Urine Metabolite Profiles after the Consumption of a Low- and a High-Digestible Protein Meal, and Comparison of Urine Normalization Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Test Meals
2.2. Urine Sample Preparation and LC-MS Analysis
2.3. Chemometrical Methods
2.3.1. LC-MS Data Processing and Matrix Pretreatment
2.3.2. Normalization Methods
2.3.3. Data Analysis
3. Results
3.1. Normalization
3.2. AComDim-ICA
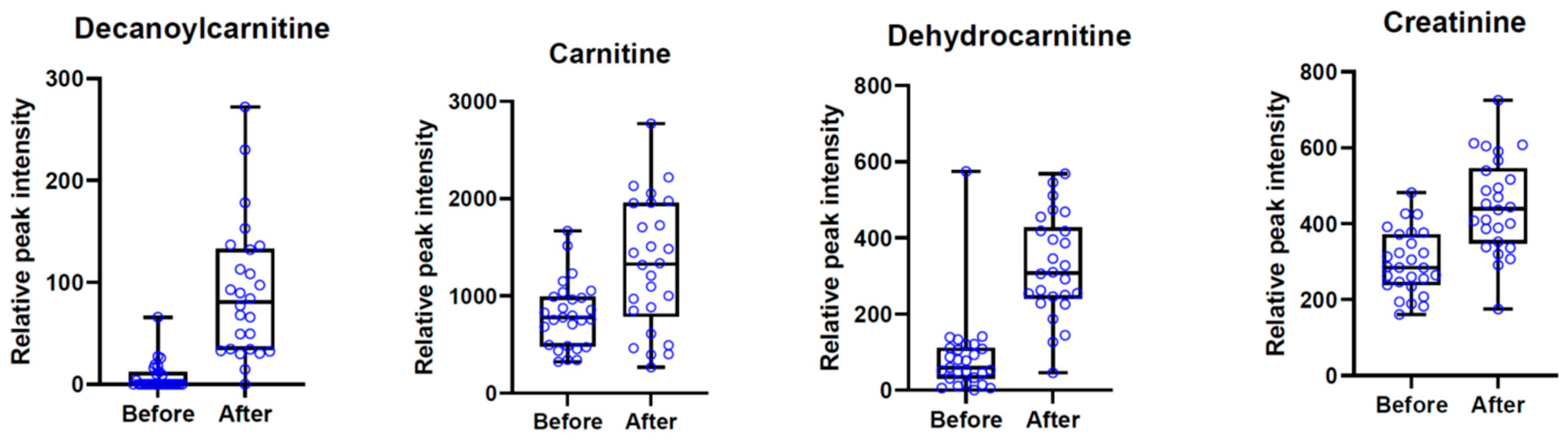
3.3. Discriminant Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schmidt, J.A.; Rinaldi, S.; Ferrari, P.; Carayol, M.; Achaintre, D.; Scalbert, A.; Cross, A.J.; Gunter, M.J.; Fensom, G.K.; Appleby, P.N.; et al. Metabolic profiles of male meat eaters, fish eaters, vegetarians, and vegans from the EPIC-Oxford cohort. Am. J. Clin. Nutr. 2015, 102, 1518–1526. [Google Scholar] [CrossRef]
- Schmidt, J.A.; Fensom, G.K.; Rinaldi, S.; Scalbert, A.; Gunter, M.J.; Holmes, M.V.; Key, T.J.; Travis, R.C. NMR Metabolite Profiles in Male Meat-Eaters, Fish-Eaters, Vegetarians and Vegans, and Comparison with MS Metabolite Profiles. Metabolites 2021, 11, 121. [Google Scholar] [CrossRef] [PubMed]
- Hovinen, T.; Korkalo, L.; Freese, R.; Skaffari, E.; Isohanni, P.; Niemi, M.; Nevalainen, J.; Gylling, H.; Zamboni, N.; Erkkola, M.; et al. Vegan diet in young children remodels metabolism and challenges the statuses of essential nutrients. EMBO Mol. Med. 2021, 13, e13492. [Google Scholar] [CrossRef] [PubMed]
- Miles, F.L.; Orlich, M.J.; Mashchak, A.; Chandler, P.D.; Lampe, J.W.; Duerksen-Hughes, P.; Fraser, G.E. The Biology of Veganism: Plasma Metabolomics Analysis Reveals Distinct Profiles of Vegans and Non-Vegetarians in the Adventist Health Study-2 Cohort. Nutrients 2022, 14, 709. [Google Scholar] [CrossRef] [PubMed]
- Miles, F.L.; Lloren, J.I.C.; Haddad, E.; Jaceldo-Siegl, K.; Knutsen, S.; Sabate, J.; Fraser, G.E. Plasma, Urine, and Adipose Tissue Biomarkers of Dietary Intake Differ Between Vegetarian and Non-Vegetarian Diet Groups in the Adventist Health Study-2. J. Nutr. 2019, 149, 667–675. [Google Scholar] [CrossRef]
