A Chemical Structure and Machine Learning Approach to Assess the Potential Bioactivity of Endogenous Metabolites and Their Association with Early Childhood Systemic Inflammation
Abstract
1. Introduction
1.1. hs-CRP as a Target for Physiological Conditions
1.2. Signals in the Metabolome
1.3. Aim of This Work
2. Materials and Methods
2.1. Cohort, Metabolomics Profiling, and hs-CRP Assessment
2.1.1. Cohort
2.1.2. Blood Collection and Metabolite Quantification
2.1.3. Assessment of hs-CRP Levels
2.2. Data Preparation and Model Building
2.2.1. Data Preparation for Metabolites
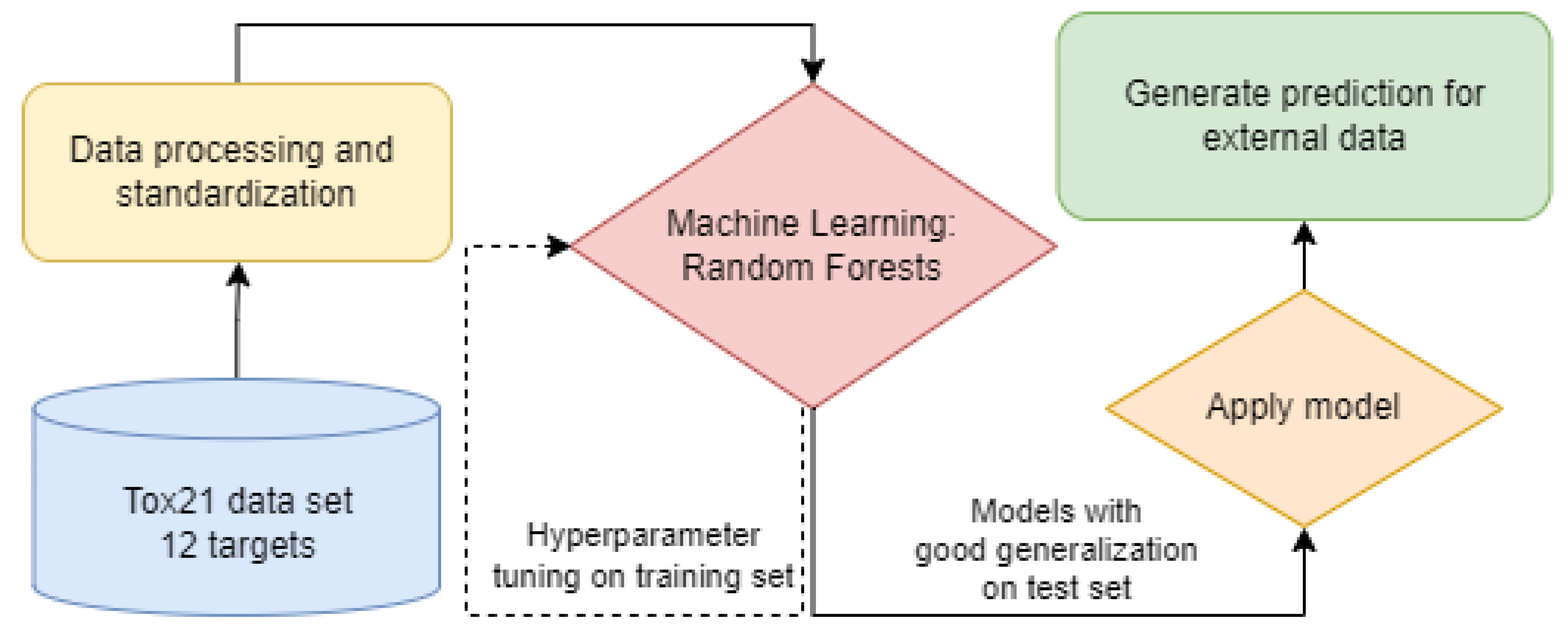
2.2.2. Bioactivity Assessment Pipeline
2.2.3. Bioactivity Data
2.2.4. Machine Learning Models
2.3. Association Testing and Toxic Unit Approach
3. Results
3.1. QSAR Model Results
3.2. Predicting Metabolite Target Activity
3.3. Association with hs-CRP
4. Discussion
Limitations and Future Perspective
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-López, Á.; López-Gonzálvez, Á.; Barker-Tejeda, T.C.; Barbas, C. A Review of Validated Biomarkers Obtained through Metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.R.; Santos, J.C.; Ribeiro, M.L.; Talarico, M.C.R.; Viana, L.R.; Derchain, S.F.M. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int. J. Mol. Sci. 2018, 19, 617. [Google Scholar] [CrossRef] [PubMed]
- Papamichael, M.M.; Katsardis, C.; Sarandi, E.; Georgaki, S.; Frima, E.-S.; Varvarigou, A.; Tsoukalas, D. Application of Metabolomics in Pediatric Asthma: Prediction, Diagnosis and Personalized Treatment. Metabolites 2021, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-Based Metabolic Phenotyping (Metabonomics/Metabolomics): The State of the Art. J. Chromatogr. B 2019, 1117, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Rago, D.; Pedersen, C.E.T.; Huang, M.; Kelly, R.S.; Gürdeniz, G.; Brustad, N.; Knihtilä, H.; Lee-Sarwar, K.A.; Morin, A.; Rasmussen, M.A.; et al. Characteristics and Mechanisms of a Sphingolipid-Associated Childhood Asthma Endotype. Am. J. Respir. Crit. Care Med. 2021, 203, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond Biomarkers and towards Mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Thomas, A. Metabolomics as a Tool to Understand Pathophysiological Processes. In Clinical Metabolomics: Methods and Protocols; Giera, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 3–28. ISBN 978-1-4939-7592-1. [Google Scholar]
- Nappo, A.; Iacoviello, L.; Fraterman, A.; Gonzalez-Gil, E.M.; Hadjigeorgiou, C.; Marild, S.; Molnar, D.; Moreno, L.A.; Peplies, J.; Sioen, I.; et al. High-Sensitivity C-Reactive Protein Is a Predictive Factor of Adiposity in Children: Results of the Identification and Prevention of Dietary- and Lifestyle-Induced Health Effects in Children and Infants (IDEFICS) Study. J. Am. Heart Assoc. 2013, 2, e000101. [Google Scholar] [CrossRef] [PubMed]
