Correction: Fadil et al. Isotope Ratio Outlier Analysis (IROA) for HPLC–TOFMS-Based Metabolomics of Human Urine. Metabolites 2022, 12, 741
Clarification
| Protocol Employed in Fadil et al. [1] | IROA TruQuant IQQ Workflow Kit Protocol before 12/2022 [8,9] | IROA TruQuant IQQ Workflow Kit Protocol after 12/2022 [10] | |
|---|---|---|---|
| Volume to reconstitute LTRS/vial | 80 µL | 40 µL | 30 µL |
| Volume to reconstitute IS/vial | 600 µL | 1200 µL | 900 µL |
| Sample volume | 20 µL | 40 µL | 30 µL |
| Sample dried | no | yes | yes |
| Volume of IS per sample | Urine sample is mixed with concentrated IS: 20 µL urine and 10 µL IS | 40 µL to reconstitute dried sample | 30 µL to reconstitute dried sample |
| Discovery Phase | Semi-Targeted IROA Search | ||
|---|---|---|---|
| M/Z tolerance, ppm | 20 | M/Z tolerance, ppm | 30 |
| Limit retention time range, min | 0.5–18 | Limit retention time range, min | 0.5–12 |
| Smoothing filter width | 9 | Smoothing filter width | 9 |
| Minimum base peak intensity | 5000 | Minimum base peak intensity | 10000 |
| Exclude peaks below intensity | 2500 | Exclude peaks below intensity | 5000 |
| Signal to noise cutoff | 3 | Signal to noise cutoff | 5 |
| Peak shape correlation cutoff | 0.3 | Peak shape correlation cutoff | 0.5 |
| Minimal score for isotope pattern quality | 30 | Minimal score for isotope pattern quality | 30 |