- Xu, J.; Yang, S.; Cai, S.; Dong, J.; Li, X.; Chen, Z. Identification of biochemical changes in lactovegetarian urine using 1H NMR spectroscopy and pattern recognition. Anal. Bioanal. Chem. 2010, 396, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, H.M.; Rådjursöga, M.; Torstensson, T.; Jansson, L.; Ellegård, L.; Winkvist, A. Urine Metabolite Profiles and Nutrient Intake Based on 4-Day Weighed Food Diary in Habitual Vegans, Vegetarians, and Omnivores. J. Nutr. 2021, 151, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Alonso, P.; Becerra-Tomás, N.; Papandreou, C.; Bulló, M.; Guasch-Ferré, M.; Toledo, E.; Ruiz-Canela, M.; Clish, C.B.; Corella, D.; Dennis, C.; et al. Plasma Metabolomics Profiles are Associated with the Amount and Source of Protein Intake: A Metabolomics Approach within the PREDIMED Study. Mol. Nutr. Food Res. 2020, 64, 2000178. [Google Scholar] [CrossRef] [PubMed]
- Gaudichon, C.; Calvez, J. Determinants of amino acid bioavailability from ingested protein in relation to gut health. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 55–61. [Google Scholar] [CrossRef]
- Khodorova, N.V.; Jouan-Rimbaud, D.; Pilard, S.; Cordella, C.; Locquet, N.; Oberli, M.; Gaudichon, C. Consumption of Boiled, but Not Grilled, Roasted, or Barbecued Beef Modifies the Urinary Metabolite Profiles in Rats. Mol. Nutr. Food Res. 2022, 66, 2100872. [Google Scholar] [CrossRef]
- Calvez, J.; Benoit, S.; Piedcoq, J.; Khodorova, N.; Azzout-Marniche, D.; Tomé, D.; Benamouzig, R.; Airinei, G.; Gaudichon, C. Very low ileal nitrogen and amino acid digestibility of zein compared to whey protein isolate in healthy volunteers. Am. J. Clin. Nutr. 2021, 113, 70–82. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Correction to the Description of Standard Normal Variate (SNV) and De-Trend (DT) Transformations in Practical Spectroscopy with Applications in Food and Beverage Analysis—2nd Edition. J. Near Infrared Spectrosc. 1993, 1, 185–186. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 1993, 78, 4281–4290. [Google Scholar] [CrossRef]
