Transcriptome in Liver of Periparturient Dairy Cows Differs between Supplementation of Rumen-Protected Niacin and Rumen-Protected Nicotinamide
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Culture of Human Hepatocyte Line LO2
2.3. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
2.4. RNA Isolation and cDNA Library Construction
2.5. Analysis of Differential Expression of Genes
2.6. Analysis of DEG Enrichment
2.7. Statistical Analysis
3. Results
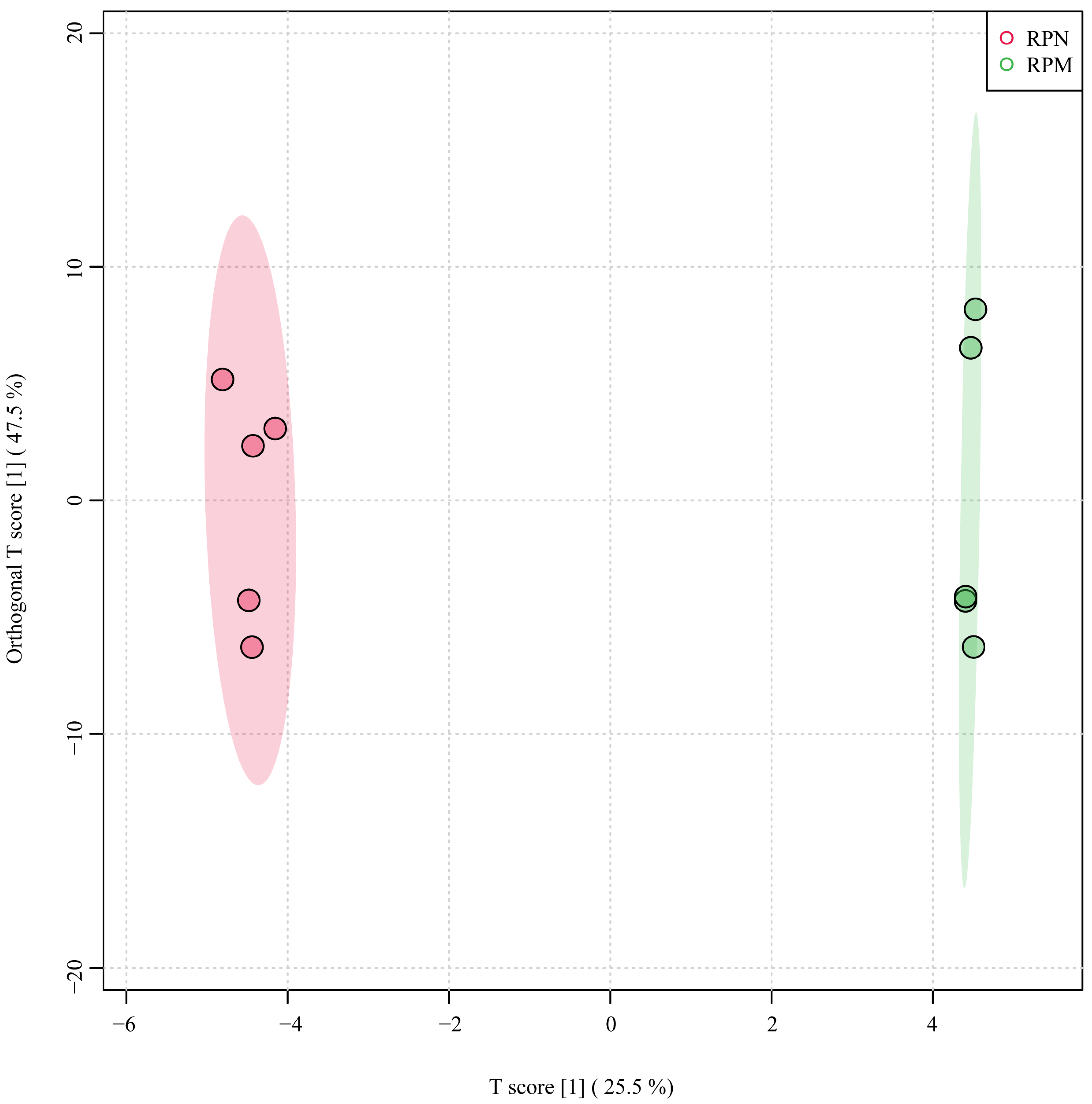
3.1. Sequencing Data Quality Control
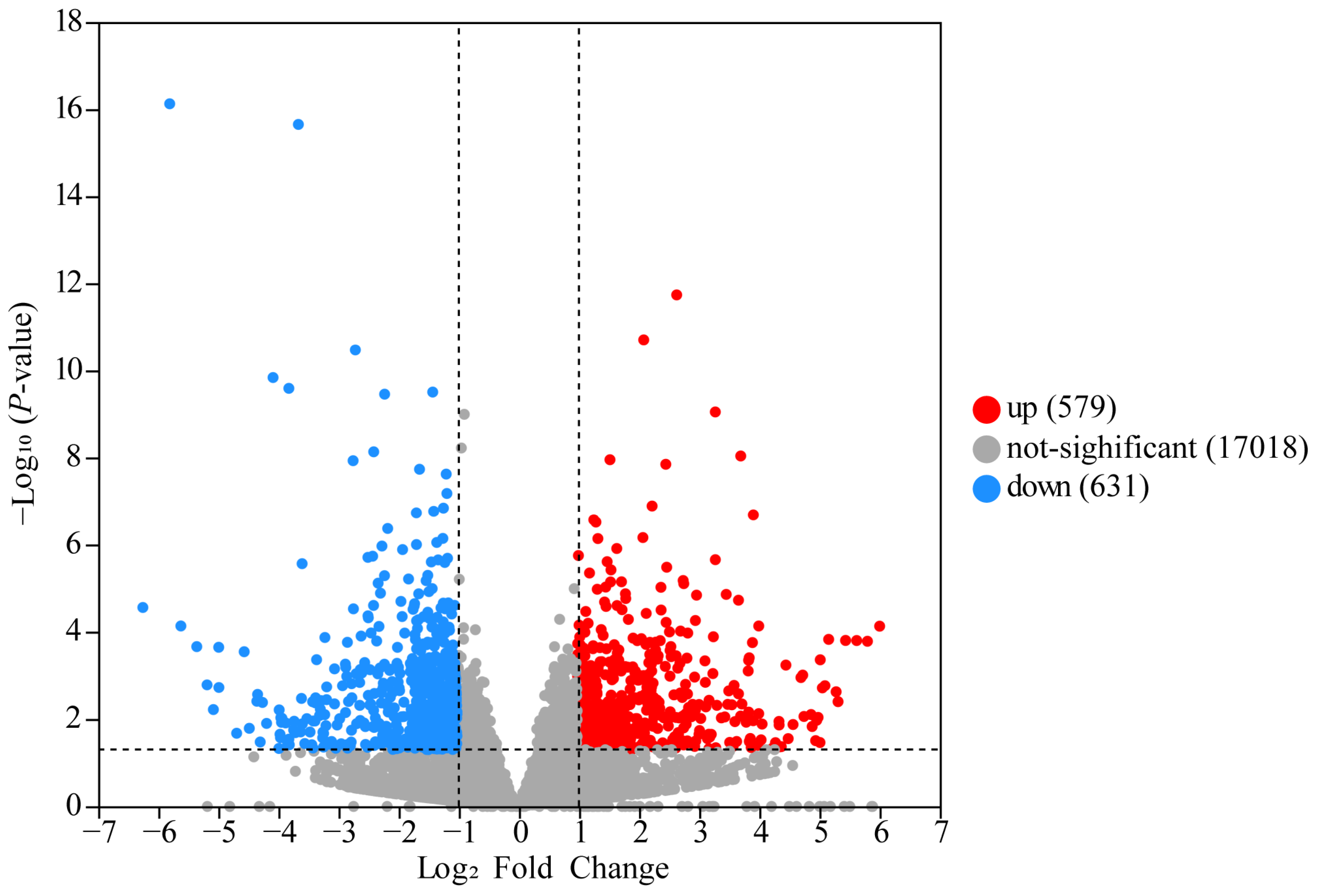
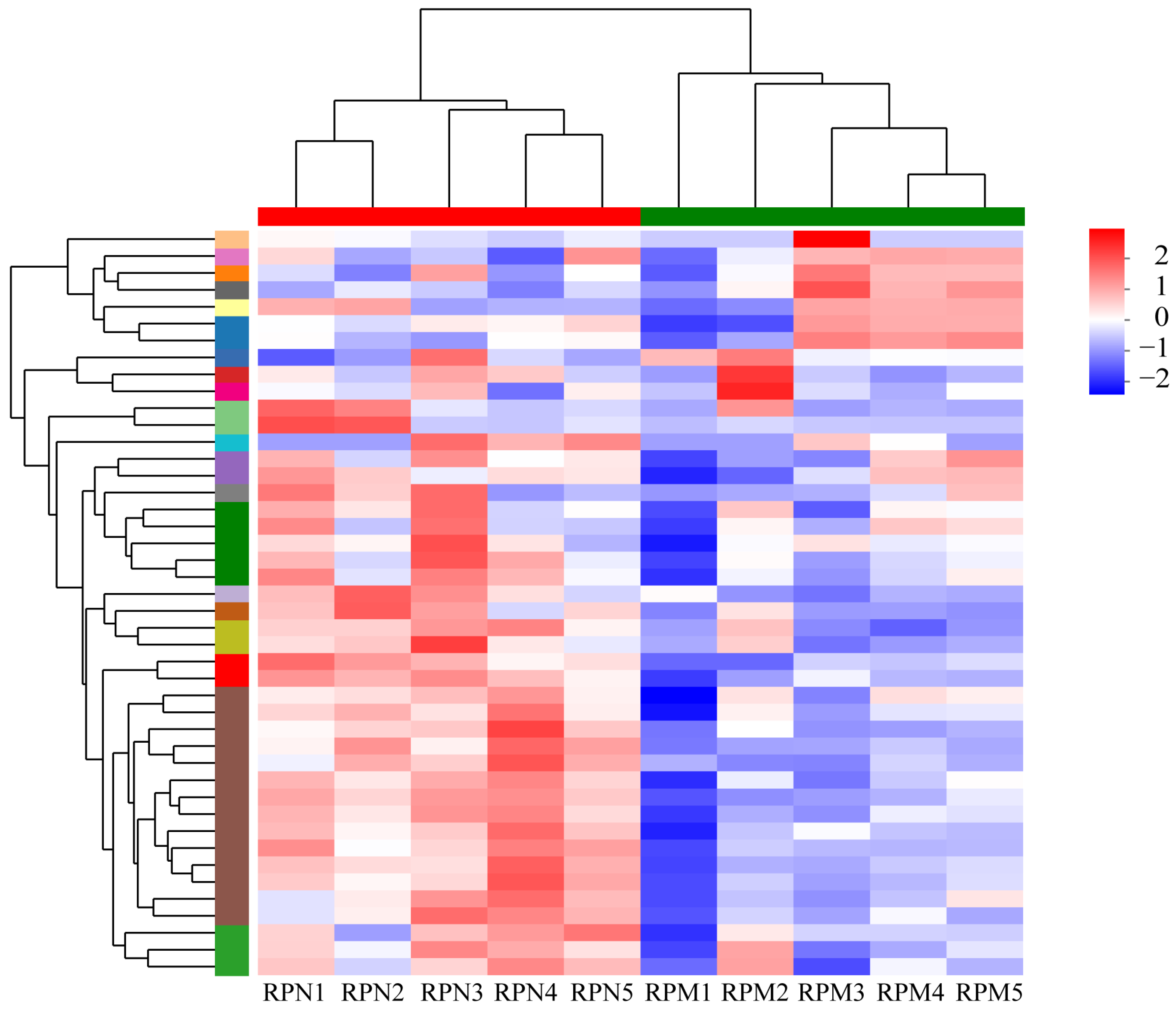
3.2. Analysis of DEGs