- Nishide, R.; Ando, M.; Funabashi, H.; Yoda, Y.; Nakano, M.; Shima, M. Association of Serum Hs-CRP and Lipids with Obesity in School Children in a 12-Month Follow-up Study in Japan. Env. Health Prev. Med. 2015, 20, 116–122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, A.; Jat, K.R.; Sankar, J.; Lakshmy, R.; Lodha, R.; Kabra, S.K. Role of High-Sensitivity C-Reactive Protein (Hs-CRP) in Assessment of Asthma Control in Children. J. Asthma 2023, 60, 1466–1473. [Google Scholar] [CrossRef]
- Chawes, B.L.; Stokholm, J.; Schoos, A.-M.M.; Fink, N.R.; Brix, S.; Bisgaard, H. Allergic Sensitization at School Age Is a Systemic Low-Grade Inflammatory Disorder. Allergy 2017, 72, 1073–1080. [Google Scholar] [CrossRef]
- Rosenberg, J.B.; Jepsen, J.R.M.; Mohammadzadeh, P.; Sevelsted, A.; Vinding, R.; Sørensen, M.E.; Horner, D.; Aagaard, K.; Fagerlund, B.; Brix, S.; et al. Maternal Inflammation during Pregnancy Is Associated with Risk of ADHD in Children at Age 10. Brain Behav. Immun. 2023, 115, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Mokkala, K.; Houttu, N.; Koivuniemi, E.; Sørensen, N.; Nielsen, H.B.; Laitinen, K. GlycA, a Novel Marker for Low Grade Inflammation, Reflects Gut Microbiome Diversity and Is More Accurate than High Sensitive CRP in Reflecting Metabolomic Profile. Metabolomics 2020, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR Modeling: Where Have You Been? Where Are You Going To? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed]
- Gramatica, P. Principles of QSAR Models Validation: Internal and External. QSAR Comb. Sci. 2007, 26, 694–701. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Nguyen, D.-T.; Zhao, T.; Sakamuru, S.; Zhao, J.; Shahane, S.A.; Rossoshek, A.; Simeonov, A. Tox21Challenge to Build Predictive Models of Nuclear Receptor and Stress Response Pathways as Mediated by Exposure to Environmental Chemicals and Drugs. Front. Environ. Sci. 2016, 3, 85. [Google Scholar] [CrossRef]
- Richard, A.M.; Huang, R.; Waidyanatha, S.; Shinn, P.; Collins, B.J.; Thillainadarajah, I.; Grulke, C.M.; Williams, A.J.; Lougee, R.R.; Judson, R.S.; et al. The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem. Res. Toxicol. 2020, 34, 189–216. [Google Scholar] [CrossRef]
- Heinze, K.; Lin, A.; Reniers, R.L.E.P.; Wood, S.J. Longer-Term Increased Cortisol Levels in Young People with Mental Health Problems. Psychiatry Res. 2016, 236, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.J.; Golden, S.H. Cortisol Dysregulation: The Bidirectional Link between Stress, Depression, and Type 2 Diabetes Mellitus. Ann. N. Y. Acad. Sci. 2017, 1391, 20–34. [Google Scholar] [CrossRef]