- Habchi, B.; Alves, S.; Jouan-Rimbaud Bouveresse, D.; Moslah, B.; Paris, A.; Lécluse, Y.; Gauduchon, P.; Lebailly, P.; Rutledge, D.N.; Rathahao-Paris, E. An innovative chemometric method for processing direct introduction high resolution mass spectrometry metabolomic data: Independent component–discriminant analysis (IC–DA). Metabolomics 2017, 13, 45. [Google Scholar] [CrossRef]
- Harrington, P.B.; Vieira, N.E.; Espinoza, J.; Nien, J.K.; Romero, R.; Yergey, A.L. Analysis of variance-principal component analysis: A soft tool for proteomic discovery. Anal. Chim. Acta 2005, 544, 118–127. [Google Scholar] [CrossRef]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.; Lamers, R.J.; van der Greef, J.; Timmerman, M.E. ANOVA-simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef]
- Qannari, M.; Wakeling, I.; Courcoux, P.; MacFie, H.J.H. Defining the underlying sensory dimensions. Food Qual. Prefer. 2000, 11, 151–154. [Google Scholar] [CrossRef]
- Tormena, C.D.; Rutledge, D.N.; Rakocevic, M.; Bruns, R.E.; Scarminio, I.S.; Marcheafave, G.G.; Pauli, E.D. Exogenous application of bioregulators in Coffea arabica beans during ripening: Investigation of UV–Visible and NIR mixture design-fingerprints using AComDim-ICA. Microchem. J. 2022, 181, 10770. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Heinzmann, S.S.; Brown, I.J.; Chan, Q.; Bictash, M.; Dumas, M.E.; Kochhar, S.; Stamler, J.; Holmes, E.; Elliott, P.; Nicholson, J.K. Metabolic profiling strategy for discovery of nutritional biomarkers: Proline betaine as a marker of citrus consumption. Am. J. Clin. Nutr. 2010, 92, 436–443. [Google Scholar] [CrossRef]
- Wilson, T.; Garcia-Perez, I.; Posma, J.M.; Lloyd, A.J.; Chambers, E.S.; Tailliart, K.; Zubair, H.; Beckmann, M.; Mathers, J.C.; Holmes, E.; et al. Spot and Cumulative Urine Samples Are Suitable Replacements for 24-Hour Urine Collections for Objective Measures of Dietary Exposure in Adults Using Metabolite Biomarkers. J. Nutr. 2019, 149, 1692–1700. [Google Scholar] [CrossRef]