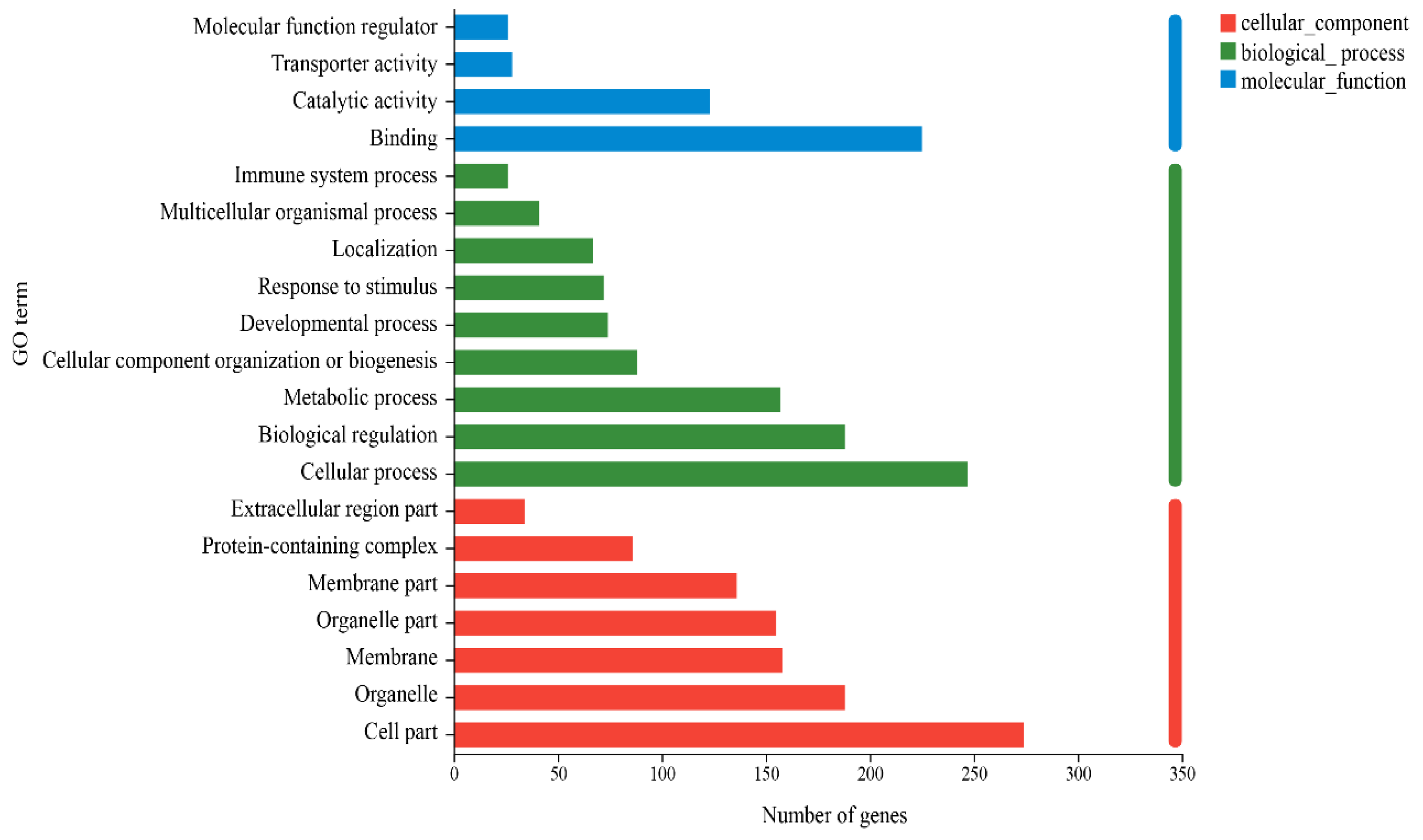
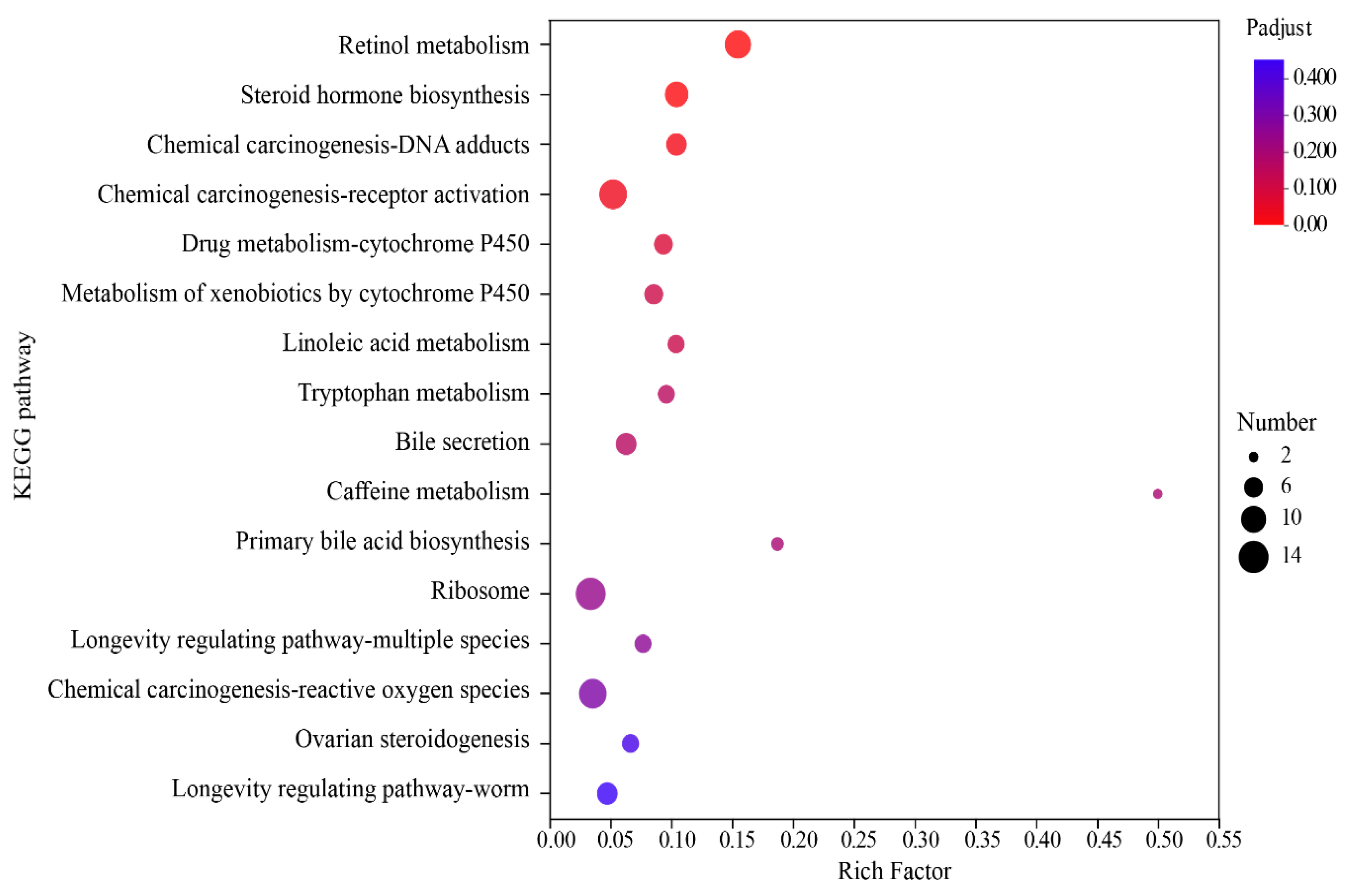
3.3. Analysis of Differential Gene Enrichment
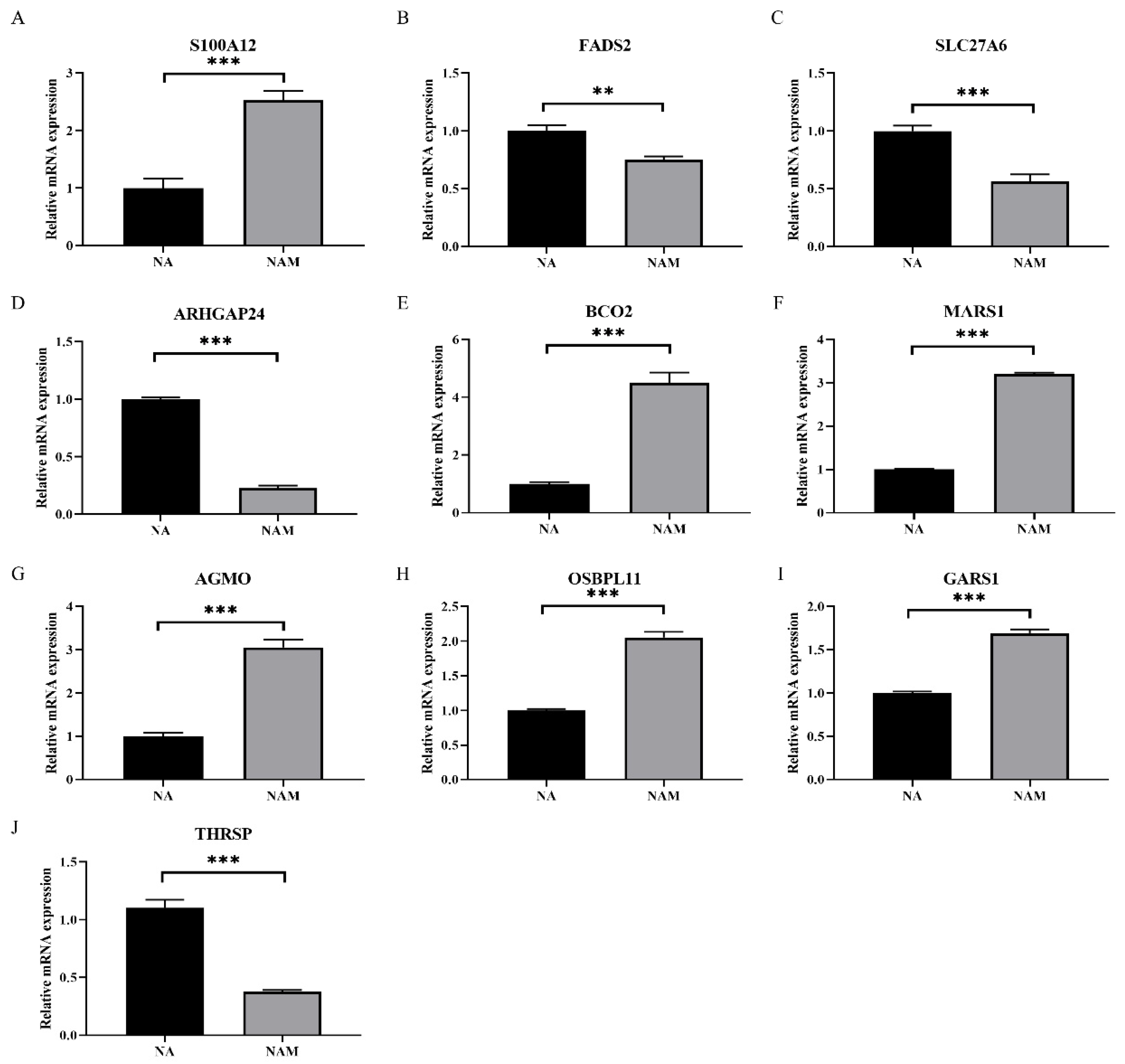
3.4. Validation of DEGs in LO2 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ishikawa, S.; Ikuta, K.; Obara, Y.; Oka, A.; Otani, Y.; Takahashi, Y.; Bai, H.; Terada, F.; Kushibiki, S. Cluster Analysis to Evaluate Disease Risk in Periparturient Dairy Cattle. Anim. Sci. J. 2020, 91, e13442. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Chiba, E.; Sugino, T.; Kawashima, K.; Kushibiki, S.; Kizaki, K.; Kim, Y.