In-Situ Metabolic Profiling of Different Kinds of Rheum palmatum L. by Laser Desorption–Dielectric Barrier Discharge Ionization Mass Spectrometry Imaging
Abstract
1. Introduction
2. Materials and Methods
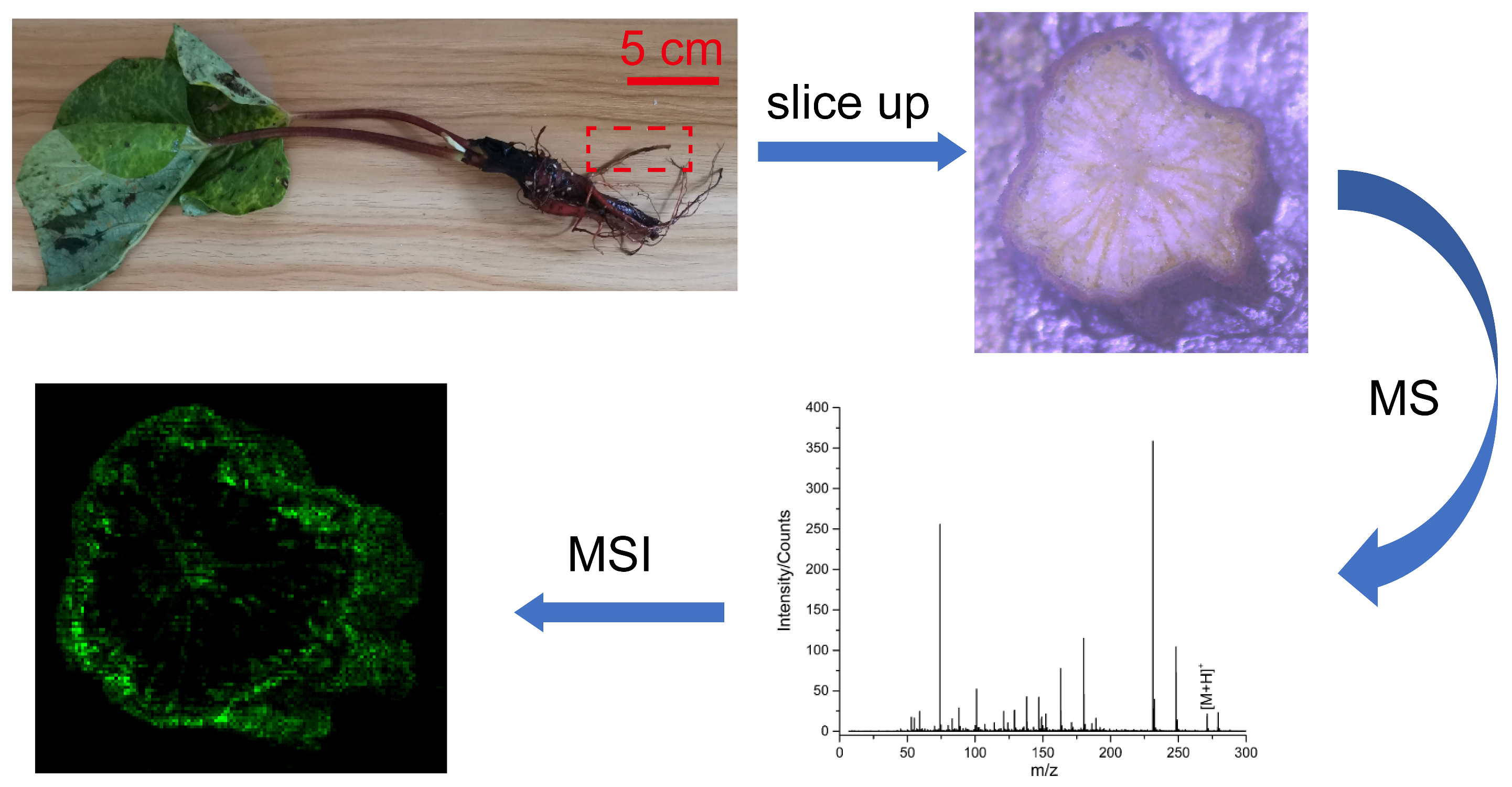
2.1. Reagents and Plant Root Slice Preparation
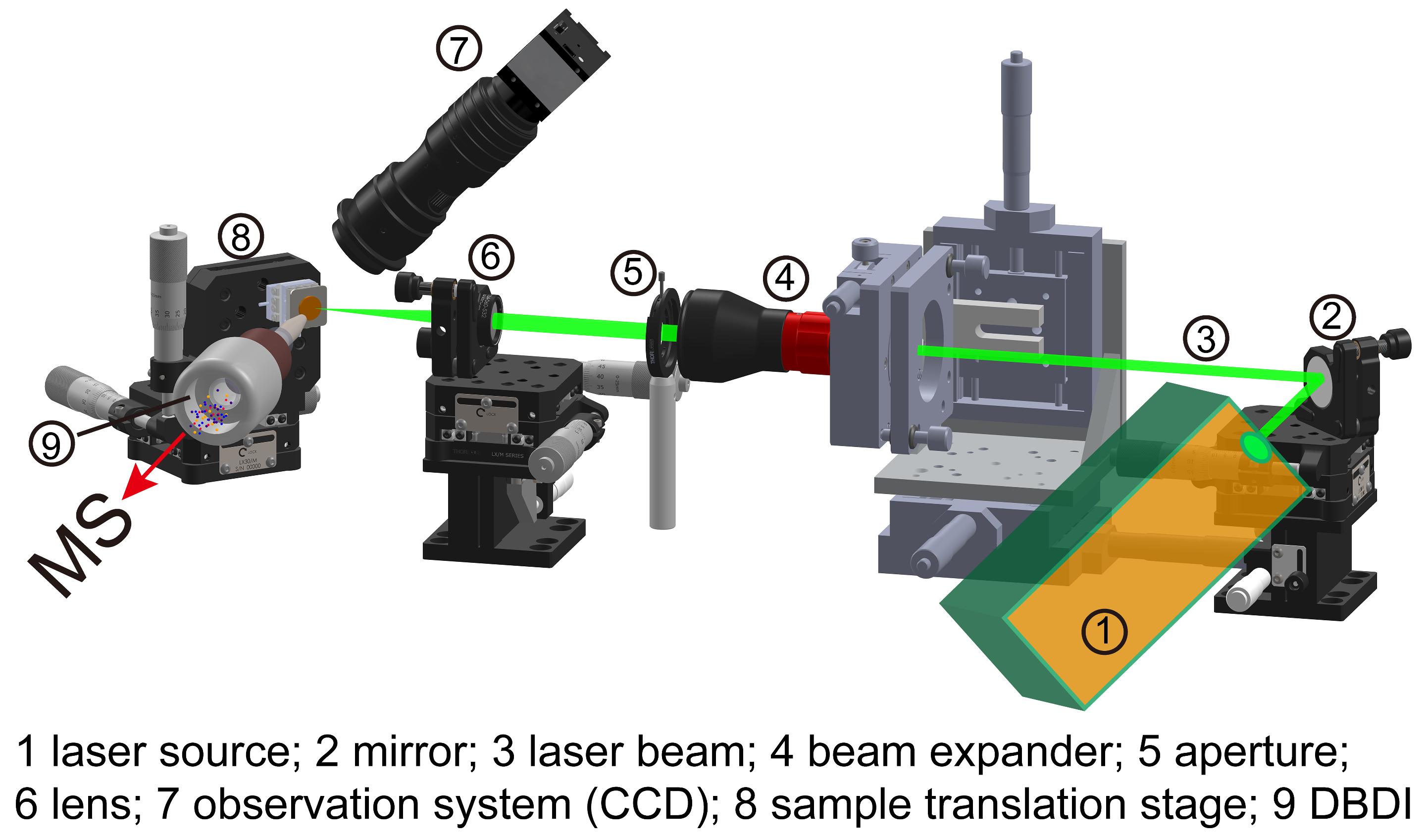
2.2. Laser Desorption System
2.3. Dielectric Barrier Discharge Ionization Source
2.4. Mass Spectrometry and Mass Spectrometry Imaging Setup
2.5. Data Processing and Drawing
3. Results
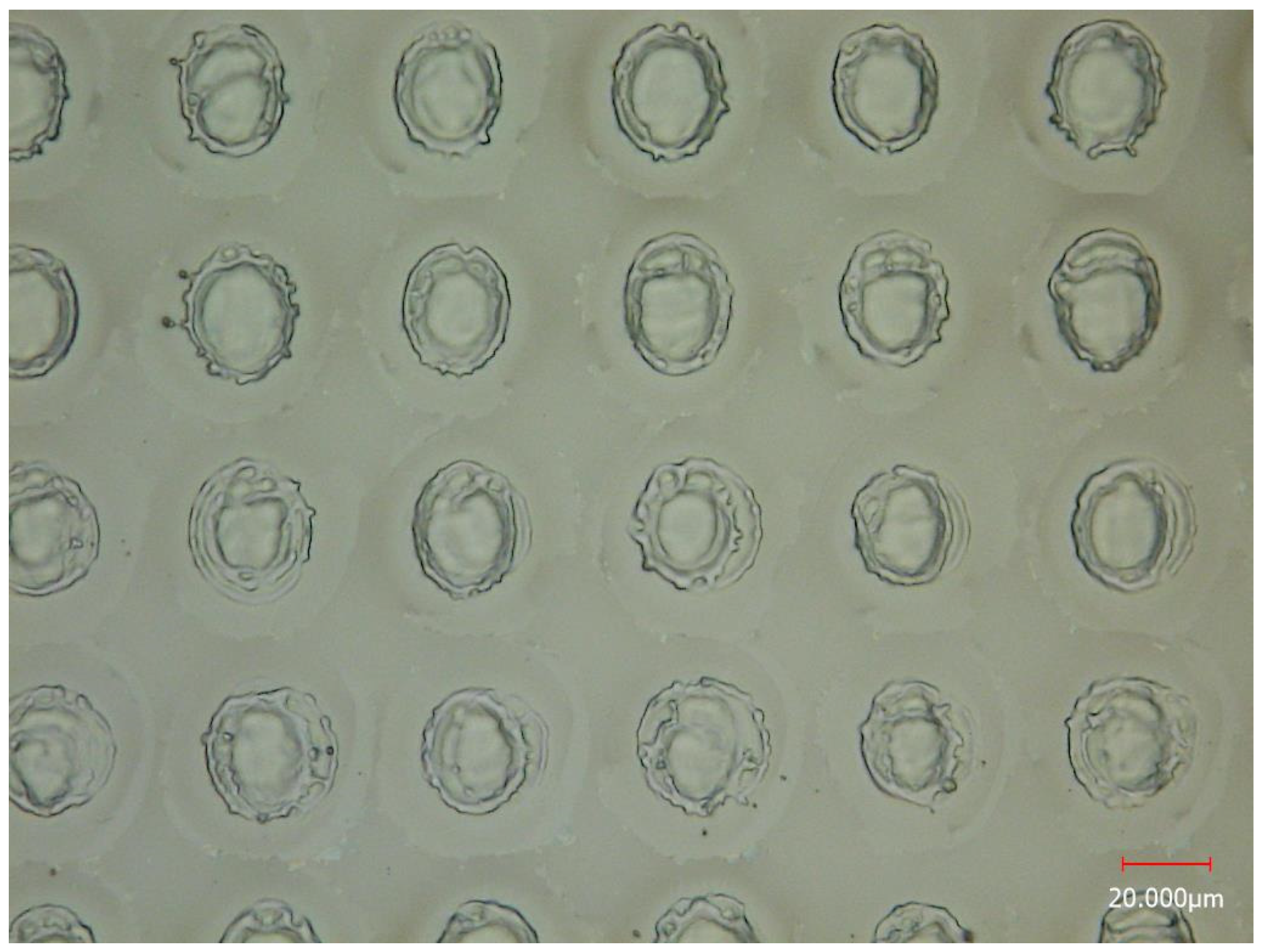
3.1. Spatial Resolution of MSI
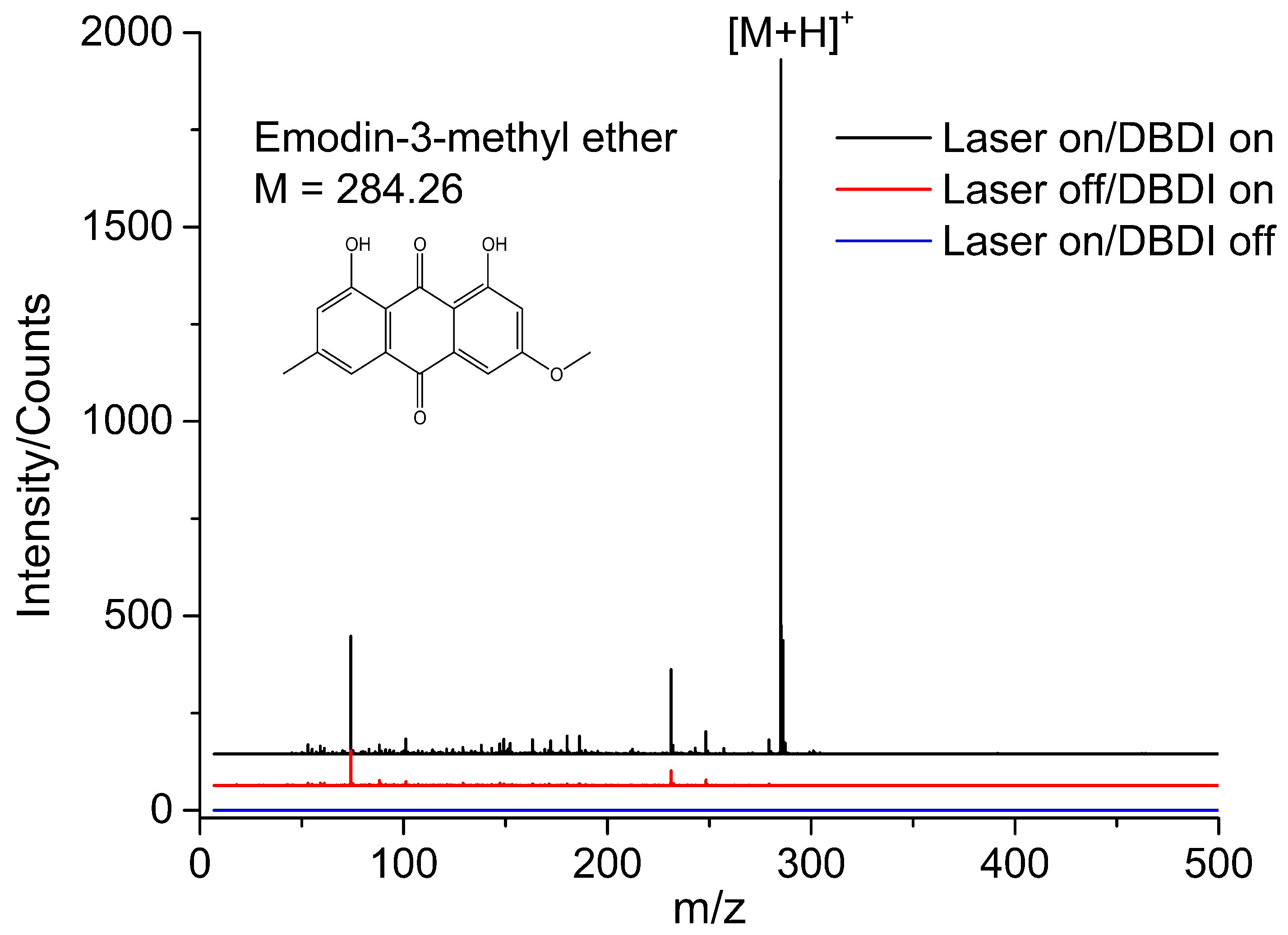
3.2. Direct Analysis of Standard Sample by LD-DBDI-MS
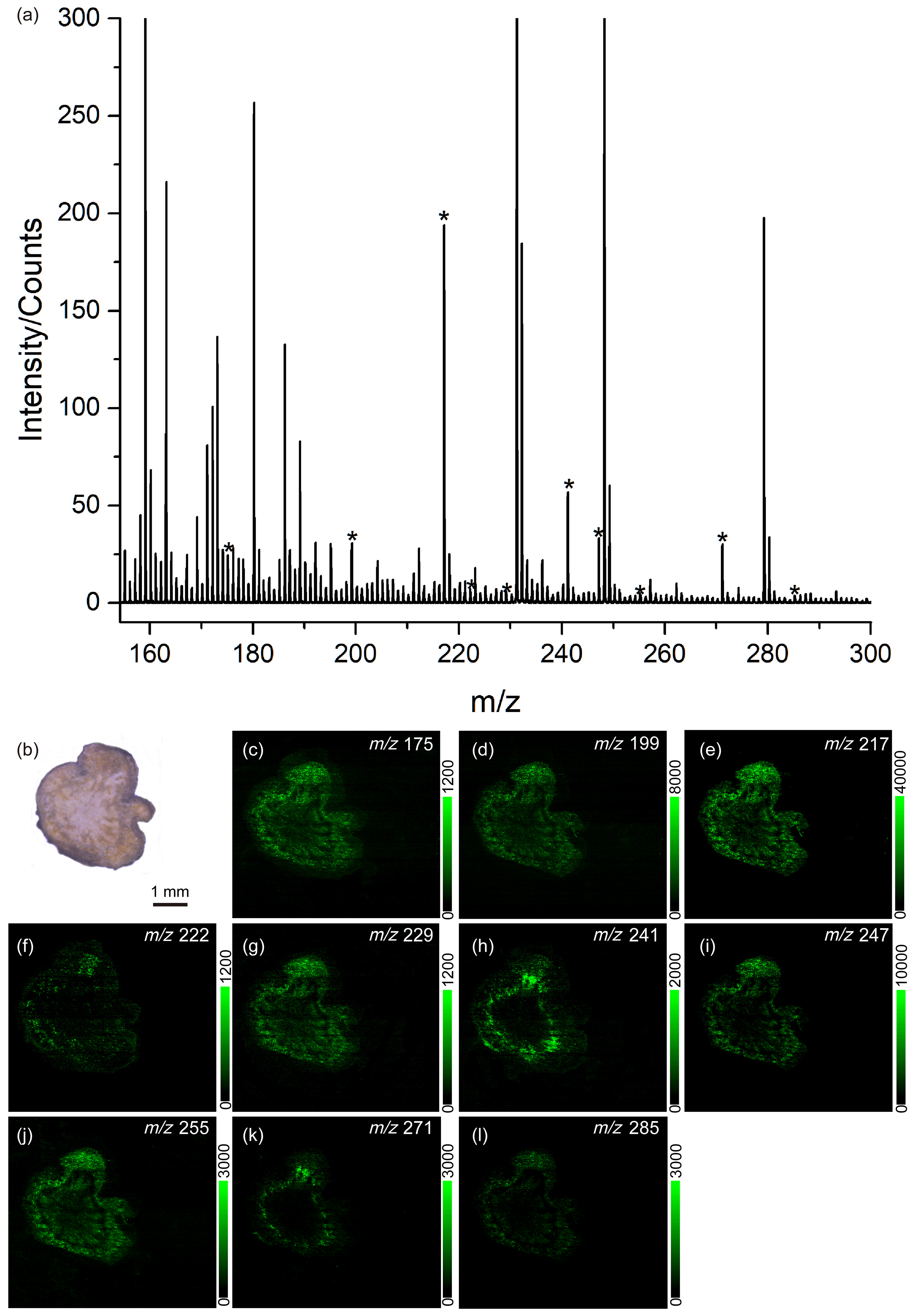
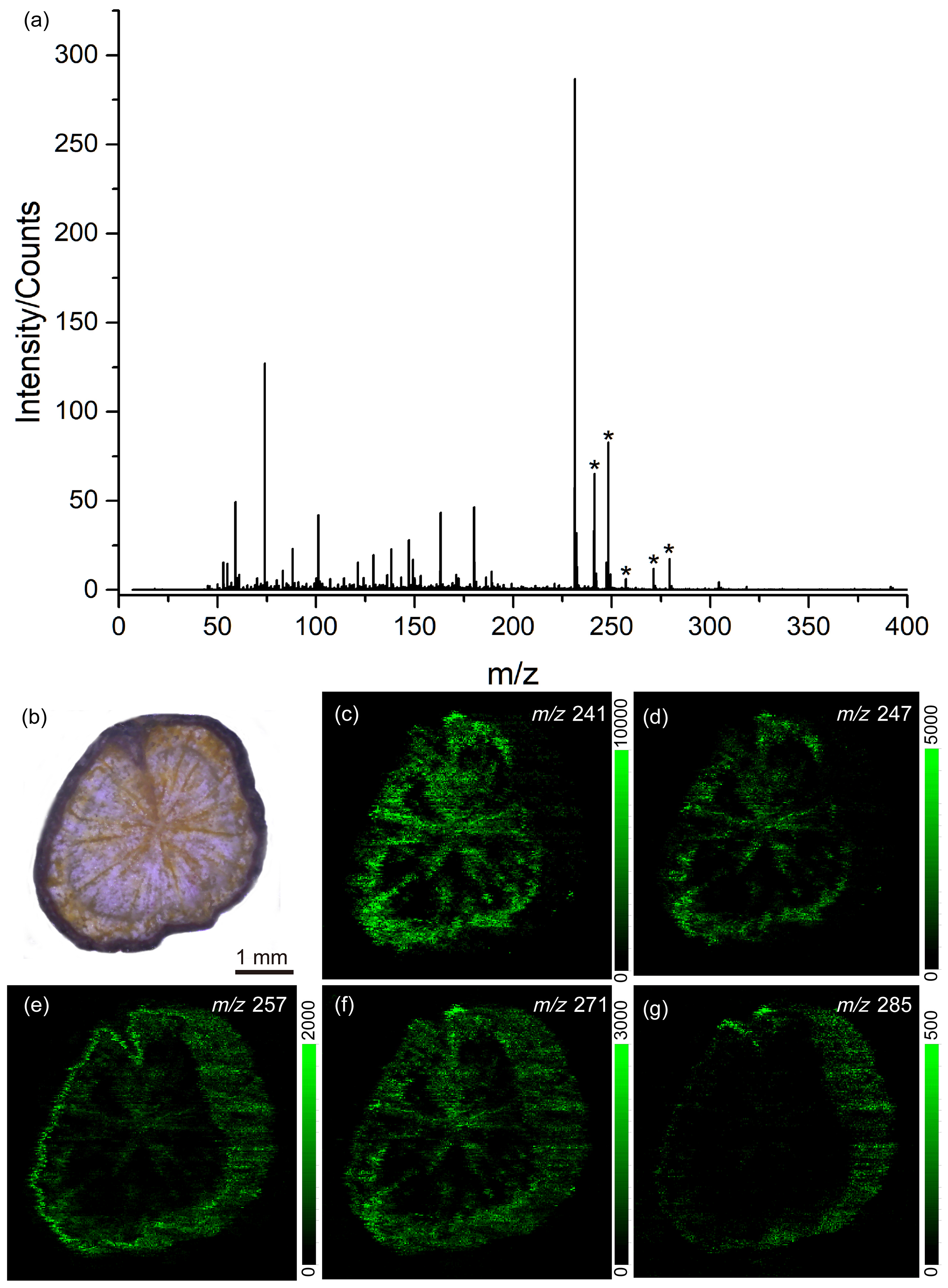
3.3. Identification and Distribution of Endogenous Molecules from Different Rheum palmatum L.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shen, S.; Zhan, C.; Yang, C.; Fernie, A.R.; Luo, J. Metabolomics-centered mining of plant metabolic diversity and function: Past decade and future perspectives. Mol. Plant 2023, 16, 43–63. [Google Scholar] [CrossRef]
- Ghini, V.; Meoni, G.; Vignoli, A.; Di Cesare, F.; Tenori, L.; Turano, P.; Luchinat, C. Fingerprinting and profiling in metabolomics of biosamples. Prog. Nucl. Magn. Reson. Spectrosc. 2023, 138–139, 105–135. [Google Scholar] [CrossRef]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef]