- Wedekind, R.; Keski-Rahkonen, P.; Robinot, N.; Mercier, F.; Engel, E.; Huybrechts, I.; Scalbert, A. Signatures of 10 Processed and Non-processed Meat Products after In Vitro Digestion. Metabolites 2020, 10, 272. [Google Scholar] [CrossRef]
- Cheung, W.; Keski-Rahkonen, P.; Assi, N.; Ferrari, P.; Freisling, H.; Rinaldi, S.; Slimani, N.; Zamora-Ros, R.; Rundle, M.; Frost, G.; et al. A metabolomic study of biomarkers of meat and fish intake. Am. J. Clin. Nutr. 2017, 105, 600–608. [Google Scholar] [CrossRef]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar]
- Vogl, F.C.; Mehrl, S.; Heizinger, L.; Schlecht, I.; Zacharias, H.U.; Ellmann, L.; Nürnberger, N.; Gronwald, W.; Leitzmann, M.F.; Rossert, J.; et al. Evaluation of dilution and normalization strategies to correct for urinary output in HPLC-HRTOFMS metabolomics. Anal. Bioanal. Chem. 2016, 408, 8483–8493. [Google Scholar] [CrossRef] [PubMed]
- Chetwynd, A.J.; Abdul-Sada, A.; Holt, S.B.; Hill, E.M. Use of a pre-analysis osmolality normalisation method to correct forvariable urine concentrations and for improved metabolomic analyses. J. Chrom. A 2016, 1431, 103–110. [Google Scholar] [CrossRef]
- Gagnebin, Y.; Tonoli, D.; Lescuyer, P.; Ponte, B.; De Seigneux, S.; Martin, P.Y.; Schappler, J.; Boccard, J.; Rudaz, S. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Anal. Chim. Acta 2017, 955, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Hertel, J.; Rotter, M.; Frenzel, S.; Zacharias, H.U.; Krumsiek, J.; Rathkolb, B.; Hrabe de Angelis, M.; Rabstein, S.; Pallapies, D.; Brüning, T.; et al. Dilution correction for dynamically influenced urinary analyte data. Anal. Chim. Acta 2018, 1032, 18–31. [Google Scholar] [CrossRef] [PubMed]
 Protein−free group,
Protein−free group,  Zein group,
Zein group,  WPI group. Note that no separation between the groups of consumed proteins was observed, for either raw or osmolality-corrected RP and HILIC data sets.
WPI group. Note that no separation between the groups of consumed proteins was observed, for either raw or osmolality-corrected RP and HILIC data sets.