-H.; Sato, S. Liver Transcriptome Response to Periparturient Hormonal and Metabolic Changes Depends on the Postpartum Occurrence of Subacute Ruminal Acidosis in Holstein Cows. Physiol. Genom. 2021, 53, 285–294. [Google Scholar] [CrossRef]
- Mellouk, N.; Rame, C.; Naquin, D.; Jaszczyszyn, Y.; Touzé, J.-L.; Briant, E.; Guillaume, D.; Ntallaris, T.; Humblot, P.; Dupont, J. Impact of the Severity of Negative Energy Balance on Gene Expression in the Subcutaneous Adipose Tissue of Periparturient Primiparous Holstein Dairy Cows: Identification of Potential Novel Metabolic Signals for the Reproductive System. PLoS ONE 2019, 14, e0222954. [Google Scholar] [CrossRef]
- Wu, Z.-L.; Chen, S.-Y.; Hu, S.; Jia, X.; Wang, J.; Lai, S.-J. Metabolomic and Proteomic Profiles Associated with Ketosis in Dairy Cows. Front. Genet. 2020, 11, 551587. [Google Scholar] [CrossRef]
- Luo, Z.; Yong, K.; Du, Z.; Huang, Y.; Zhou, T.; Ma, L.; Yao, X.; Shen, L.; Yu, S.; Yan, Z.; et al. Association between Tryptophan Metabolism and Inflammatory Biomarkers in Dairy Cows with Ketosis. Metabolites 2023, 13, 333. [Google Scholar] [CrossRef]
- Niacin for Dairy Cattle: A Review|British Journal of Nutrition|Cambridge Core. Available online: https://www.cambridge.org/core/journals/british-journal-of-nutrition/article/niacin-for-dairy-cattle-a-review/CBDD2F19CE0BF0E5F9A1036CE028E4EB (accessed on 25 July 2023).