- Maia, M.; Figueiredo, A.; Cordeiro, C.; Sousa Silva, M. FT-ICR-MS-based metabolomics: A deep dive into plant metabolism. Mass Spectrom. Rev. 2023, 42, 1535–1556. [Google Scholar] [CrossRef]
- Zumsteg, J.; Bossard, E.; Gourguillon, L.; Villette, C.; Heintz, D. Comparison of nocturnal and diurnal metabolomes of rose flowers and leaves. Metabolomics 2023, 20, 4. [Google Scholar] [CrossRef]
- Chevalier, Q.; Gallé, J.-B.; Wasser, N.; Mazan, V.; Villette, C.; Mutterer, J.; Elustondo, M.M.; Girard, N.; Elhabiri, M.; Schaller, H.; et al. Unravelling the Puzzle of Anthranoid Metabolism in Living Plant Cells Using Spectral Imaging Coupled to Mass Spectrometry. Metabolites 2021, 11, 571. [Google Scholar] [CrossRef]
- Hu, W.; Han, Y.; Sheng, Y.; Wang, Y.; Pan, Q.; Nie, H. Mass spectrometry imaging for direct visualization of components in plants tissues. J. Sep. Sci. 2021, 44, 3462–3476. [Google Scholar] [CrossRef] [PubMed]
- Qi, K.; Wu, L.; Liu, C.; Pan, Y. Recent Advances of Ambient Mass Spectrometry Imaging and Its Applications in Lipid and Metabolite Analysis. Metabolites 2021, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, C.; Meng, Y. Mass spectrometry imaging advances and application in pharmaceutical research. Acta Mater. Med. 2022, 1, 507–533. [Google Scholar] [CrossRef]
- Mao, H.; Wang, W.; Xiang, X.; Li, Y.; Zhao, J.; Huang, Y.; Di, S.; Zhuo, Q.; Nie, H. Analysis of Metabolite Distribution in Rat Liver of High-Fat Model by Mass Spectrometry Imaging. Metabolites 2023, 13, 411. [Google Scholar] [CrossRef] [PubMed]