 Protein−free group,
Protein−free group,  Zein group,
Zein group,  WPI group. Note that no separation between the groups of consumed proteins was observed, for either raw or osmolality-corrected RP and HILIC data sets.
WPI group. Note that no separation between the groups of consumed proteins was observed, for either raw or osmolality-corrected RP and HILIC data sets.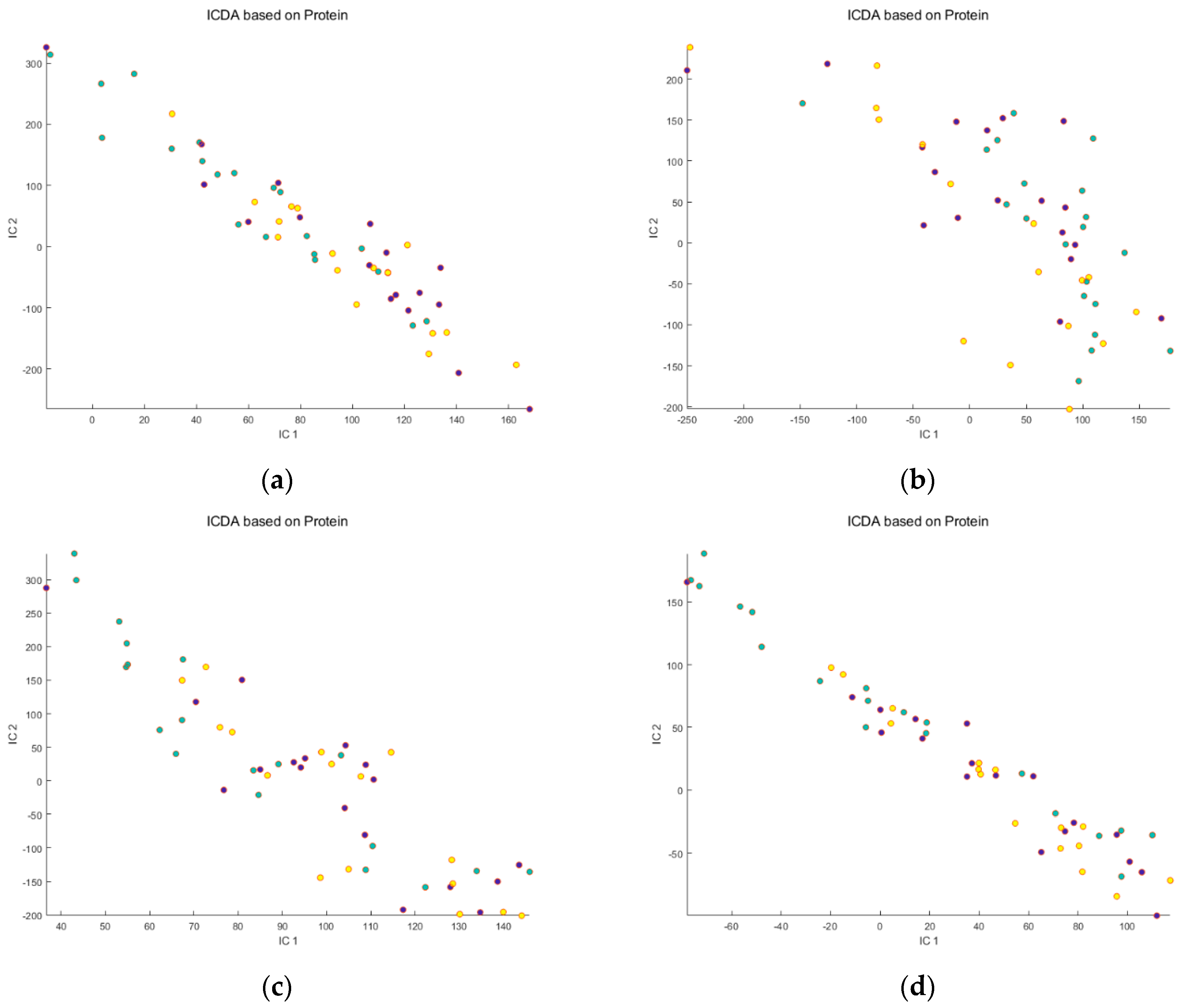
 Before meal ingestion,
Before meal ingestion,  9 h after meal ingestion. Note that osmolality correction did not significantly reduce the dispersion of the individuals, nor increase the separation between the “before” and “after” meal ingestion groups.
9 h after meal ingestion. Note that osmolality correction did not significantly reduce the dispersion of the individuals, nor increase the separation between the “before” and “after” meal ingestion groups.
 Before meal ingestion,
Before meal ingestion,  9 h after meal ingestion. Note that osmolality correction did not significantly reduce the dispersion of the individuals, nor increase the separation between the “before” and “after” meal ingestion groups.
9 h after meal ingestion. Note that osmolality correction did not significantly reduce the dispersion of the individuals, nor increase the separation between the “before” and “after” meal ingestion groups.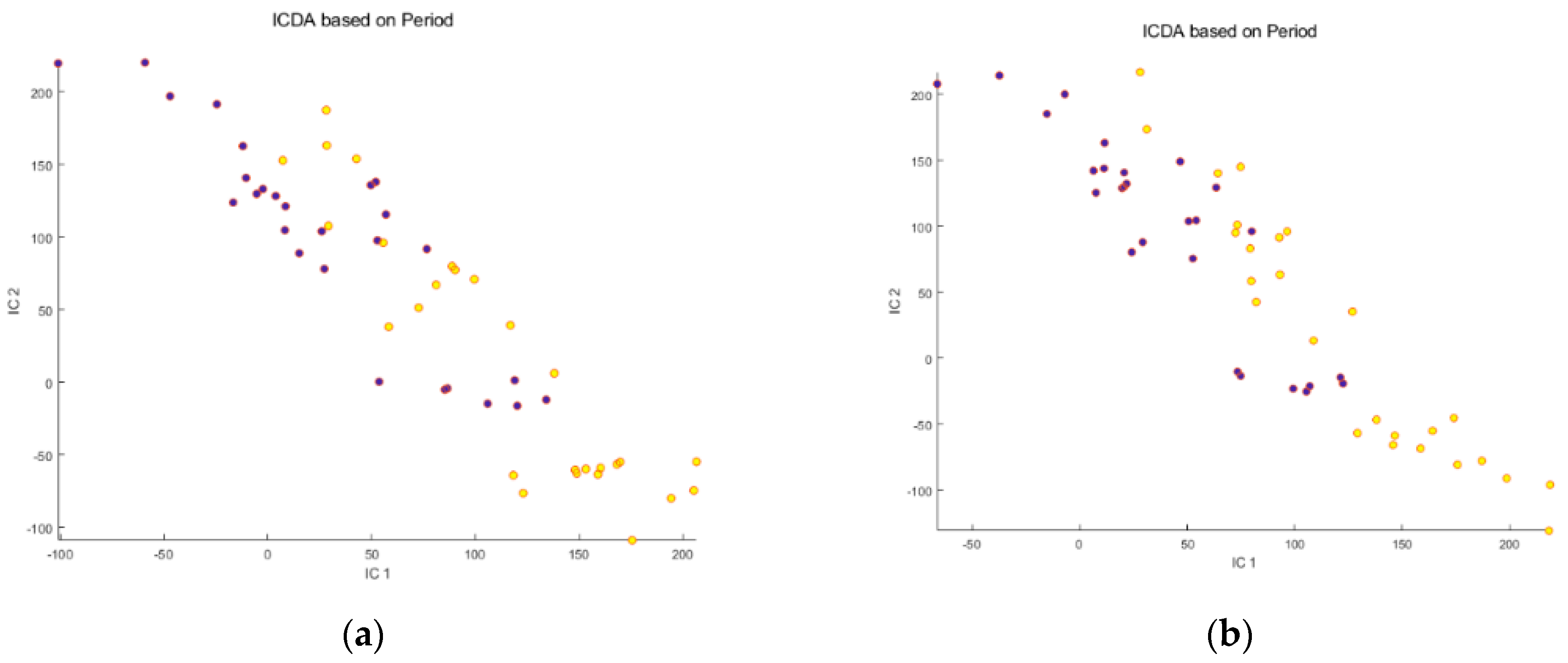
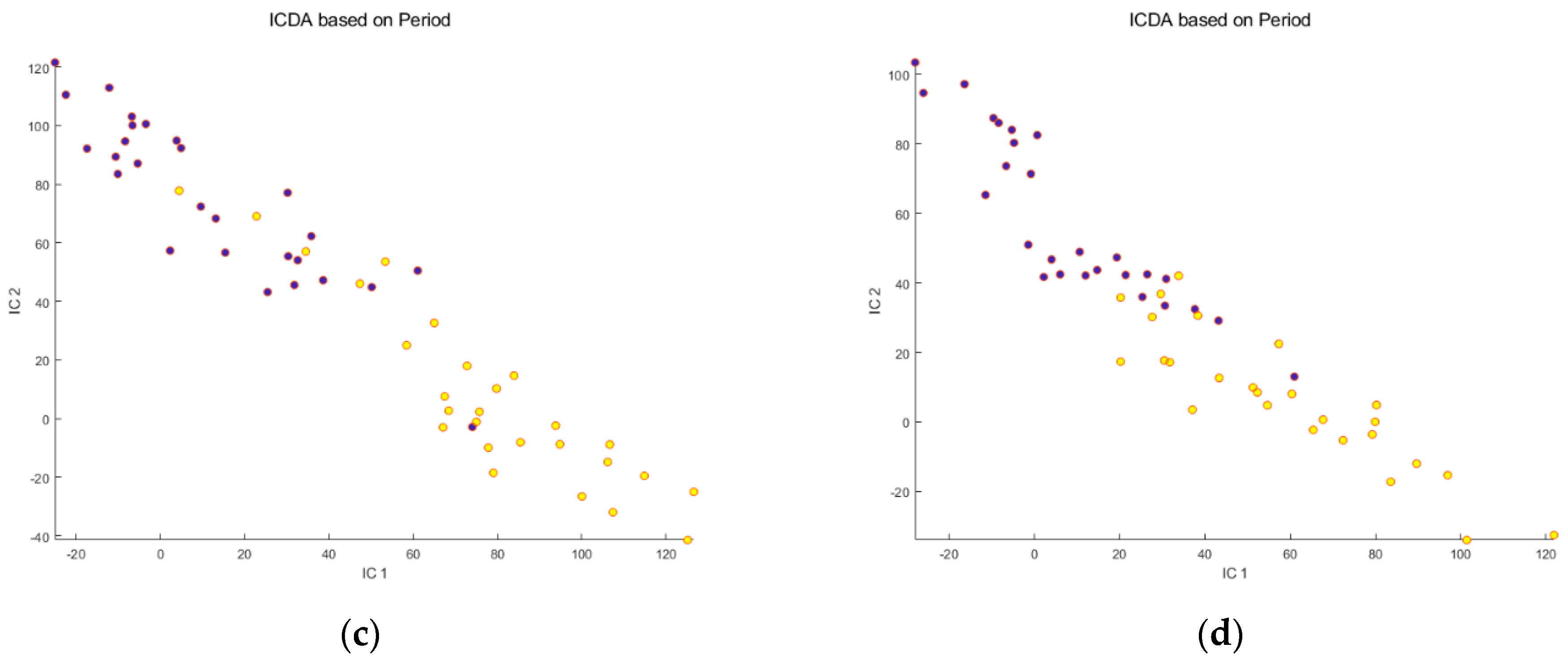
 Before meal ingestion,
Before meal ingestion,  9 h after meal ingestion. Note that SNV correction improves the separation between the groups and decreases the dispersion of the samples within each group.
9 h after meal ingestion. Note that SNV correction improves the separation between the groups and decreases the dispersion of the samples within each group.
 Before meal ingestion,
Before meal ingestion,  9 h after meal ingestion. Note that SNV correction improves the separation between the groups and decreases the dispersion of the samples within each group.