- van Spronsen, F.J.; Hoeksma, M.; Reijngoud, D.-J. Brain Dysfunction in Phenylketonuria: Is Phenylalanine Toxicity the Only Possible Cause? J. Inherit. Metab. Dis. 2009, 32, 46–51. [Google Scholar] [CrossRef]
- Lee, N.; Spears, M.E.; Carlisle, A.E.; Kim, D. Endogenous Toxic Metabolites and Implications in Cancer Therapy. Oncogene 2020, 39, 5709–5720. [Google Scholar] [CrossRef]
- Chawes, B.L.; Bønnelykke, K.; Stokholm, J.; Vissing, N.H.; Bjarnadóttir, E.; Schoos, A.-M.M.; Wolsk, H.M.; Pedersen, T.M.; Vinding, R.K.; Thorsteinsdóttir, S.; et al. Effect of Vitamin D3 Supplementation During Pregnancy on Risk of Persistent Wheeze in the Offspring: A Randomized Clinical Trial. JAMA 2016, 315, 353–361. [Google Scholar] [CrossRef]
- Bisgaard, H.; Stokholm, J.; Chawes, B.L.; Vissing, N.H.; Bjarnadóttir, E.; Schoos, A.-M.M.; Wolsk, H.M.; Pedersen, T.M.; Vinding, R.K.; Thorsteinsdóttir, S.; et al. Fish Oil–Derived Fatty Acids in Pregnancy and Wheeze and Asthma in Offspring. N. Engl. J. Med. 2016, 375, 2530–2539. [Google Scholar] [CrossRef] [PubMed]
- Gürdeniz, G.; Ernst, M.; Rago, D.; Kim, M.; Courraud, J.; Stokholm, J.; Bønnelykke, K.; Björkbom, A.; Trivedi, U.; Sørensen, S.J.; et al. Neonatal Metabolome of Caesarean Section and Risk of Childhood Asthma. Eur. Respir. J. 2022, 59, 2102406. [Google Scholar] [CrossRef] [PubMed]
- Rago, D.; Rasmussen, M.A.; Lee-Sarwar, K.A.; Weiss, S.T.; Lasky-Su, J.; Stokholm, J.; Bønnelykke, K.; Chawes, B.L.; Bisgaard, H. Fish-Oil Supplementation in Pregnancy, Child Metabolomics and Asthma Risk. eBioMedicine 2019, 46, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Olarini, A.; Ernst, M.; Gürdeniz, G.; Kim, M.; Brustad, N.; Bønnelykke, K.; Cohen, A.; Hougaard, D.; Lasky-Su, J.; Bisgaard, H.; et al. Vertical Transfer of Metabolites Detectable from Newborn’s Dried Blood Spot Samples Using UPLC-MS: A Chemometric Study. Metabolites 2022, 12, 94. [Google Scholar] [CrossRef] [PubMed]
- Fink, N.R.; Chawes, B.; Bønnelykke, K.; Thorsen, J.; Stokholm, J.; Rasmussen, M.A.; Brix, S.; Bisgaard, H. Levels of Systemic Low-Grade Inflammation in Pregnant Mothers and Their Offspring Are Correlated. Sci. Rep. 2019, 9, 3043. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Godzien, J.; Alonso-Herranz, V.; López-Gonzálvez, Á.; Barbas, C. Missing Value Imputation Strategies for Metabolomics Data. Electrophoresis 2015, 36, 3050–3060. [Google Scholar] [CrossRef] [PubMed]
