, 125.0230 | 8-Dimethylallyleriodictyol/6-Dimethylallyleriodictyol | ++ | ||
| 63. | 277.18008 | C17H24O3 | 28.84 | 2.28 | 177.0912, 145.0649, 137.0598 | 6-Shogaol | +++ | ++ | |
| 64. | 355.15480[M-H]− | C21H24O5 | 29.81 | −1.06 | 323.1284, 233.1176, 207.1017, 135.0438, 125.0230, 109.0280 | Isopentadienyl glycyrrhizoflavone | ++ | ||
| 65. | 367.11790[M-H]− | C21H20O6 | 29.53 | −2.00 | 309.0400, 297.0400, 284.0325, 203.0702 | Glycycoum-arim/Licocoumarione | +++ | ||
| 66. | 321.11262[M-H]− | C20H18O4 | 30.07 | −1.93 | 306.0892, 174.9549 | Licoflavone A | + | ||
| 67. | 353.10290[M-H]− | C20H18O6 | 30.17 | −1.33 | 339.1187, 321.1126, 295.0613, 283.0614, 270.0535 | Isolicoflanonol | +++ | ++ | |
| 68. | 353.13782 | C21H20O5 | 30.22 | −1.59 | 299.0906, 297.0857, 267.0653, 199.0758, 147.0441, 135.0441 | Gancaonin M | ++ | ||
| 69. | 383.11273[M-H]− | C21H20O7 | 30.39 | −2.33 | 338.2439, 247.1310, 227.0704, 207.1015, 155.0337, 140.0101 | Licopyranocoumarin | + | ||
| 70. | 383.14828 | C22H22O6 | 30.66 | −1.50 | 327.0859, 299.0913, 191.0704 | Glycyrin | ++ | ++ | |
| 71. | 355.15320 | C21H22O5 | 30.94 | −2.01 | 289.0549, 287.0553, 191.1067, 153.0548, 69.0708 | Licobenzofuran/liconeolignan | +++ | ||
| 72. | 337.10780[M-H]− | C20H18O5 | 31.02 | −1.07 | 314.0428, 282.0531 | Licoflavone C | ++ | ||
| 73. | 365.10239[M-H]− | C21H18O6 | 30.17 | −1.63 | 307.0244, 295.0245, 282.0169 | Isoglycyrol | 4.84 | ++ | |
| 74. | 365.10236[M-H]− | C21H18O6 | 31.12 | −1.99 | 307.0242, 295.0243, 282.0167 | Glycyrol | 5.04 | +++ | |
| 75. | 333.24170 | C21H32O3 | 34.17 | −1.99 | 177.0911, 145.0649, 137.0598 | 10-Shogaol | ++ | ||
| 76. | 279.23264[M-H]− | C18H32O2 | 38.27 | 1.13 | 261.2219, 199.8500 | Linoleic acid | ++ | ||
| Saponins | |||||||||
| 77. | 879.40173[M-H]− 881.41516 705.38361[M+H-glcA]+ 511.34122[agl+H-H2O]+ | C44H64O18 | 24.011 | −0.67 −1.38 | (−) 351.0557, 193.0342, 113.0229 (+) 511.3408, 493.3279, 451.3188, 141.0183 | Uralsaponin M | ++ | ||
| 78. | 837.39105[M-H]− 839.40466 469.33072[gal+H-H2O]+ | C42H62O17 | 25.49 | −0.79 −1.32 | (−) 351.05603, 289.05652, 193.03430, 175.02340, 113.02294 (+) 469.3304, 487.3415, 451.3209, 141.0184 | Yunganoside K2 | ++ | ||
| 79. | 837.39178[M-H]− 839.40491 469.33084[agl+H-H2O]+ | C42H62O17 | 26.07 | −0.84 −1.07 | (−) 351.05557, 289.05621, 193.03413, 175.02360, 113.02285 (+) 469.3304, 487.3413, 451.3198, 141.0183 | Licoricesaponin G2 | + | ||
| 80. | 471.34613 | C30H46O4 | 26.62 | −1.41 | 453.33508, 425.34262, 317.21100, 235.16887, 189.16374 | Glycyrrhetinic acid (enoxolone) * | + | ||