9 h after meal ingestion. Note that SNV correction improves the separation between the groups and decreases the dispersion of the samples within each group.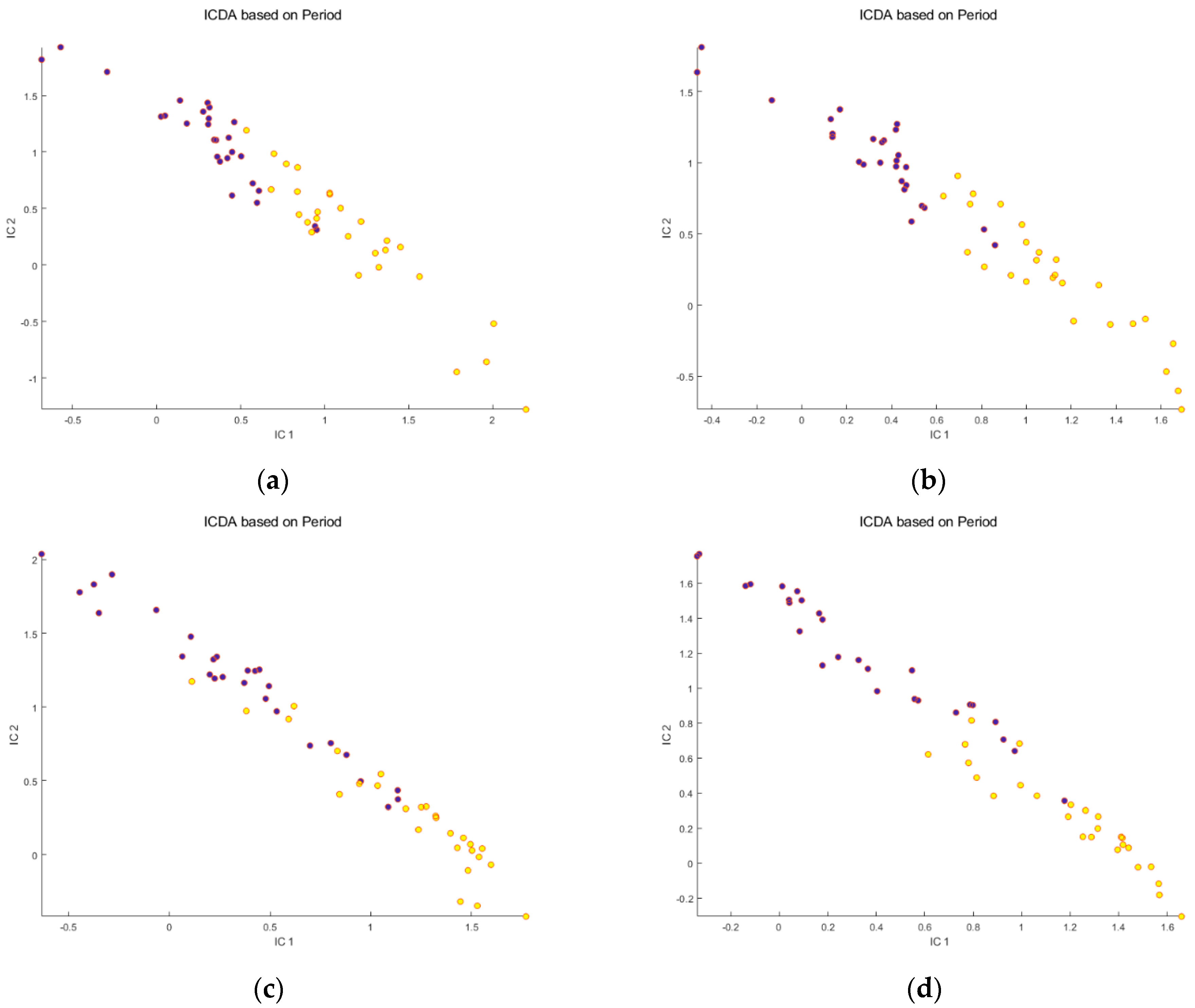
 Before meal ingestion,
Before meal ingestion,  9 h after meal ingestion. Note that PQN correction increases the dispersion of the samples within the groups.
9 h after meal ingestion. Note that PQN correction increases the dispersion of the samples within the groups.
 Before meal ingestion,
Before meal ingestion,  9 h after meal ingestion. Note that PQN correction increases the dispersion of the samples within the groups.