- Lovrić, M.; Horner, D.; Chen, L.; Brustad, N.; Malby Schoos, A.-M.; Lasky-Su, J.; Chawes, B.; Rasmussen, M.A. Vertical Metabolome Transfer from Mother to Child: An Explainable Machine Learning Method for Detecting Metabolomic Heritability. Metabolites 2024, 14, 136. [Google Scholar] [CrossRef]
- Fernandez, M.; Ban, F.; Woo, G.; Hsing, M.; Yamazaki, T.; Leblanc, E.; Rennie, P.S.; Welch, W.J.; Cherkasov, A. Toxic Colors: The Use of Deep Learning for Predicting Toxicity of Compounds Merely from Their Graphic Images. J. Chem. Inf. Model. 2018, 58, 1533–1543. [Google Scholar] [CrossRef]
- Hemmerich, J.; Asilar, E.; Ecker, G.F. Conformational Oversampling as Data Augmentation for Molecules. In Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2019; Volume 11731, pp. 788–792. [Google Scholar] [CrossRef]
- Idakwo, G.; Thangapandian, S.; Luttrell, J.; Li, Y.; Wang, N.; Zhou, Z.; Hong, H.; Yang, B.; Zhang, C.; Gong, P. Structure–Activity Relationship-Based Chemical Classification of Highly Imbalanced Tox21 Datasets. J. Cheminformatics 2020, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Hochreiter, S. DeepTox: Toxicity Prediction Using Deep Learning. Front. Environ. Sci. 2016, 3, 80. [Google Scholar] [CrossRef]
- Klimenko, K.; Rosenberg, S.A.; Dybdahl, M.; Wedebye, E.B.; Nikolov, N.G. QSAR Modelling of a Large Imbalanced Aryl Hydrocarbon Activation Dataset by Rational and Random Sampling and Screening of 80,086 REACH Pre-Registered and/or Registered Substances. PLoS ONE 2019, 14, e0213848. [Google Scholar] [CrossRef] [PubMed]
- Lovrić, M.; Đuričić, T.; Tran, H.T.N.; Hussain, H.; Lacić, E.; Rasmussen, M.A.; Kern, R. Should We Embed in Chemistry? A Comparison of Unsupervised Transfer Learning with PCA, UMAP, and VAE on Molecular Fingerprints. Pharmaceuticals 2021, 14, 758. [Google Scholar] [CrossRef] [PubMed]
- Lovrić, M.; Molero, J.M.; Kern, R. PySpark and RDKit: Moving towards Big Data in Cheminformatics. Mol. Inform. 2019, 38, e1800082. [Google Scholar] [CrossRef] [PubMed]
- Fourches, D.; Muratov, E.; Tropsha, A. Trust, but Verify: On the Importance of Chemical Structure Curation in Cheminformatics and QSAR Modeling Research. J. Chem. Inf. Model. 2010, 50, 1189–1204. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics Software. Available online: http://rdkit.org/ (accessed on 25 April 2024).
- Gütlein, M.; Kramer, S. Filtered Circular Fingerprints Improve Either Prediction or Runtime Performance While Retaining Interpretability. J. Cheminformatics 2016, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Landrum, G. RDKit: Colliding Bits III. Available online: http://rdkit.blogspot.com/2016/02/colliding-bits-iii.html (accessed on 23 December 2019).