| 81. | 821.39630[M-H]− 823.40936 647.37744[M+H-glcA]+ 453.33554[agl+H-H2O]+ | C42H62O16 | 26.64 | 1.08 −1.70 | (−) 351.05573, 193.03406, 175.02338, 113.02288 (+) 453.3354, 471.3451, 435.3259 | Glycyrrhizic acid * | ++++ | ||
| 82. | 821.39612[M-H]− 823.40936 647.37787[M+H-glcA]+ 453.33585[agl+H-H2O]+ | C42H62O16 | 27.65 | 0.86 −0.47 | (−) 351.05640, 193.03404, 175.02319, 113.02289 (+) 453.3354, 435.3257 | Uralsaponin B or Licoricesaponine K2/H2 | ++ | ||
| ID | [M+H]+ (m/z) | Formula | tR (min) | Error (ppm) | ms/ms | Composition Change | Identification | ClogP | Area | |
|---|---|---|---|---|---|---|---|---|---|---|
| Urine | Plasma | |||||||||
| M1. | 410.25302 | C22H35NO6 | 3.72 | −1.69 | 392.2425, 374.2317, 360.2165, 342.2054 | +O | Hydroxy karakolidine | ++ | ||
| M2. | 374.23212 | C22H31NO4 | 7.55 | −1.25 | 356.2212, 338.2106, 198.1122 | +O | Hydroxy songorine | ++ | ||
| M3. | 394.25812 | C22H35NO5 | 7.76 | −1.85 | 376.2476, 358.2362, 98.0971, 58.0611 | +O | Hydroxy karakoline | ++ | ||
| M4. | 438.28433 | C24H39NO6 | 10.76 | 0.67 | 406.2588, 388.2476, 374.230, 356.2226 | +O | 10-Hydroxy talatizamine | ++ | ||
| M5. | 632.30560 | C33H45NO11 | 22.61 | −1.50 | 572.2853, 540.2590, 512.2641, 508.2310, 480.2390, 358.2004, 354.1703, 105.0341 | +O | Hydroxy hypaconitine | ++ | ||
| M6. | 392.24268 | C22H33NO5 | 8.28 | −1.21 | 374.2315, 344.2221, 312.1962, 114.0916 | -H2 | Dehydrogenated karakolidine | +++ | ||
| M7. | 392.24249 | C22H33NO5 | 8.95 | −2.15 | 374.2325, 344.2240 | -H2 | Dehydrogenated karakolidine | +++ | ||
| M8. | 452.26361 | C24H37NO7 | 9.85 | −1.48 | 434.2529, 416.2419, 204.2270, 384.2155 | -H2 | Dehydrogenated fuziline | +++ | ||
| M9. | 376.24780 | C22H33NO4 | 10.38 | −1.82 | 358.2373, 98.0969 | -H2 | Dehydrogenated karakoline | ++ | ||
| M10. | 376.24756 | C22H33NO4 | 10.96 | −1.15 | 358.2375, 234.0137, 98.0970 | -H2 | Dehydrogenated karakoline | +++ | ||
| M11. | 436.26895 | C24H37NO6 | 10.24 | −0.95 | 418.2581, 400.2475, 386.2315, 358.2355, 340.2265 | -H2 | Dehydrogenated neoline | +++ | + | |
| M12. | 436.26907 | C24H37NO6 | 10.64 | −0.67 | 418.2585, 400.2473, 386.2303, 358.2383 | -H2 | Dehydrogenated neoline | +++ | ||
| M13. | 420.27393 | C24H37NO5 | 13.60 | −1.33 | 388.2477, 370.2375, 98.0972 | -H2 | 14-Dehydrogenated talatizamine | +++ | ||
| M14. | 364.24744 | C21H33NO4 | 7.39 | −2.20 | 346.2374, 328.2268 | -CH2 | Demethyl karakoline | +++ | + | |
| M15. | 364.24740 | C21H33NO4 | 7.86 | −2.20 | 346.2370, 328.2266 | -CH2 | Demethyl karakoline | ++++ | + | |
| M16. | 424.26892 | C23H37NO6 | 10.68 | −1.05 | 406.2583, 374.2327, 356.2211, 342.2069, 154.1226 | -CH2 | Demethyl neoline | +++ | + | |