9 h after meal ingestion. Note that PQN correction increases the dispersion of the samples within the groups.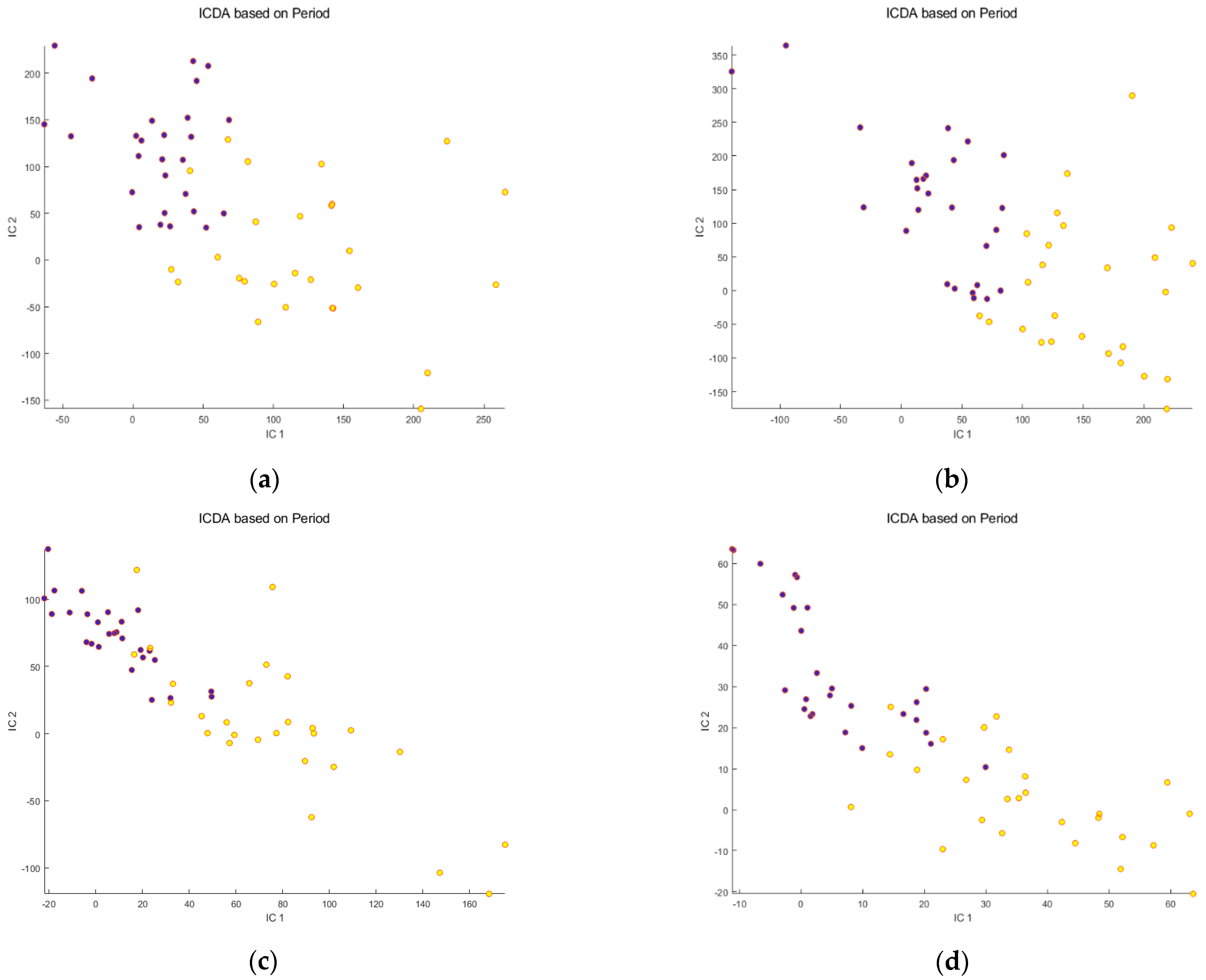

 : Before meal ingestion ;
: Before meal ingestion ;  : After meal ingestion. Note the better separation of the groups for non-corrected, raw RP data, whereas for HILIC there is little difference between osmolality-corrected, SNV-pretreated data and the non-corrected, raw data.
: After meal ingestion. Note the better separation of the groups for non-corrected, raw RP data, whereas for HILIC there is little difference between osmolality-corrected, SNV-pretreated data and the non-corrected, raw data.
 : Before meal ingestion ;
: Before meal ingestion ;  : After meal ingestion. Note the better separation of the groups for non-corrected, raw RP data, whereas for HILIC there is little difference between osmolality-corrected, SNV-pretreated data and the non-corrected, raw data.