- Morey, S.D.; Mamedova, L.K.; Anderson, D.E.; Armendariz, C.K.; Titgemeyer, E.C.; Bradford, B.J. Effects of Encapsulated Niacin on Metabolism and Production of Periparturient Dairy Cows. J. Dairy Sci. 2011, 94, 5090–5104. [Google Scholar] [CrossRef]
- Chen, J.; Yang, Z.; Dong, G. Niacin Nutrition and Rumen-Protected Niacin Supplementation in Dairy Cows: An Updated Review. Br. J. Nutr. 2019, 122, 1103–1112. [Google Scholar] [CrossRef]
- Ye, L.; Cao, Z.; Lai, X.; Wang, W.; Guo, Z.; Yan, L.; Wang, Y.; Shi, Y.; Zhou, N. Niacin Fine-Tunes Energy Homeostasis through Canonical GPR109A Signaling. FASEB J. 2019, 33, 4765–4779. [Google Scholar] [CrossRef] [PubMed]
- Dhuguru, J.; Dellinger, R.W.; Migaud, M.E. Defining NAD(P)(H) Catabolism. Nutrients 2023, 15, 3064. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Patil, S.; Chauhan, B.; Guo, S.; Powell, D.R.; Le, J.; Klotsas, A.; Matika, R.; Xiao, X.; Franks, R.; et al. FoxO1 Regulates Multiple Metabolic Pathways in the Liver: Effects on Gluconeogenic, Glycolytic, and Lipogenic Gene Expression. J. Biol. Chem. 2006, 281, 10105–10117. [Google Scholar] [CrossRef] [PubMed]
- Lukasova, M.; Hanson, J.; Tunaru, S.; Offermanns, S. Nicotinic Acid (Niacin): New Lipid-Independent Mechanisms of Action and Therapeutic Potentials. Trends Pharmacol. Sci. 2011, 32, 700–707. [Google Scholar] [CrossRef]
- Kinoshita, A.; Kenéz, Á.; Locher, L.; Meyer, U.; Dänicke, S.; Rehage, J.; Huber, K. Insulin Signaling in Liver and Adipose Tissues in Periparturient Dairy Cows Supplemented with Dietary Nicotinic Acid. PLoS ONE 2016, 11, e0147028. [Google Scholar] [CrossRef][Green Version]
- Wei, X.; Yin, Q.; Zhao, H.; He, J.; Cao, Y.; Yao, J. Nicotinamide Supplementation during Postpartum and Peripartum Modulates Hepatic Energy and Lipid Metabolism, Oxidative Status, and Metabolomics Profile, as Well as Lipids in the Adipose Tissue of Goats. Anim. Feed Sci. Technol. 2021, 274, 114849. [Google Scholar] [CrossRef]
- Luo, C.; Yang, C.; Wang, X.; Chen, Y.; Liu, X.; Deng, H. Nicotinamide Reprograms Adipose Cellular Metabolism and Increases Mitochondrial Biogenesis to Ameliorate Obesity. J. Nutr. Biochem. 2022, 107, 109056. [Google Scholar] [CrossRef]
- Jaster, E.H.; Ward, N.E. Supplemental Nicotinic Acid or Nicotinamide for Lactating Dairy Cows. J. Dairy Sci. 1990, 73, 2880–2887. [Google Scholar] [CrossRef] [PubMed]
- Aragona, K.M.; Rice, E.M.; Engstrom, M.; Erickson, P.S. Supplementation of Nicotinic Acid to Prepartum Holstein Cows Increases Colostral Immunoglobulin G, Excretion of Urinary Purine Derivatives, and Feed Efficiency in Calves. J. Dairy Sci. 2020, 103, 2287–2302. [Google Scholar] [CrossRef] [PubMed]
- Petrović, K.; Djoković, R.; Cincović, M.; Hristovska, T.; Lalović, M.; Petrović, M.; Majkić, M.; Došenović Marinković, M.; Anđušić, L.; Devečerski, G.; et al. Niacin Status Indicators and Their Relationship with Metabolic Parameters in Dairy Cows during Early Lactation. Animals 2022, 12, 1524. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Shaver, R.D.; Bertics, S.J.; Espineira, M.; Grummer, R.R. Effect of Rumen-Protected Niacin on Lipid Metabolism, Oxidative Stress, and Performance of Transition Dairy Cows. J. Dairy Sci. 2012, 95, 2673–2679. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, Y.; Che, H.; Sun, Y.; Wang, S.; Zhan, Y.; Cai, D.; Chen, S. Metabolic Engineering of Bacillus Licheniformis for the Bioproduction of Nicotinamide Riboside from Nicotinamide and Glucose. ACS Sustain. Chem. Eng. 2023, 11, 6201–6210. [Google Scholar] [CrossRef]
- Shen, T.; Xu, F.; Fang, Z.; Loor, J.J.; Ouyang, H.; Chen, M.; Jin, B.; Wang, X.; Shi, Z.; Zhu, Y.; et al. Hepatic Autophagy and Mitophagy Status in Dairy Cows with Subclinical and Clinical Ketosis. J. Dairy Sci. 2021, 104, 4847–4857. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Weigel, N.L.; Moore, N.L. Kinases and Protein Phosphorylation as Regulators of Steroid Hormone Action. Nucl Recept Signal 2007, 5, nrs.05005. [Google Scholar] [CrossRef]
- Hayaishi, O. Oxygenases. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 371–374. ISBN 978-0-12-378631-9. [Google Scholar]
- Ren, J.; Xu, N.; Zheng, H.; Tian, W.; Li, H.; Li, Z.; Wang, Y.; Tian, Y.; Kang, X.; Liu, X. Expression of Thyroid Hormone Responsive SPOT 14 Gene Is Regulated by Estrogen in Chicken (Gallus Gallus). Sci. Rep. 2017, 7, 10243. [Google Scholar] [CrossRef]
- Tsatsos, N.G.; Augustin, L.B.; Anderson, G.W.; Towle, H.C.; Mariash, C.N. Hepatic Expression of the SPOT 14 (S14) Paralog S14-Related (Mid1 Interacting Protein) Is Regulated by Dietary Carbohydrate. Endocrinology 2008, 149, 5155–5161. [Google Scholar] [CrossRef][Green Version]