- Lovrić, M.; Pavlović, K.; Žuvela, P.; Spataru, A.; Lučić, B.; Kern, R.; Wong, M.W. Machine Learning in Prediction of Intrinsic Aqueous Solubility of Drug-like Compounds: Generalization, Complexity, or Predictive Ability? J. Chemom. 2021, 35, e3349. [Google Scholar] [CrossRef]
- Pedregosa, F.; Michel, V.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Vanderplas, J.; Cournapeau, D.; Varoquaux, G.; Gramfort, A.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal Classifier for Imbalanced Data Using Matthews Correlation Coefficient Metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef] [PubMed]
- Lovrić, M.; Malev, O.; Klobučar, G.; Kern, R.; Liu, J.J.; Lučić, B. Predictive Capability of QSAR Models Based on the CompTox Zebrafish Embryo Assays: An Imbalanced Classification Problem. Molecules 2021, 26, 1617. [Google Scholar] [CrossRef]
- Lučić, B.; Batista, J.; Bojović, V.; Lovrić, M.; Sović Kržić, A.; Bešlo, D.; Nadramija, D.; Vikić-Topić, D. Estimation of Random Accuracy and Its Use in Validation of Predictive Quality of Classification Models within Predictive Challenges. Croat. Chem. Acta 2019, 92, 379–391. [Google Scholar] [CrossRef]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. In Proceedings of the NIPS 2012, Lake Tahoe, NV, USA, 13 June 2012; Volume 4, pp. 2951–2959. [Google Scholar]
- Ginebreda, A.; Kuzmanovic, M.; Guasch, H.; de Alda, M.L.; López-Doval, J.C.; Muñoz, I.; Ricart, M.; Romaní, A.M.; Sabater, S.; Barceló, D. Assessment of Multi-Chemical Pollution in Aquatic Ecosystems Using Toxic Units: Compound Prioritization, Mixture Characterization and Relationships with Biological Descriptors. Sci. Total Environ. 2014, 468–469, 715–723. [Google Scholar] [CrossRef]
- Vallat, R. Pingouin: Statistics in Python. JOSS 2018, 3, 1026. [Google Scholar] [CrossRef]
- Lovrić, M. Modelling Data and Model Results Tox21 (0.1) [Data Set]. Zenodo. Available online: https://zenodo.org/records/10888738 (accessed on 25 April 2024).
- Plebani, M. Why C-Reactive Protein Is One of the Most Requested Tests in Clinical Laboratories? Clin. Chem. Lab. Med. 2023, 61, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Windgassen, E.B.; Funtowicz, L.; Lunsford, T.N.; Harris, L.A.; Mulvagh, S.L. C-Reactive Protein and High-Sensitivity C-Reactive Protein: An Update for Clinicians. Postgrad. Med. 2011, 123, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, F.A.H.; de Oliveira Izar, M.C. High-Sensitivity C-Reactive Protein and Cardiovascular Disease Across Countries and Ethnicities. Clinics 2016, 71, 235–242. [Google Scholar] [CrossRef]
- Hod, K.; Ringel-Kulka, T.; Martin, C.F.; Maharshak, N.; Ringel, Y. High-Sensitive C-Reactive Protein as a Marker for Inflammation in Irritable Bowel Syndrome. J. Clin. Gastroenterol. 2016, 50, 227–232. [Google Scholar] [CrossRef]
- Valkanova, V.; Ebmeier, K.P.; Allan, C.L. CRP, IL-6 and Depression: A Systematic Review and Meta-Analysis of Longitudinal Studies. J. Affect. Disord. 2013, 150, 736–744. [Google Scholar] [CrossRef]