: After meal ingestion. Note the better separation of the groups for non-corrected, raw RP data, whereas for HILIC there is little difference between osmolality-corrected, SNV-pretreated data and the non-corrected, raw data.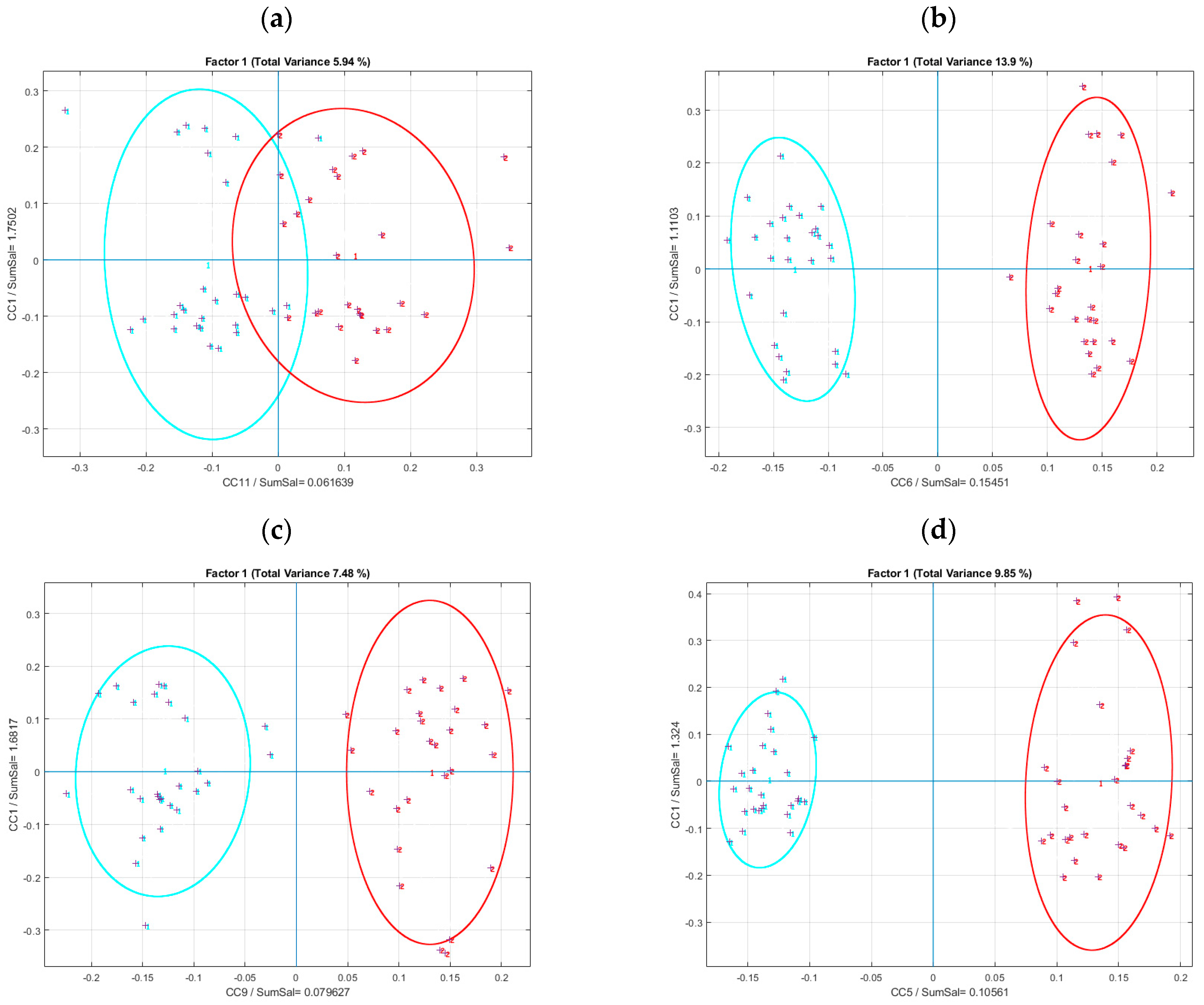

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khodorova, N.; Calvez, J.; Pilard, S.; Benoit, S.; Gaudichon, C.; Rutledge, D.N. Urine Metabolite Profiles after the Consumption of a Low- and a High-Digestible Protein Meal, and Comparison of Urine Normalization Techniques. Metabolites 2024, 14, 177. https://doi.org/10.3390/metabo14040177
Khodorova N, Calvez J, Pilard S, Benoit S, Gaudichon C, Rutledge DN. Urine Metabolite Profiles after the Consumption of a Low- and a High-Digestible Protein Meal, and Comparison of Urine Normalization Techniques. Metabolites. 2024; 14(4):177. https://doi.org/10.3390/metabo14040177
Chicago/Turabian StyleKhodorova, Nadezda, Juliane Calvez, Serge Pilard, Simon Benoit, Claire Gaudichon, and Douglas N. Rutledge. 2024. "Urine Metabolite Profiles after the Consumption of a Low- and a High-Digestible Protein Meal, and Comparison of Urine Normalization Techniques" Metabolites 14, no. 4: 177. https://doi.org/10.3390/metabo14040177
APA StyleKhodorova, N., Calvez, J., Pilard, S., Benoit, S., Gaudichon, C., & Rutledge, D. N. (2024). Urine Metabolite Profiles after the Consumption of a Low- and a High-Digestible Protein Meal, and Comparison of Urine Normalization Techniques. Metabolites, 14(4), 177. https://doi.org/10.3390/metabo14040177