- Boughton, B.A.; Thinagaran, D.; Sarabia, D.; Bacic, A.; Roessner, U. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2016, 15, 445–488. [Google Scholar] [CrossRef]
- Hong, Y.; Birse, N.; Quinn, B.; Li, Y.; Jia, W.; van Ruth, S.; Elliott, C.T. MALDI-ToF MS and chemometric analysis as a tool for identifying wild and farmed salmon. Food Chem. 2024, 432, 137279. [Google Scholar] [CrossRef]
- Qin, L.; Han, J.; Wang, C.; Xu, B.; Tan, D.; He, S.; Guo, L.; Bo, X.; Xie, J. Key defatting tissue pretreatment protocol for enhanced MALDI MS Imaging of peptide biomarkers visualization in the castor beans and their attribution applications. Front. Plant Sci. 2022, 13, 1083901. [Google Scholar] [CrossRef]
- Beneito-Cambra, M.; Gilbert-López, B.; Moreno-González, D.; Bouza, M.; Franzke, J.; García-Reyes, J.F.; Molina-Díaz, A. Ambient (desorption/ionization) mass spectrometry methods for pesticide testing in food: A review. Anal. Methods 2020, 12, 4831–4852. [Google Scholar] [CrossRef]
- Liao, Y.; Fu, X.; Zhou, H.; Rao, W.; Zeng, L.; Yang, Z. Visualized analysis of within-tissue spatial distribution of specialized metabolites in tea (Camellia sinensis) using desorption electrospray ionization imaging mass spectrometry. Food Chem. 2019, 292, 204–210. [Google Scholar] [CrossRef]