| M17. | 424.26890 | C23H37NO6 | 11.08 | −0.98 | 406.2581, 374.2317, 356.2222, 342.2076, 154.1228 | -CH2 | Demethyl neoline | ++++ | ||
| M18. | 408.27386 | C23H37NO5 | 9.87 | −1.44 | 376.2475, 358.2365, 326.2136 | -CH2 | 18-o-Demethyl talatizamine | −0.78 | ++++ | ++ |
| M19. | 408.27393 | C23H37NO5 | 11.17 | −1.29 | 376.2478, 358.2373, 326.2129 | -CH2 | 16-o-Demethyl talatizamine | −0.73 | ++++ | ++ |
| M20. | 602.29486 | C32H43NO10 | 21.31 | −1.70 | 542.2742, 510.2477, 482.2540, 478.2212, 324.1592, 105.0339 | -CH2 | Demethyl hypaconitine | ++++ | + | |
| M21. | 618.28992 | C32H43NO11 | 22.17 | −1.22 | 558.2684, 526.2423, 508.2394, 354.1695, 105.0341 | -CH2 | Demethyl mesaconitine | ++ | ||
| M22. | 330.20581 | C20H27NO3 | 8.19 | −1.70 | 312.1954 | -C2H4 | Deethyl songorine | +++ | ||
| M23. | 350.23181 | C20H31NO4 | 9.05 | −2.21 | 332.2215, 314.2106, 300.1958, 234.9901, 158.9743 | -C2H4 | Deethyl karakoline | +++ | ||
| M24. | 410.25314 | C22H35NO6 | 10.97 | −1.40 | 392.2423, 378.2271, 360.2163, 328.1906 | -C2H4 | Deethyl neoline | ++ | ||
| M25. | 408.27374 | C23H37NO5 | 8.96 | −1.74 | 390.2631, 372.2537, 358.2369 | -CH2O | Demethyl-deoxy neoline | ++++ | ++ | |
| M26. | 392.27896 | C23H37NO4 | 13.38 | −1.46 | 360.2527, 342.2436, 328.2265 | -CH2O | 16-O-Demethyl-14-deoxy Talatizamine | +++ | ||
| M27. | 602.29529 | C32H43NO10 | 23.87 | −1.20 | 542.2773, 510.2486, 478.2222, 324.1592, 105.0341 | -CH2O | Demethyl-deoxy mesaconitine | +++ | ||
| M28. | 360.25272 | C22H33NO3 | 9.38 | −1.68 | 342.2422, 324.2325, 121.0651 | -H2O | Dehydrated karakoline | +++ | + | |
| M29. | 360.25250 | C22H33NO3 | 9.92 | −2.11 | 342.2422, 324.2307 | -H2O | Dehydrated karakoline | ++++ | + | |
| M30. | 614.2517 | C33H43NO10 | 24.17 | −1.57 | 544.2743, 522.2518, 494.2534, 372.2162, 344.2215 | -H2O | Dehydrated mesaconitine | ++ | ||
| M31. | 380.24277 | C22H33NO5 | 8.81 | −1.01 | 362.2316, 344.2046, 330.2065 | -CH2+O | Demethyl-hydroxy karakoline | ++ | ||
| M32. | 438.24811 | C23H35NO7 | 12.09 | −1.06 | 420.2374, 402.2265, 392.2440, 374.2317 | -CH2-H2 | Dehydrogenated-demethyl fuziline | ++ | + | |
| M33. | 438.24890 | C23H35NO7 | 12.21 | 0.49 | 420.2367, 402.2283, 392.2442, 374.2323 | -CH2-H2 | Dehydrogenated-demethyl fuziline | ++ | + | |
| M34. | 362.23172 | C21H31NO4 | 8.15 | −0.45 | 344.2223, 185.0710 | -CH2-H2 | Dehydrogenated-demethyl karakoline | +++ | ||
| M35. | 422.25327 | C23H35NO6 | 10.99 | −1.07 | 390.2268, 406.2597, 390.2268, 374.2324 | -CH2-H2 | Dehydrogenated-demethyl neoline | +++ | + | |
| M36. | 406.25839 | C23H35NO5 | 10.70 | −1.01 | 388.2477, 370.2368, 328.2266 | -CH2-H2 | 14-Dehydrogenated-16-O-demethyl talatizamine | +++ | ||