| Precursor isolation window, Da | 0.2 | Precursor isolation window, Da | 0.2 |
| MSMS precursor M/Z tolerance, ppm | 50 | MSMS precursor M/Z tolerance, ppm | 50 |
| Relative intensity cutoff for MSMS lookup (0–1) | 0.3 | Relative intensity cutoff for MSMS lookup (0–1) | 0.3 |
| %13C incorporation in sample | 95 | %13C incorporation in sample | 95 |
| %13C incorporation in control | 5 | %13C incorporation in Control | 1.1 |
| Maximal charge to report | 2 | Require at least two 12C isotopes | no |
| Max. shift between 12C and 13C envelopes, min | 0.1 | Require at least two 13C isotopes | no |
| RT window for peak binning, min | 0.2 | RT window for peak binning | 0.25 |
| Peak width range, min | 0.05–1 | Max error in relative IROA isotopes intensity | 30% |
| Smoothing window multiplier | 1.5 | Calculate complete isotopic pattern score | no |
| Create orphaned clusters | no | Try to identify unkowns | no |
| Calculate complete isotopic pattern score | no | Keep compound IDs from source for unkown compounds | yes |
| Run reintegration | yes | ||
| Creatinine [mM] | 6.5 mM CF4 | 2 mM CF4 | 6.5 mM MZmine | 2 mM MZmine | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method Used | Raw | Ratios | Suppression Corrected | Raw | Ratios | Suppression Corrected | Raw | Ratios | Raw | Ratios |