- Chawes, B.L.K.; Stokholm, J.; Bønnelykke, K.; Brix, S.; Bisgaard, H. Neonates with Reduced Neonatal Lung Function Have Systemic Low-Grade Inflammation. J. Allergy Clin. Immunol. 2015, 135, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Rahman Fink, N.; Chawes, B.L.; Thorsen, J.; Stokholm, J.; Krogfelt, K.A.; Schjørring, S.; Kragh, M.; Bønnelykke, K.; Brix, S.; Bisgaard, H. Neonates Colonized with Pathogenic Bacteria in the Airways Have a Low-Grade Systemic Inflammation. Allergy 2018, 73, 2150–2159. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, T.; Obase, Y.; Kishikawa, R.; Iwanaga, T. Serum High-Sensitivity C-Reactive Protein Can Be an Airway Inflammation Predictor in Bronchial Asthma. Allergy Asthma Proc. 2015, 36, e23–e28. [Google Scholar] [CrossRef]
- Kurosaki, K.; Wu, R.; Uesawa, Y. A Toxicity Prediction Tool for Potential Agonist/Antagonist Activities in Molecular Initiating Events Based on Chemical Structures. Int. J. Mol. Sci. 2020, 21, 7853. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, N.C.; Kyratzi, E.; Lamprokostopoulou, A.; Chrousos, G.P.; Charmandari, E. Stress, the Stress System and the Role of Glucocorticoids. Neuroimmunomodulation 2014, 22, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Hos, D.; Saban, D.R.; Bock, F.; Regenfuss, B.; Onderka, J.; Masli, S.; Cursiefen, C. Suppression of Inflammatory Corneal Lymphangiogenesis by Application of Topical Corticosteroids. Arch. Ophthalmol. 2011, 129, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Shimba, A.; Ikuta, K. Control of Immunity by Glucocorticoids in Health and Disease. Semin. Immunopathol. 2020, 42, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Rueggeberg, R.; Wrosch, C.; Miller, G.E.; McDade, T.W. Associations between Health-Related Self-Protection, Diurnal Cortisol, and C-Reactive Protein in Lonely Older Adults. Psychosom. Med. 2012, 74, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.E.; Chen, E.; Sze, J.; Marin, T.; Arevalo, J.M.G.; Doll, R.; Ma, R.; Cole, S.W. A Functional Genomic Fingerprint of Chronic Stress in Humans: Blunted Glucocorticoid and Increased NF-κB Signaling. Biol. Psychiatry 2008, 64, 266–272. [Google Scholar] [CrossRef]
- Raison, C.L.; Miller, A.H. When Not Enough Is Too Much: The Role of Insufficient Glucocorticoid Signaling in the Pathophysiology of Stress-Related Disorders. FOC 2011, 9, 399–410. [Google Scholar] [CrossRef]
- De Dios, O.; Navarro, P.; Ortega-Senovilla, H.; Herrero, L.; Gavela-Pérez, T.; Soriano-Guillen, L.; Lasunción, M.A.; Garcés, C. Plasma Retinol Levels and High-Sensitivity C-Reactive Protein in Prepubertal Children. Nutrients 2018, 10, 1257. [Google Scholar] [CrossRef] [PubMed]
- García, O.P.; Ronquillo, D.; Del Carmen Caamaño, M.; Martínez, G.; Camacho, M.; López, V.; Rosado, J.L. Zinc, Iron and Vitamins A, C and E Are Associated with Obesity, Inflammation, Lipid Profile and Insulin Resistance in Mexican School-Aged Children. Nutrients 2013, 5, 5012–5030. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.W.; Walker, B.R.; Shaw, R.J.; Honour, J.W.; Jessop, D.S.; Lightman, S.L.; Zumla, A.; Rook, G.A. Increased Cortisol: Cortisone Ratio in Acute Pulmonary Tuberculosis. Am. J. Respir. Crit. Care Med. 2000, 162, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Topete, D.; Cidlowski, J.A. One Hormone, Two Actions: Anti- and pro-Inflammatory Effects of Glucocorticoids. Neuroimmunomodulation 2015, 22, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, P.; Graham, T.E.; Kahn, B.B.; Altomare, A.; Funanage, V.; George, D. Reduction of Elevated Serum Retinol Binding Protein in Obese Children by Lifestyle Intervention: Association with Subclinical Inflammation. J. Clin. Endocrinol. Metab. 2007, 92, 1971–1974. [Google Scholar] [CrossRef] [PubMed]