- Zeng, N.; Huang, R.; Li, N.; Jiang, H.; Li, R.; Wang, F.; Chen, W.; Xia, M.; Wang, Q. MiR-451a Attenuates Free Fatty Acids–Mediated Hepatocyte Steatosis by Targeting the Thyroid Hormone Responsive Spot 14 Gene. Mol. Cell. Endocrinol. 2018, 474, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Gudas, L.J. Retinoid Metabolism: New Insights. J. Mol. Endocrinol. 2022, 69, T37–T49. [Google Scholar] [CrossRef]
- Strychalski, J.; Gugołek, A.; Antoszkiewicz, Z.; Fopp-Bayat, D.; Kaczorek-Łukowska, E.; Snarska, A.; Zwierzchowski, G.; Król-Grzymała, A.; Matusevičius, P. The Effect of the BCO2 Genotype on the Expression of Genes Related to Carotenoid, Retinol, and α-Tocopherol Metabolism in Rabbits Fed a Diet with Aztec Marigold Flower Extract. Int. J. Mol. Sci. 2022, 23, 10552. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Liu, C.; Hu, K.-Q.; Smith, D.E.; Wang, X.-D. Ablation of Carotenoid Cleavage Enzymes (BCO1 and BCO2) Induced Hepatic Steatosis by Altering the Farnesoid X Receptor/miR-34a/Sirtuin 1 Pathway. Arch. Biochem. Biophys. 2018, 654, 1–9. [Google Scholar] [CrossRef]
- Tian, R.; Cullen, N.G.; Morris, C.A.; Fisher, P.J.; Pitchford, W.S.; Bottema, C.D.K. Major Effect of Retinal Short-Chain Dehydrogenase Reductase (RDHE2) on Bovine Fat Colour. Mamm. Genome 2012, 23, 378–386. [Google Scholar] [CrossRef]
- Wu, L.; Lu, P.; Guo, X.; Song, K.; Lyu, Y.; Bothwell, J.; Wu, J.; Hawkins, O.; Clarke, S.L.; Lucas, E.A.; et al. β-Carotene Oxygenase 2 Deficiency-Triggered Mitochondrial Oxidative Stress Promotes Low-Grade Inflammation and Metabolic Dysfunction. Free Radic. Biol. Med. 2021, 164, 271–284. [Google Scholar] [CrossRef]
- Zachut, M.; Contreras, G.A. Symposium Review: Mechanistic Insights into Adipose Tissue Inflammation and Oxidative Stress in Periparturient Dairy Cows. J. Dairy Sci. 2022, 105, 3670–3686. [Google Scholar] [CrossRef]
- Dong, X.; Tan, P.; Cai, Z.; Xu, H.; Li, J.; Ren, W.; Xu, H.; Zuo, R.; Zhou, J.; Mai, K.; et al. Regulation of FADS2 Transcription by SREBP-1 and PPAR-α Influences LC-PUFA Biosynthesis in Fish. Sci. Rep. 2017, 7, 40024. [Google Scholar] [CrossRef]
- Young, R.S.E.; Bowman, A.P.; Williams, E.D.; Tousignant, K.D.; Bidgood, C.L.; Narreddula, V.R.; Gupta, R.; Marshall, D.L.; Poad, B.L.J.; Nelson, C.C.; et al. Apocryphal FADS2 Activity Promotes Fatty Acid Diversification in Cancer. Cell Rep. 2021, 34, 108738. [Google Scholar] [CrossRef]
- Walle, P.; Männistö, V.; de Mello, V.D.; Vaittinen, M.; Perfilyev, A.; Hanhineva, K.; Ling, C.; Pihlajamäki, J. Liver DNA Methylation of FADS2 Associates with FADS2 Genotypex. Clin. Epigenetics 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.M.; Stahl, A. SLC27 Fatty Acid Transport Proteins. Mol. Asp. Med. 2013, 34, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Kojder, K.; Jeżewski, D.; Simińska, D.; Tomasiak, P.; Tarnowski, M.; Chlubek, D.; Baranowska-Bosiacka, I. Reduced Expression of Very-Long-Chain Acyl-CoA Synthetases SLC27A4 and SLC27A6 in the Glioblastoma Tumor Compared to the Peritumoral Area. Brain Sci. 2023, 13, 771. [Google Scholar] [CrossRef] [PubMed]
- Stahl, A. A Current Review of Fatty Acid Transport Proteins (SLC27). Pflug. Arch. Eur. J. Physiol. 2004, 447, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Wilken-Schmitz, A.; Hernandez-Olmos, V.; Proschak, E.; Stark, H.; Fleming, I.; Weigert, A.; Thurn, M.; Hofmann, M.; Werner, E.R.; et al. AGMO Inhibitor Reduces 3T3-L1 Adipogenesis. Cells 2021, 10, 1081. [Google Scholar] [CrossRef] [PubMed]
- Sailer, S.; Lackner, K.; Pras-Raves, M.L.; Wever, E.J.M.; van Klinken, J.B.; Dane, A.D.; Geley, S.; Koch, J.; Golderer, G.; Werner-Felmayer, G.; et al. Adaptations of the 3T3-L1 Adipocyte Lipidome to Defective Ether Lipid Catabolism upon Agmo Knockdown. J. Lipid Res. 2022, 63, 100222. [Google Scholar] [CrossRef] [PubMed]
- Sailer, S.; Keller, M.A.; Werner, E.R.; Watschinger, K. The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification. Life 2021, 11, 88. [Google Scholar] [CrossRef]
- Vormittag, S.; Hüsler, D.; Haneburger, I.; Kroniger, T.; Anand, A.; Prantl, M.; Barisch, C.; Maaß, S.; Becher, D.; Letourneur, F.; et al. Legionella- and Host-Driven Lipid Flux at LCV-ER Membrane Contact Sites Promotes Vacuole Remodeling. EMBO Rep. 2023, 24, e56007. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Cui, Z.; Zhang, R.; Zhao, K.; Wang, L.; Yao, J.; Liu, S.; Cai, C.; Cao, Y. The Effects of Rumen-Protected Choline and Rumen-Protected Nicotinamide on Liver Transcriptomics in Periparturient Dairy Cows. Metabolites 2023, 13, 594. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, W.; Shen, W.; Wang, L.; Zuo, Y. ARHGAP24 Ameliorates Inflammatory Response through Inactivating Rac1/Akt/NF-κB Pathway in Acute Pneumonia Model of Rat. Ann. Transl. Med. 2020, 8, 1289. [Google Scholar] [CrossRef]
- Sreejit, G.; Flynn, M.C.; Patil, M.; Krishnamurthy, P.; Murphy, A.J.; Nagareddy, P.R. Chapter Six—S100 Family Proteins in Inflammation and Beyond. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 98, pp. 173–231. [Google Scholar]