| arginine | 0.43 | 0.88 | 0.88 | 0.48 | 0.67 | 0.67 | 0.66 | 0.55 | 0.43 | 0.73 |
| asparagine | 0.55 | 0.62 | 0.62 | 0.45 | 0.61 | 0.61 | 0.55 | 0.54 | 0.58 | 0.65 |
| aspartate | 0.57 | 0.13 | 0.13 | 0.53 | 0.11 | 0.11 | 0.49 | 0.10 | 0.57 | 0.12 |
| glutamine | 0.71 | 0.60 | 0.60 | 0.75 | 0.69 | 0.69 | 0.70 | 0.60 | 0.77 | 0.73 |
| histidine | 0.74 | 0.84 | 0.84 | 0.79 | 0.82 | 0.82 | 0.79 | 0.82 | 0.67 | 0.61 |
| lysine | 0.85 | 0.95 | 0.95 | 0.78 | 0.95 | 0.95 | 0.79 | 0.88 | 0.73 | 0.87 |
| methionine | 0.79 | 0.80 | 0.80 | 0.81 | 0.79 | 0.79 | 0.77 | 0.78 | 0.79 | 0.82 |
| phenylalanine | 0.89 | 0.92 | 0.92 | 0.91 | 0.90 | 0.90 | 0.89 | 0.90 | 0.91 | 0.89 |
| serine | 0.52 | 0.70 | 0.70 | 0.60 | 0.71 | 0.71 | 0.53 | 0.71 | 0.59 | 0.70 |
| threonine | 0.84 | 0.52 | 0.52 | 0.87 | 0.75 | 0.75 | 0.85 | 0.46 | 0.88 | 0.74 |
| tryptophan | 0.88 | 0.93 | 0.93 | 0.95 | 0.93 | 0.93 | 0.90 | 0.95 | 0.95 | 0.93 |
| tyrosine | 0.95 | 0.93 | 0.93 | 0.94 | 0.96 | 0.96 | 0.96 | 0.93 | 0.94 | 0.96 |
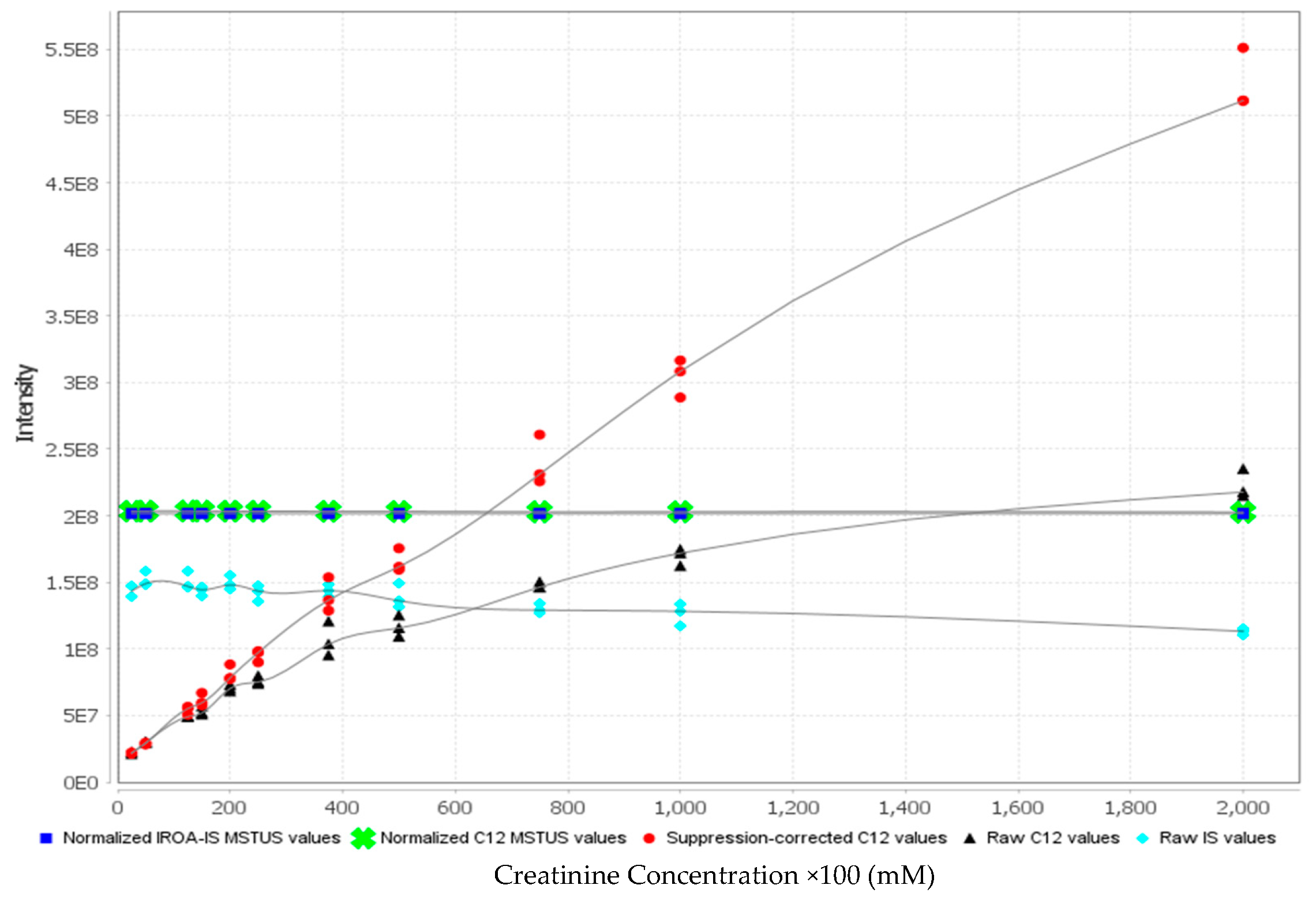
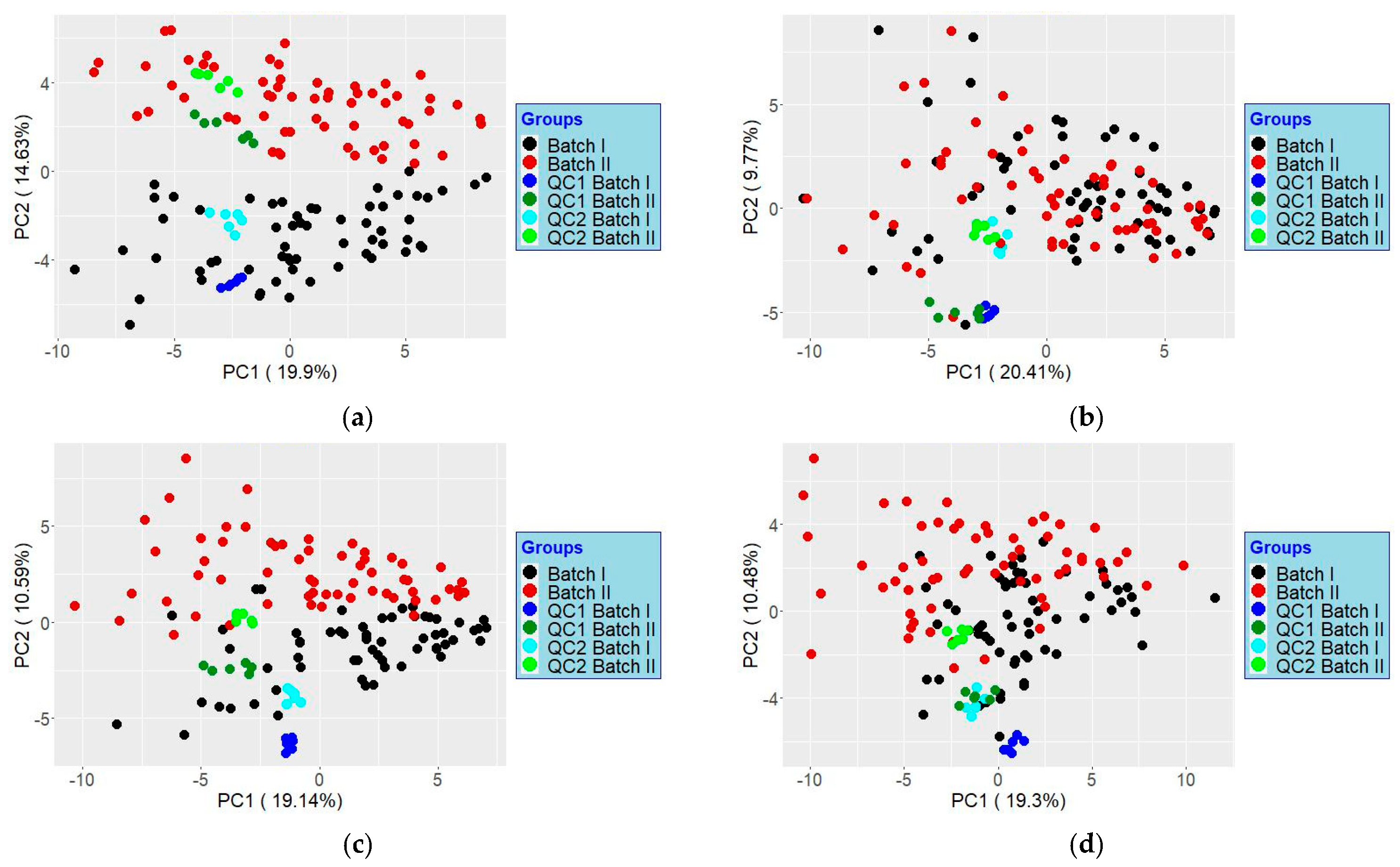
- 27.
- Sigma-Aldrich Co., LLC. Product Information, IROA TruQuant IQQ Workflow Kit. Supplied by IROA Technologies, LLC.: Catalog Number WORKFLOW. 2019. Available online: https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/362/924/workflowpis.pdf (accessed on 14 November 2019).
