Integrated Transcriptomic and Metabolomic Analysis Reveals Possible Molecular Mechanisms of Leaf Growth and Development in Disanthus cercidifolius var. longipes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants
2.2. Leaf Age Determination
2.3. Detection of Leaf Chemical Composition
- (a)
- Free amino acids: After the samples were ground to a powder in liquid nitrogen, approximately 0.1 g was added to 1 mL of 10% acetic acid solution, fully homogenized, and extracted for 15 min in a boiling water bath. The sample was cooled and centrifuged at 8000× g, and the supernatant was diluted for testing. In total, 10 μL of the test solution, 120 μL of an acetic acid–sodium acetate buffer solution, 100 μL of a 3% ninhydrin solution, and 10 μL of a 0.3% ascorbic acid solution were added to a tube. After mixing, the tube was sealed and placed in a boiling water bath for 15 min. After cooling, the tube was centrifuged at 8000× g for 5 min. The supernatant was used to determine the absorbance (Spectra Max Single Mode Reader ABS Plus, Molecular Devices, Shanghai, China) at 570 nm.
- (b)
- Soluble protein: Approximately 0.05 g of fresh sample was added to 1 mL of distilled water and then homogenized in an ice bath. The resulting sample was centrifuged, and the supernatant was used for testing. In total, 20 μL of the supernatant solution was added to 200 μL of Coomassie brilliant blue G-250 solution and mixed well, and the absorbance at 595 nm was determined.
- (c)
- Total soluble sugar: Approximately 0.05 g of fresh sample was ground into a homogenate, heated and cooled in a water bath, and then brought to a volume of 1.25 mL. After centrifugation, the supernatant was diluted at room temperature (25 °C) to be tested. In total, 40 μL of the supernatant was added to 40 μL of distilled water, 20 μL of an anthrone–sulfuric acid solution, and 200 μL of concentrated sulfuric acid. After mixing well and cooling, 200 μL was used to measure the absorbance at 620 nm.
- (d)
- Total flavonoids: Approximately 0.05 g of fresh sample was added to 1 mL of a 60% ethanol solution. After ultrasonic extraction and centrifugation, the supernatant (diluted) was used for testing. In total, 20 μL of the supernatant was added to 6 μL of 5% sodium nitrite, mixed, and allowed to rest. Next, 6 μL of a 10% Al(NO3)3 solution was added and mixed well, followed by 80 μL of a 4% NaOH solution and 88 μL of distilled water. Finally, the absorption value at 510 nm was determined.
- (e)
- Total phenols: Approximately 0.1 g of fresh sample was added to 1 mL of 60% ethanol, fully homogenized, and ultrasonicated for 30 min. In total, 10 μL of the resulting supernatant was added to 50 μL of Folin’s reagent, and 50 μL of a 12% Na2CO3 solution and 90 μL of distilled water were added. The absorbance at 760 nm was determined.
- (f)
- Total alkaloids: Approximately 0.02 g of dry sample was extracted with concentrated ammonia. Next, 1 mL of a chloroform–methanol mixed solution (chloroform/methanol = 4:1) was added. After ultrasonication, the resulting solution was centrifuged for 10 min. In total, 20 μL of the supernatant was dried at 80 °C. Next, 100 μL of a 0.2 mol/L potassium hydrogen phthalate buffer and 80 μL of bromocresol green solution were added. The mixture was shaken vigorously, after which 300 μL of chloroform was added, followed by more shaking. Finally, after 30 min, 200 μL of the lower chloroform layer was used to determine absorbance at 410 nm.
- (g)
- Total triterpenes: Approximately 0.05 g of dry sample was placed into a 2 mL centrifuge tube. In total, 1 mL chloroform was added, ultrasonicated, and centrifuged. In total, 50 μL of the supernatant was steam dried at 60 °C. Next, 20 μL of a 5% vanillin-acetic acid solution was added to dissolve the precipitate, and then 50 μL of perchloric acid was added. The solution was mixed well, incubated in a 60 °C water bath for 15 min, and cooled in an ice bath. Next, 250 μL of glacial acetic acid was mixed and incubated for 15 min. Finally, 200 μL of the resulting supernatant was used to measure the absorbance at 548 nm.