- Li, N.; Dong, J.; Dong, C.; Han, Y.; Liu, H.; Du, F.; Nie, H. Spatial distribution of endogenous molecules in coffee beans by atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 2020, 31, 2503–2510. [Google Scholar] [CrossRef]
- Ding, X.; Liu, K.; Shi, Z. Laser desorption/ablation postionization mass spectrometry: Recent progress in bioanalytical applications. Mass Spectrom. Rev. 2020, 40, 566–605. [Google Scholar] [CrossRef]
- Nie, W.; Lu, Q.; Hu, T.; Xie, M.; Hu, Y. Visualizing the distribution of curcumin in the root of Curcuma longa via VUV-postionization mass spectrometric imaging. Analyst 2023, 148, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Song, X.; Zare, R.N. Laser ablation electrospray ionization achieves 5 μm resolution using a microlensed fiber. Anal. Chem. 2022, 94, 10278–10282. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Zhang, J.; Chang, C.; Li, L.; Li, M.; Xiong, X.; Guo, C.; Tang, F.; Bai, Y.; Liu, H. Ambient mass spectrometry imaging: Plasma assisted laser desorption ionization mass spectrometry imaging and its applications. Anal. Chem. 2014, 86, 4164–4169. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Xu, Z.; You, X.; Ma, S.; Zenobi, R. Atmospheric pressure mass spectrometry imaging using laser ablation, followed by dielectric barrier discharge ionization. Anal. Chem. 2021, 93, 6232–6238. [Google Scholar] [CrossRef]