- Nazari, A.; Khorramdelazad, H.; Hassanshahi, G.; Day, A.S.; Sardoo, A.M.; Fard, E.T.; Abedinzadeh, M.; Nadimi, A.E. S100A12 in Renal and Cardiovascular Diseases. Life Sci. 2017, 191, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yan, W.X.; Cai, H.; Tedla, N.; Armishaw, C.; Di Girolamo, N.; Wang, H.W.; Hampartzoumian, T.; Simpson, J.L.; Gibson, P.G.; et al. S100A12 Provokes Mast Cell Activation: A Potential Amplification Pathway in Asthma and Innate Immunity. J. Allergy Clin. Immunol. 2007, 119, 106–114. [Google Scholar] [CrossRef]
- Carvalho, A.; Lu, J.; Francis, J.D.; Moore, R.E.; Haley, K.P.; Doster, R.S.; Townsend, S.D.; Johnson, J.G.; Damo, S.M.; Gaddy, J.A. S100A12 in Digestive Diseases and Health: A Scoping Review. Gastroenterol. Res. Pract. 2020, 2020, e2868373. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.I.; Kwon, N.H.; Lim, B.J.; Nahm, J.H.; Park, J.S.; Kang, C.M.; Park, S.R.; Kang, C.M.; Park, S.R.; Lee, S.Y.; et al. New Staining Method Using Methionyl-tRNA Synthetase 1 Antibody for Brushing Cytology of Bile Duct Cancer. Gastrointest. Endosc. 2020, 92, 310–319. [Google Scholar] [CrossRef]
- Nie, J.; Liu, T.; Mao, T.; Yang, H.; Deng, W.; Liu, X.; Fu, B. Transcriptome Sequencing and Single-Cell Sequencing Analysis Identify GARS1 as a Potential Prognostic and Immunotherapeutic Biomarker for Multiple Cancers, Including Bladder Cancer. Front. Immunol. 2023, 14, 1169588. [Google Scholar] [CrossRef]
- Oprescu, S.N.; Griffin, L.B.; Beg, A.A.; Antonellis, A. Predicting the Pathogenicity of Aminoacyl-tRNA Synthetase Mutations. Methods 2017, 113, 139–151. [Google Scholar] [CrossRef]
- Yang, S.J.; Choi, J.M.; Kim, L.; Park, S.E.; Rhee, E.J.; Lee, W.Y.; Oh, K.W.; Park, S.W.; Park, C.-Y. Nicotinamide Improves Glucose Metabolism and Affects the Hepatic NAD-Sirtuin Pathway in a Rodent Model of Obesity and Type 2 Diabetes. J. Nutr. Biochem. 2014, 25, 66–72. [Google Scholar] [CrossRef] [PubMed]
| Prepartum | Postpartum | |
|---|---|---|
| Ingredients, DM% | ||
| Corn silage | 22.67 | 30.36 |
| Oat hay | 8.12 | 4.55 |
| Straw | 39.35 | 0.00 |
| Alfalfa hay | 0.00 | 14.43 |
| Flaking corn | 0.00 | 15.92 |
| Soybean meal | 0.00 | 3.06 |
| Cottonseed meal | 0.00 | 4.54 |
| NaHCO3 | 0.00 | 0.95 |
| 1512 1 | 29.86 | 0.00 |
| 1818 2 | 0.00 | 16.10 |
| 1979 3 | 0.00 | 10.08 |
| 4 Chemical composition, DM% | ||
| NEL (Mcal/kg) | 1.30 | 1.69 |
| CP | 10.97 | 17.49 |
| EE | 3.48 | 4.22 |
| Ash | 8.11 | 8.71 |
| NDF | 52.10 | 36.39 |
| ADF | 28.34 | 20.53 |
| Starch | 14.00 | 25.00 |
| Ca | 0.87 | 0.93 |
| p | 0.56 | 0.57 |
| Target Gene | Forward (5′-3′) | Forward (5′-3′) |
|---|---|---|
| S100A12 | GCTCAGTGCCCTTCACCACT | AGCCTTCAGCGCAATGGCTA |
| FADS2 | ACCGTGACTGGTTCAGTAGC | CGGGGCGATCTTGTGTAAGT |
| SLC27A6 | GAAGAGAAGGACGCTGGTGG | AAGTGCAGGACGACCATTCC |
| ARHGAP24 | GTATCGCCAATGCAGGATGC | CGTGGGCATGGTTTAGCAAG |
| BCO2 | GGCTGATGGAACGATCTGGT | CCCCACTAAATGCCGAAAGC |
| MARS1 | TCCATGCTGACATCTACCGC | CAGTGCTCACATCGCAGTTG |
| AGMO | ACTACATTCTGGGCCACACC | GGGAACTTCTTTGCCGGTGA |
| OSBPL11 | ATGAAGCTGGGCTGTTGGAG | GTGCTGTCGCTCTTTTGCAT |
| GARS1 | TCTCCCCTCGATCTGGACTG | TGACCTGGGCTTTTGCTGAA |
| GAPDH | AAGGTCGGAGTGAACGGATTC | ATTGATGGCGACGATGTCCA |
| THRSP | TGAGGCCCCTGATCTCTACA | CTTCCTCTGTCTCTGCGGTT |
| Gene Name | Gene Description | Log2FC (RPN/RPM) | p-Value |
|---|---|---|---|
| THRSP | Thyroid hormone responsive | 5.7793 | 7.50 × 10−17 |
| CYP26A1 | Cytochrome P450, family 26, subfamily A, Polypeptide 1 | 3.6386 | 2.23 × 10−16 |
| MT1E | Metallothionein 1E | −2.6549 | 1.84 × 10−12 |
| ARMH1 | Armadillo like helical domain containing 1 | −2.1066 | 1.98 × 10−11 |
| CYP2C19 | Cytochrome P450, family 2, subfamily C, Polypeptide 19 | 2.6907 | 3.39 × 10−11 |
| SEC14L3 | SEC14 like lipid binding 3 | 4.0599 | 1.46 × 10−10 |
| CES1 | Carboxylesterase 1 (monocyte/macrophage serine esterase 1) | 1.4039 | 3.15 × 10−10 |
| KCNN2 | Potassium calcium-activated channel Subfamily N member 2 | 2.2039 | 3.52 × 10−10 |
| S100A12 | S100 calcium binding protein A12 | −3.2975 | 9.00 × 10−10 |
| ATP6V1C2 | ATPase H+ transporting V1 subunit C2 | −3.7203 | 9.22 × 10−9 |
| PLXNB1 | plexin B1 | −1.5450 | 1.12 × 10−8 |
| FADS2 | Fatty acid desaturase 2 | 2.7283 | 1.18 × 10−8 |
| SLC27A6 | Solute carrier family 27 member 6 | 1.6232 | 1.86 × 10−8 |
| HSD17B6 | Hydroxysteroid (17-beta) dehydrogenase 6 | 1.1792 | 2.40 × 10−8 |
| ARHGAP24 | Rho gtpase activating protein 24 | 1.1679 | 6.69 × 10−8 |
| MUSTN1 | Musculoskeletal, embryonic nuclear protein 1 | −2.2457 | 1.30 × 10−7 |
| BCO2 | Beta-carotene oxygenase 2 | 1.2256 | 1.45 × 10−7 |
| ANO1 | Anoctamin 1 | 1.3874 | 1.72 × 10−7 |
| TRPM2 | Transient receptor potential cation channel subfamily M member 2 | 1.6755 | 1.87 × 10−7 |
| NT5DC2 | 5’-nucleotidase domain containing 2 | −1.2762 | 2.71 × 10−7 |
| ECD | Ecdysoneless cell cycle regulator | −1.3113 | 3.05 × 10−7 |