| M37. | 346.20090 | C20H27NO4 | 7.79 | −1.12 | 328.1904, 296.1645, 268.1701, 251.1437 | -C2H4+O | N-Deethyl-hydroxy songorine | ++ | + | |
| M38. | 346.20071 | C20H27NO4 | 8.49 | −1.65 | 328.1903, 296.1650, 268.1699, 251.1429 | -C2H4+O | N-Deethyl-hydroxy songorine | +++ | ||
| M39. | 378.26337 | C22H35NO4 | 10.98 | −1.43 | 346.2371, 328.2279 | -C2H4-O | N-Deethyl-14-deoxy talatizamine | 0.88 | +++ | + |
| M40. | 378.26334 | C22H35NO4 | 11.23 | −1.45 | 346.2371, 328.2267 | -C2H4-O | N-Deethyl-8-deoxy talatizamine | 1.15 | +++ | |
| ID | [M+H]+ (m/z) | Formula | tR (min) | Error (ppm) | ms/ms | Composition Change | Identification | Area | |
|---|---|---|---|---|---|---|---|---|---|
| Urine Plasma | |||||||||
| M41. | 259.09701 | C15H14O4 | 17.57 | 2.53 | 153.0548, 135.0441, 107.0496 | +H2 | Hydrogenated liquiritigenin | ++++ | ++ |
| M42. | 259.09689 | C15H14O4 | 20.78 | 1.83 | 153.0549, 107.0497 | +H2 | Hydrogenated isoliquiritigenin | ++++ | +++ |
| M43. | 297.20602 | C17H28O4 | 22.11 | −0.23 | 177.0912, 163.0755, 137.0598, 131.0494 | +H2 | Hydrogenated 6-gingerol | ++++ | ++ |
| M44. | 269.08170[M-H]− | C16H14O4 | 26.82 | −0.03 | 254.0582, 153.0178, 135.0073, 91.0173 | +H2 | Hydrogenated formononetin | ++++ | ++ |
| M45. | 367.11792[M-H]− | C21H20O6 | 29.54 | −2.01 | 352.0936, 309.0400, 310.0434, 284.0325 | +H2 | Hydrogenated glycyrol | +++ | + |
| M46. | 355.15311 | C21H22O5 | 30.71 | −2.57 | 337.1065, 299.0912, 189.0911, 177.0546, 151.0393 | +H2 | Hydrogenated gancaonin M | ++ | |
| M47. | 309.20578 | C18H28O4 | 26.17 | −0.70 | 163.0756, 137.0599, 131.0494 | +CH2 | Methyl 6-gingerol | ++ | |
| M48. | 309.20572 | C18H28O4 | 26.80 | −0.92 | 179.0704, 150.068, 137.0598, 83.0864 | +CH2 | Methyl 6-gingerol | ++ | |
| M49. | 285.07587 | C16H12O5 | 18.49 | 0.51 | 270.0525, 253.0499, 299.0866, 225.0546, 123.0443 | +O | Hydroxy formononetin | +++ | |
| M50. | 273.07593 | C15H12O5 | 19.06 | 0.72 | 255.066, 179.0339, 153.0184, 147.0442, 123.044, 119.0496 | +O | Hydroxy liquiritigenin/isoliquiritigenin | +++ | ++ |
| M51. | 369.13266 | C21H20O6 | 29.18 | −1.71 | 351.1222, 229.0860, 193.0497, 165.0548, 151.0389 | +O | Hydroxy gancaonin M | +++ | + |
| M52. | 313.20038 | C17H28O5 | 15.07 | −0.96 | 203.1066, 163.0754, 137.0598 | +H2O | Hydrated 6-gingerol | +++ | |
| M53. | 273.07629[M-H]− | C15H14O5 | 20.08 | −1.36 | 255.0661, 167.0337, 109.0279 | +H2O | Hydrated liquiritigenin | +++ | |
| M54. | 273.07660[M-H]− | C15H14O5 | 23.05 | −0.68 | 255.0655, 151.0387, 135.0072, 109.0280 | +H2O | Hydrated isoliquiritigenin | +++ | |