- Lu, Q.; Lin, R.; Du, C.; Meng, Y.; Yang, M.; Zenobi, R.; Hang, W. Metal probe microextraction coupled to dielectric barrier discharge ionization–mass spectrometry for detecting drug residues in organisms. Anal. Chem. 2020, 92, 5921–5928. [Google Scholar] [CrossRef] [PubMed]
- Foest, D.; Knodel, A.; Ahrends, R.; Coman, C.; Franzke, J.; Brandt, S. Flexible Microtube Plasma for the Consecutive-Ionization of Cholesterol in Nano-Electrospray Mass Spectrometry. Anal. Chem. 2023, 95, 8423–8432. [Google Scholar] [CrossRef]
- Lu, Q.; Guan, X.; You, X.; Xu, Z.; Zenobi, R. High-spatial resolution atmospheric pressure mass spectrometry imaging using fiber probe laser ablation-dielectric barrier discharge ionization. Anal. Chem. 2021, 93, 14694–14700. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Hang, W.; Zare, R.N. Microlensed fiber allows subcellular imaging by laser-based mass spectrometry. Nat. Protoc. 2023, 18, 2558–2578. [Google Scholar] [CrossRef]
- Wang, J.; Liu, F.; Mo, Y.; Wang, Z.; Zhang, S.; Zhang, X. A new instrument of VUV laser desorption/ionization mass spectrometry imaging with micrometer spatial resolution and low level of molecular fragmentation. Rev. Sci. Instrum. 2017, 88, 114102. [Google Scholar] [CrossRef]
- Nudnova, M.M.; Sigg, J.; Wallimann, P.; Zenobi, R. Plasma ionization source for atmospheric pressure mass spectrometry imaging using near-field optical laser ablation. Anal. Chem. 2015, 87, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, I.; Filevich, J.; Dong, F.; Woolston, M.; Chao, W.; Anderson, E.H.; Bernstein, E.R.; Crick, D.C.; Rocca, J.J.; Menoni, C.S. Three-dimensional nanoscale molecular imaging by extreme ultraviolet laser ablation mass spectrometry. Nat. Commun. 