- Wilmanski, T.; Rappaport, N.; Earls, J.C.; Magis, A.T.; Manor, O.; Lovejoy, J.; Omenn, G.S.; Hood, L.; Gibbons, S.M.; Price, N.D. Blood Metabolome Predicts Gut Microbiome α-Diversity in Humans. Nat. Biotechnol. 2019, 37, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Zouiouich, S.; Loftfield, E.; Huybrechts, I.; Viallon, V.; Louca, P.; Vogtmann, E.; Wells, P.M.; Steves, C.J.; Herzig, K.-H.; Menni, C.; et al. Markers of Metabolic Health and Gut Microbiome Diversity: Findings from Two Population-Based Cohort Studies. Diabetologia 2021, 64, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Torres-Barrán, A.; Alonso, Á.; Dorronsoro, J.R. Regression Tree Ensembles for Wind Energy and Solar Radiation Prediction. Neurocomputing 2019, 326–327, 151–160. [Google Scholar] [CrossRef]
- Fava, F.; Rizzetto, L.; Tuohy, K.M. Gut Microbiota and Health: Connecting Actors across the Metabolic System. Proc. Nutr. Soc. 2019, 78, 177–188. [Google Scholar] [CrossRef]
- Kolmar, S.S.; Grulke, C.M. The Effect of Noise on the Predictive Limit of QSAR Models. J. Cheminformatics 2021, 13, 92. [Google Scholar] [CrossRef] [PubMed]
- Myshkin, E.; Brennan, R.; Khasanova, T.; Sitnik, T.; Serebriyskaya, T.; Litvinova, E.; Guryanov, A.; Nikolsky, Y.; Nikolskaya, T.; Bureeva, S. Prediction of Organ Toxicity Endpoints by QSAR Modeling Based on Precise Chemical-Histopathology Annotations. Chem. Biol. Drug Des. 2012, 80, 406–416. [Google Scholar] [CrossRef] [PubMed]


| Endpoint | MCC Train | BACC Train | MCC CV | MCC Test | BACC Test | Num. of Feat. | N(1)/N(0,1) |
|---|---|---|---|---|---|---|---|
| SR-HSE | 0.47 | 0.82 | 0.27 | 0.23 | 0.69 | 38 | 340/6402 |
| * NR-AR | 0.63 | 0.95 | 0.58 | 0.7 | 0.97 | 31 | 306/7060 |
| * SR-ARE | 0.77 | 0.94 | 0.35 | 0.34 | 0.76 | 38 | 945/5758 |
| NR-Aromatase | 0.6 | 0.88 | 0.2 | 0.15 | 0.63 | 38 | 304/5652 |
| * NR-ER-LBD | 0.62 | 0.89 | 0.47 | 0.56 | 0.84 | 38 | 343/6817 |
| * NR-AhR | 0.73 | 0.9 | 0.51 | 0.45 | 0.78 | 38 | 756/6459 |
| * SR-MMP | 0.83 | 0.91 | 0.59 | 0.58 | 0.79 | 38 | 903/5715 |
| * NR-ER | 0.64 | 0.91 | 0.41 | 0.41 | 0.83 | 38 | 806/6056 |
| NR-PPAR-gamma | 0.59 | 0.92 | 0.17 | 0.09 | 0.6 | 13 | 197/6355 |
| SR-p53 | 0.41 | 0.63 | 0.25 | 0.24 | 0.58 | 38 | 423/6643 |
| * SR-ATAD5 | 0.52 | 0.76 | 0.3 | 0.31 | 0.66 | 38 | 275/6926 |
| * NR-AR-LBD | 0.61 | 0.81 | 0.58 | 0.55 | 0.82 | 38 | 234/6617 |
| SR-ATAD5 | NR-AR-LBD | NR-AR | NR-ER-LBD | NR-ER | SR-ARE | NR-AhR | SR-MMP | |
|---|---|---|---|---|---|---|---|---|
| cortisol | 0.12 | * 0.97 | * 0.95 | 0.07 | 0.35 | 0.2 | 0 | 0.13 |
| cortisone | 0.13 | * 0.96 | * 0.82 | 0.04 | 0.36 | 0.24 | 0.02 | 0.19 |
| androsterone sulfate | 0.12 | * 0.88 | * 0.67 | 0.43 | * 0.59 | 0.48 | 0.02 | 0.33 |
| dehydroepiandrosterone sulfate | 0.13 | * 0.92 | * 0.66 | 0.46 | * 0.67 | * 0.54 | 0.01 | 0.29 |
| 16a-hydroxy dhea 3-sulfate | 0.14 | * 0.93 | * 0.63 | 0.37 | * 0.57 | 0.42 | 0.02 | 0.44 |
| Active Metabolites | Active Metabolites | Active Metabolites |