| MARS1 | Methionyl-trna synthetase 1 | −2.0929 | 6.92 × 10−7 |
| BACE1 | Beta-secretase 1 | −1.3440 | 7.30 × 10−7 |
| AGMO | Alkylglycerol monooxygenase | 1.3366 | 8.96 × 10−7 |
| ODF2L | Outer dense fiber of sperm tails 2 like | 1.6741 | 1.00 × 10−6 |
| OSBPL11 | Oxysterol binding protein like 11 | 2.2486 | 1.08 × 10−6 |
| WFS1 | Wolframin ER transmembrane glycoprotein | −1.6581 | 1.24 × 10−6 |
| JARID2 | Jumonji and AT-rich interaction domain containing 2 | 1.9052 | 1.31 × 10−6 |
| GARS1 | Glycyl-trna synthetase 1 | −1.0204 | 1.79 × 10−6 |
| SPOCK1 | SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 | 2.4819 | 1.97 × 10−6 |
| GO ID | Category | Description | p-Value | Counts |
|---|---|---|---|---|
| GO:0006706 | BP | Steroid catabolic process | 1.29 × 10−7 | 6 |
| GO:0006805 | BP | Xenobiotic metabolic process | 3.08 × 10−7 | 11 |
| GO:0008395 | MF | Steroid hydroxylase activity | 1.43 × 10−7 | 10 |
| GO:0004497 | MF | Monooxygenase activity | 1.80 × 10−7 | 15 |
| GO:0020037 | MF | Heme binding | 2.34 × 10−7 | 16 |
| GO:0005506 | MF | Iron ion binding | 3.34 × 10−7 | 17 |
| GO:0046906 | MF | Tetrapyrrole binding | 4.29 × 10−7 | 16 |
| GO:0016705 | MF | Oxidoreductase activity, acting on paired Donors, with incorporation or reduction in molecular oxygen | 4.77 × 10−7 | 18 |
| GO:0008202 | BP | Steroid metabolic process | 8.64 × 10−7 | 13 |
| GO:0120254 | BP | Olefinic compound metabolic process | 1.16 × 10−6 | 10 |
| GO:0016491 | MF | Oxidoreductase activity | 2.03 × 10−6 | 36 |
| GO:0044281 | BP | Small molecule metabolic process | 4.08 × 10−6 | 40 |
| GO:0044237 | BP | Cellular metabolic process | 5.54 × 10−6 | 145 |
| GO:0001523 | BP | Retinoid metabolic process | 5.76 × 10−6 | 7 |
| GO:0016101 | BP | Diterpenoid metabolic process | 7.35 × 10−6 | 7 |
| GO:0070988 | BP | Demethylation | 8.27 × 10−6 | 7 |
| GO:0008152 | BP | Metabolic process | 1.03 × 10−5 | 157 |
| GO:0046872 | MF | Metal ion binding | 1.02 × 10−5 | 87 |
| GO:0016712 | MF | Oxidoreductase activity, acting on paired donors, with incorporation or reduction in Molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | 1.03 × 10−5 | 6 |
| GO:0006721 | BP | Terpenoid metabolic process | 1.77 × 10−5 | 7 |
| Pathway ID | KEGG Pathway | p-Value | Counts |
|---|---|---|---|
| map00830 | Retinol metabolism | 3.12 × 10−9 | 11 |
| map00140 | Steroid hormone biosynthesis | 2.66 × 10−6 | 9 |
| map05204 | Chemical carcinogenesis-DNA adducts | 3.54 × 10−5 | 7 |
| map05207 | Chemical carcinogenesis-receptor activation | 7.76 × 10−5 | 12 |
| map00982 | Drug metabolism-cytochrome P450 | 0.00024 | 6 |
| map00980 | Metabolism of xenobiotics by cytochrome P450 | 0.00039 | 6 |
| map00591 | Linoleic acid metabolism | 0.00048 | 5 |
| map00380 | Tryptophan metabolism | 0.00070 | 5 |
| map04976 | Bile secretion | 0.00083 | 7 |
| map00232 | Caffeine metabolism | 0.00109 | 2 |
| map00120 | Primary bile acid biosynthesis | 0.00124 | 3 |
| map03010 | Ribosome | 0.00168 | 14 |
| map04213 | Longevity regulating pathway-multiple species | 0.00194 | 5 |
| map05208 | Chemical carcinogenesis-reactive oxygen species | 0.00236 | 12 |
| map04913 | Ovarian steroidogenesis | 0.00363 | 5 |
| map04212 | Longevity regulating pathway-worm | 0.00412 | 7 |
| map00983 | Drug metabolism-other enzymes | 0.00587 | 5 |
| map01040 | Biosynthesis of unsaturated fatty acids | 0.00932 | 3 |
| map04923 | Regulation of lipolysis in adipocytes | 0.00964 | 4 |
| map04978 | Mineral absorption | 0.01155 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, R.; Du, X.; Cui, Z.; Jiang, X.; Wang, L.; Yao, J.; Liu, S.; Wang, J.; Cai, C.; et al. Transcriptome in Liver of Periparturient Dairy Cows Differs between Supplementation of Rumen-Protected Niacin and Rumen-Protected Nicotinamide. Metabolites 2024, 14, 150. https://doi.org/10.3390/metabo14030150
Zhang Y, Li R, Du X, Cui Z, Jiang X, Wang L, Yao J, Liu S, Wang J, Cai C, et al. Transcriptome in Liver of Periparturient Dairy Cows Differs between Supplementation of Rumen-Protected Niacin and Rumen-Protected Nicotinamide. Metabolites. 2024; 14(3):150. https://doi.org/10.3390/metabo14030150
Chicago/Turabian StyleZhang, Yuanjie, Rongrong Li, Xue’er Du, Zhijie Cui, Xingwei Jiang, Lamei Wang, Junhu Yao, Shimin Liu, Jianguo Wang, Chuanjiang Cai, and et al. 2024. "Transcriptome in Liver of Periparturient Dairy Cows Differs between Supplementation of Rumen-Protected Niacin and Rumen-Protected Nicotinamide" Metabolites 14, no. 3: 150. https://doi.org/10.3390/metabo14030150
APA StyleZhang, Y., Li, R., Du, X., Cui, Z., Jiang, X., Wang, L., Yao, J., Liu, S., Wang, J., Cai, C., & Cao, Y. (2024). Transcriptome in Liver of Periparturient Dairy Cows Differs between Supplementation of Rumen-Protected Niacin and Rumen-Protected Nicotinamide. Metabolites, 14(3), 150. https://doi.org/10.3390/metabo14030150