2015, 6, 6944. [Google Scholar] [CrossRef]
- Niziol, J.; Sekula, J.; Ruman, T. Visualizing spatial distribution of small molecules in the rhubarb stalk (Rheum rhabarbarum) by surface-transfer mass spectrometry imaging. Phytochemistry 2017, 139, 72–80. [Google Scholar] [CrossRef]
- Moon, J.H.; Yoon, S.; Bae, Y.J.; Kim, M.S. Formation of gas-phase peptide ions and their dissociation in MALDI: Insights from kinetic and ion yield studies. Mass Spectrom. Rev. 2015, 34, 94–115. [Google Scholar] [CrossRef]
- Taylor, M.J.; Lukowski, J.K.; Anderton, C.R. Spatially Resolved Mass Spectrometry at the Single Cell: Recent Innovations in Proteomics and Metabolomics. J. Am. Soc. Mass Spectrom. 2021, 32, 872–894. [Google Scholar] [CrossRef]
- Lu, Q.; Hu, Y.; Chen, J.; Jin, S. Laser desorption postionization mass spectrometry imaging of folic acid molecules in tumor tissue. Anal. Chem. 2017, 89, 8238–8243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, S.; Lv, Y.; Zhang, X. Atomization of hydride with a low-temperature, atmospheric pressure dielectric barrier discharge and its application to arsenic speciation with atomic absorption spectrometry. Anal. Chem. 2006, 78, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Habib, A.; Chen, L.; Xu, T.; Wen, L. Ultra-trace