|---|---|---|
| taurochenodeoxycholic acid 3-sulfate | taurochenodeoxycholate | chenodeoxycholate |
| 4-cholesten-3-one | ursodeoxycholate | 2-aminophenol sulfate |
| gamma-CEHC | pregnenediol sulfate | taurohyocholate |
| 5alpha-pregnan-3beta,20alpha-diol disulfate | 16a-hydroxy DHEA 3-sulfate | 5alpha-androstan-3beta,17alpha-diol disulfate |
| cortisone | lithocholate sulfate | taurocholate |
| taurocholenate sulfate | pregnenetriol sulfate | tauroursodeoxycholate |
| cholesterol | taurodeoxycholate | glycoursodeoxycholate |
| alpha-tocopherol | 4-hydroxychlorothalonil | N6-methyladenosine |
| glycodeoxycholate | hyocholate | glycocholate |
| pregnenolone sulfate | campesterol | 3beta-hydroxy-5-cholestenoate |
| taurolithocholate 3-sulfate | glycochenodeoxycholate | androsterone glucuronide |
| glycochenodeoxycholate 3-sulfate | isoursodeoxycholate | glycolithocholate |
| glycolithocholate sulfate | tauro-beta-muricholate | gamma-tocopherol/beta-tocopherol |
| androsterone sulfate | cortisol | 7-HOCA |
| glycohyocholate | solanidine | beta-cryptoxanthin |
| caffeine | dehydroepiandrosterone sulfate | piperine |
| retinol (Vitamin A) | cholate | glycocholenate sulfate |
| taurochenodeoxycholic acid 3-sulfate | deoxycholate |
| Metabolite | Super_Pathway | Sub_Pathway | coef. | p-Value |
|---|---|---|---|---|
| cortisone | Lipid | Corticosteroids | −0.91 | 0.000 |
| retinol (Vitamin A) | Cofactors and Vitamins | Vitamin A Metabolism | −1.39 | 0.000 |
| cortisol | Lipid | Corticosteroids | 1.026 | 0.000 |
| 3beta-Hydroxy-5-cholestenoate | Lipid | Sterol (or primary bile acid biosynthesis) | −0.64 | 0.000 |
| glycocholenate sulfate | Lipid | Secondary Bile Acid Metabolism | 0.581 | 0.002 |
| intercept | 2.976 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lovrić, M.; Wang, T.; Staffe, M.R.; Šunić, I.; Časni, K.; Lasky-Su, J.; Chawes, B.; Rasmussen, M.A. A Chemical Structure and Machine Learning Approach to Assess the Potential Bioactivity of Endogenous Metabolites and Their Association with Early Childhood Systemic Inflammation. Metabolites 2024, 14, 278. https://doi.org/10.3390/metabo14050278
Lovrić M, Wang T, Staffe MR, Šunić I, Časni K, Lasky-Su J, Chawes B, Rasmussen MA. A Chemical Structure and Machine Learning Approach to Assess the Potential Bioactivity of Endogenous Metabolites and Their Association with Early Childhood Systemic Inflammation. Metabolites. 2024; 14(5):278. https://doi.org/10.3390/metabo14050278
Chicago/Turabian StyleLovrić, Mario, Tingting Wang, Mads Rønnow Staffe, Iva Šunić, Kristina Časni, Jessica Lasky-Su, Bo Chawes, and Morten Arendt Rasmussen. 2024. "A Chemical Structure and Machine Learning Approach to Assess the Potential Bioactivity of Endogenous Metabolites and Their Association with Early Childhood Systemic Inflammation" Metabolites 14, no. 5: 278. https://doi.org/10.3390/metabo14050278
APA StyleLovrić, M., Wang, T., Staffe, M. R., Šunić, I., Časni, K., Lasky-Su, J., Chawes, B., & Rasmussen, M. A. (2024). A Chemical Structure and Machine Learning Approach to Assess the Potential Bioactivity of Endogenous Metabolites and Their Association with Early Childhood Systemic Inflammation. Metabolites, 14(5), 278. https://doi.org/10.3390/metabo14050278