References
- Fadil, F.; Samol, C.; Berger, R.S.; Kellermeier, F.; Gronwald, W.; Oefner, P.J.; Dettmer, K. Isotope Ratio Outlier Analysis (IROA) for HPLC–TOFMS-Based Metabolomics of Human Urine. Metabolites 2022, 12, 741. [Google Scholar] [CrossRef] [PubMed]
- Mendez, R.; Del Carmen Piqueras, M.; Raskind, A.; de Jong, F.A.; Beecher, C.; Bhattacharya, S.K.; Banerjee, S. Quantitative Metabolomics Using Isotope Residue Outlier Analysis (IROA®) with Internal Standards; Springer: New York, NY, USA, 2019. [Google Scholar]
- Beecher, C.; de Jong, F.A. Isotopic Ratio Outlier Analysis (IROA) for Quantitative Analysis; Springer: New York, NY, USA, 2019. [Google Scholar]
- Piqueras, M.D.C.; Myer, C.; Junk, A.; Bhattacharya, S.K. Isotopic Ratio Outlier Analysis (IROA) of Aqueous Humor for Metabolites; Springer: New York, NY, USA, 2019. [Google Scholar]
- Myer, C.; Abdelrahman, L.; Banerjee, S.; Khattri, R.B.; Merritt, M.E.; Junk, A.K.; Lee, R.K.; Bhattacharya, S.K. Aqueous humor metabolite profile of pseudoexfoliation glaucoma is distinctive. Mol. Omics 2020, 16, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Myer, C.; Perez, J.; Abdelrahman, L.; Mendez, R.; Khattri, R.B.; Junk, A.K.; Bhattacharya, S.K. Differentiation of soluble aqueous humor metabolites in primary open angle glaucoma and controls. Exp. Eye Res. 2020, 194, 108024. [Google Scholar] [CrossRef] [PubMed]
- Pallikkuth, S.; Mendez, R.; Russell, K.; Sirupangi, T.; Kvistad, D.; Pahwa, R.; Villinger, F.; Banerjee, S.; Pahwa, S. Age Associated Microbiome and Microbial Metabolites Modulation and Its Association With Systemic Inflammation in a Rhesus Macaque Model. Front. Immunol. 2021, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Sigma-Aldrich Co., LLC. Product Information, IROA TruQuant IQQ Workflow Kit. Supplied by IROA Technologies, LLC.: Catalog Number WORKFLOW. 2019. Available online: https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/362/924/workflowpis.pdf (accessed on 14 November 2019).
- IROA Technologies LLC. TruQuant Yeast Extract Workflow KIT. Catalog Number: WORKFLOW: 022022IROAWORKFLOW. 2022. Available online: https://www.iroatech.com/wp-content/uploads/2022/02/TruQuant-Yeast-Extract-QC-Workflow-Kit-USER-MANUAL_022022.pdf (accessed on 15 September 2022).
- IROA Technologies LLC. TruQuant Yeast Extract Semi-Targeted QC Workflow Kit. Cat. No. WORKFLOW: 12172022IROAWORKFLOW. 2022. Available online: https://www.iroatech.com/wp-content/uploads/2022/12/TruQuant-Yeast-Extract-Semi-Targeted-QC-Workflow-Kit-USER-MANUAL_FINAL.pdf (accessed on 27 March 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fadil, F.; Samol, C.; Berger, R.S.; Kellermeier, F.; Gronwald, W.; Oefner, P.J.; Dettmer, K. Correction: Fadil et al. Isotope Ratio Outlier Analysis (IROA) for HPLC–TOFMS-Based Metabolomics of Human Urine. Metabolites 2022, 12, 741. Metabolites 2024, 14, 293. https://doi.org/10.3390/metabo14060293
Fadil F, Samol C, Berger RS, Kellermeier F, Gronwald W, Oefner PJ, Dettmer K. Correction: Fadil et al. Isotope Ratio Outlier Analysis (IROA) for HPLC–TOFMS-Based Metabolomics of Human Urine. Metabolites 2022, 12, 741. Metabolites. 2024; 14(6):293. https://doi.org/10.3390/metabo14060293
Chicago/Turabian StyleFadil, Fadi, Claudia Samol, Raffaela S. Berger, Fabian Kellermeier, Wolfram Gronwald, Peter J. Oefner, and Katja Dettmer. 2024. "Correction: Fadil et al. Isotope Ratio Outlier Analysis (IROA) for HPLC–TOFMS-Based Metabolomics of Human Urine. Metabolites 2022, 12, 741" Metabolites 14, no. 6: 293. https://doi.org/10.3390/metabo14060293
APA StyleFadil, F., Samol, C., Berger, R. S., Kellermeier, F., Gronwald, W., Oefner, P. J., & Dettmer, K. (2024). Correction: Fadil et al. Isotope Ratio Outlier Analysis (IROA) for HPLC–TOFMS-Based Metabolomics of Human Urine. Metabolites 2022, 12, 741. Metabolites, 14(6), 293. https://doi.org/10.3390/metabo14060293