level detection of nonvolatile compounds studied by ultrasonic cutter blade coupled with dielectric barrier discharge ionization-mass spectrometry. Talanta 2021, 222, 121673. [Google Scholar] [CrossRef] [PubMed]
- Nudnova, M.M.; Zhu, L.; Zenobi, R. Active capillary plasma source for ambient mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ge, W.; Wang, T.; Lan, J.; Martínez-Jarquín, S.; Wolfrum, C.; Stoffel, M.; Zenobi, R. High-Throughput Single-Cell Mass Spectrometry Reveals Abnormal Lipid Metabolism in Pancreatic Ductal Adenocarcinoma. Angew. Chem. Int. Ed. 2021, 60, 24534–24542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Frankevich, V.; Ding, J.; Ma, Y.; Chingin, K.; Chen, H. Direct mass spectrometry analysis of exhaled human breath in real-time. Mass Spectrom. Rev. 2023. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Jia, L.; Hou, Q.; Zhao, X.; Sun, K. Analysis of Endophyte Diversity of Rheum palmatum from Different Production Areas in Gansu Province of China and the Association with Secondary Metabolite. Microorganisms 2021, 9, 978. [Google Scholar] [CrossRef] [PubMed]
- Dannhorn, A.; Doria, M.L.; McKenzie, J.; Inglese, P.; Swales, J.G.; Hamm, G.; Strittmatter, N.; Maglennon, G.; Ghaem-Maghami, S.; Goodwin, R.J.A.; et al. Targeted Desorption Electrospray Ionization Mass Spectrometry Imaging for Drug Distribution, Toxicity, and Tissue Classification Studies. Metabolites 2023, 13, 377. [Google Scholar] [CrossRef]