| M55. | 293.17447 | C17H24O4 | 16.57 | −0.91 | 163.0756, 137.0598, 99.0811 | -H2 | Dehydrogenated 6-gingerol | ++ | + |
| M56. | 255.06552 | C15H10O4 | 18.49 | 0.41 | 227.0703, 199.0756, 137.0234 | -H2 | Dehydrogenated liquiritigenin | +++ | + |
| M57. | 255.06550 | C15H10O4 | 21.29 | −0.16 | 227.0699, 199.0755, 137.0234 | -H2 | Dehydrogenated isoliquiritigenin | ++++ | + |
| M58. | 307.15466[M-H]− | C17H24O5 | 26.17 | −1.41 | 275.1288, 171.1014, 153.0907, 121.0280, 111.0799 | -H2+O | Dehydrogenated-hydroxy 6-gingerol | ++ | |
| M59. | 277.18039 | C17H24O3 | 28.85 | 2.07 | 189.0914, 177.09123, 145.05493, 137.0597 | -H2-O | Dehydrated 6-gingerol | ++++ | ++ |
| M60. | 285.07648[M-H]− | C16H14O5 | 22.03 | −0.35 | 270.0533, 153.0180, 149.0594, 135.0073, 134.0358, 91.0174 | +CH2+O | Methyl-hydroxy liquiritigenin | +++ | + |
| M61. | 285.07645[M-H]− | C16H14O5 | 26.88 | −1.06 | 270.0535, 153.0180, 149.0595, 135.0073, 91.0174 | +CH2+O | Methyl-hydroxy isoliquiritigenin | ++ | |
| M62. | 299.09170 | C17H14O5 | 26.99 | 0.69 | 284.0680, 243.1061, 166.0268 | +CH2+O | Methyl-hydroxy formononetin | +++ | + |
| M63. | 335.02261[M-H]− | C15H12O7S | 18.62 | −1.33 | 255.0661, 199.0064, 135.0073, 119.0487 | +SO3 | Liquiritigenin sulfate | ++++ | +++ |
| M64. | 335.02271[M-H]− | C15H12O7S | 23.32 | −0.94 | 255.0663, 199.0055, 135.0073, 119.0486 | +SO3 | Isoliquiritigenin sulfate | ++++ | ++++ |
| M65. | 347.02263[M-H]− | C16H12O7S | 23.56 | −1.41 | 267.0664, 252.0427 | +SO3 | Formononetin sulfate | ++++ | +++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Zhan, Y.; Peng, M.; Xu, W.; Deng, G. Metabolite Profiling Analysis of the Tongmai Sini Decoction in Rats after Oral Administration through UHPLC-Q-Exactive-MS/MS. Metabolites 2024, 14, 333. https://doi.org/10.3390/metabo14060333
Zheng X, Zhan Y, Peng M, Xu W, Deng G. Metabolite Profiling Analysis of the Tongmai Sini Decoction in Rats after Oral Administration through UHPLC-Q-Exactive-MS/MS. Metabolites. 2024; 14(6):333. https://doi.org/10.3390/metabo14060333
Chicago/Turabian StyleZheng, Xianhui, Yingying Zhan, Mengling Peng, Wen Xu, and Guanghai Deng. 2024. "Metabolite Profiling Analysis of the Tongmai Sini Decoction in Rats after Oral Administration through UHPLC-Q-Exactive-MS/MS" Metabolites 14, no. 6: 333. https://doi.org/10.3390/metabo14060333
APA StyleZheng, X., Zhan, Y., Peng, M., Xu, W., & Deng, G. (2024). Metabolite Profiling Analysis of the Tongmai Sini Decoction in Rats after Oral Administration through UHPLC-Q-Exactive-MS/MS. Metabolites, 14(6), 333. https://doi.org/10.3390/metabo14060333