- Lohse, M.; Haag, R.; Lippold, E.; Vetterlein, D.; Reemtsma, T.; Lechtenfeld, O.J. Direct Imaging of Plant Metabolites in the Rhizosphere Using Laser Desorption Ionization Ultra-High Resolution Mass Spectrometry. Front. Plant Sci. 2021, 12, 753812. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Guan, X.; Xu, Z.; Lu, Q. In-Situ Metabolic Profiling of Different Kinds of Rheum palmatum L. by Laser Desorption–Dielectric Barrier Discharge Ionization Mass Spectrometry Imaging. Metabolites 2024, 14, 131. https://doi.org/10.3390/metabo14030131
Xiao X, Guan X, Xu Z, Lu Q. In-Situ Metabolic Profiling of Different Kinds of Rheum palmatum L. by Laser Desorption–Dielectric Barrier Discharge Ionization Mass Spectrometry Imaging. Metabolites. 2024; 14(3):131. https://doi.org/10.3390/metabo14030131
Chicago/Turabian StyleXiao, Xue, Xiaokang Guan, Zhouyi Xu, and Qiao Lu. 2024. "In-Situ Metabolic Profiling of Different Kinds of Rheum palmatum L. by Laser Desorption–Dielectric Barrier Discharge Ionization Mass Spectrometry Imaging" Metabolites 14, no. 3: 131. https://doi.org/10.3390/metabo14030131
APA StyleXiao, X., Guan, X., Xu, Z., & Lu, Q. (2024). In-Situ Metabolic Profiling of Different Kinds of Rheum palmatum L. by Laser Desorption–Dielectric Barrier Discharge Ionization Mass Spectrometry Imaging. Metabolites, 14(3), 131. https://doi.org/10.3390/metabo14030131