2.4. Metabolite Analysis
2.5. RNA-Seq and Identification of Differentially Expressed Genes
2.6. Evaluation of Gene Expression Using Quantitative Reverse Transcription-PCR (qRT-PCR)
2.7. Statistical Analysis
3. Results
3.1. Differences in the Chemical Composition of Leaves During Different Developmental Stages
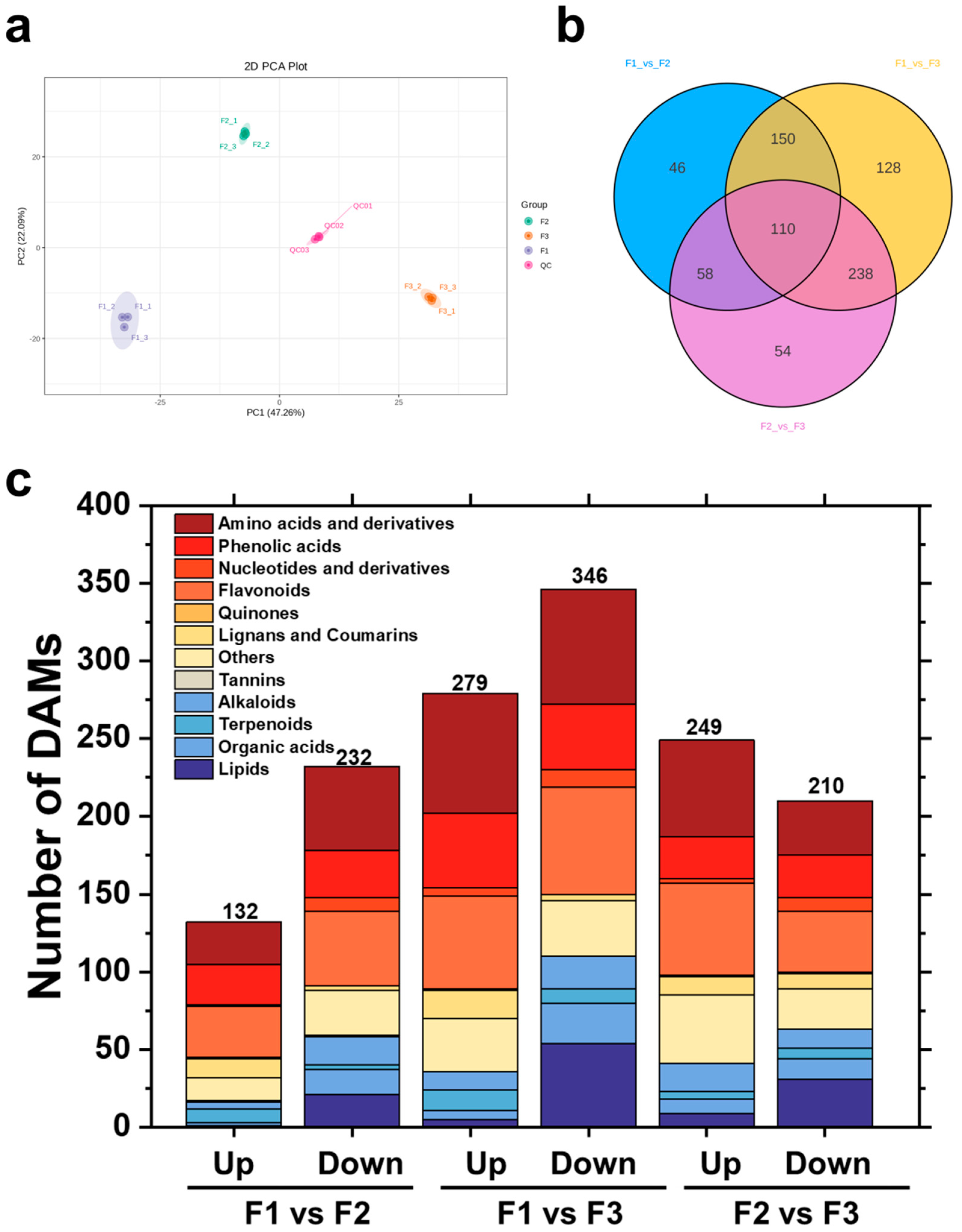
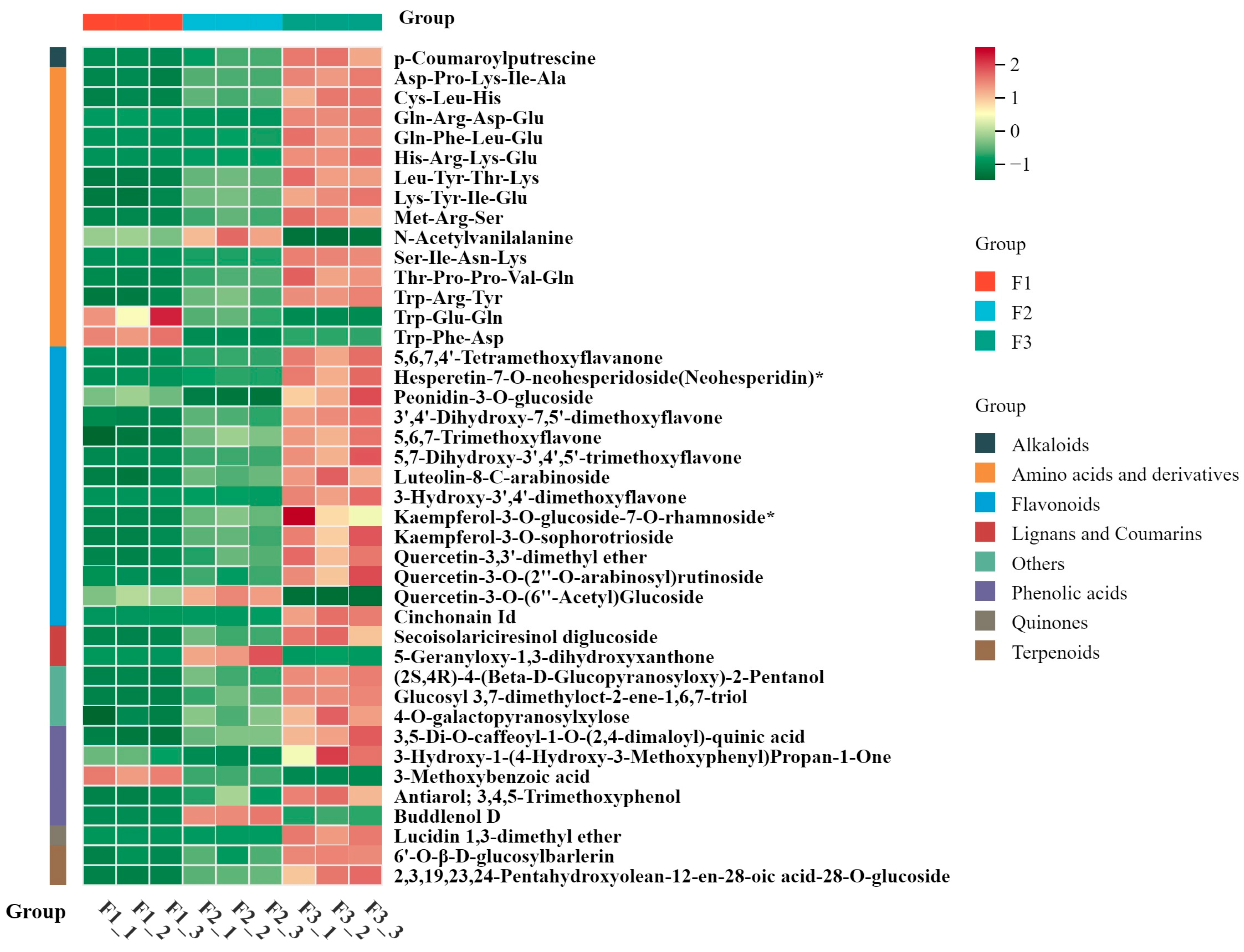
3.2. Differential Metabolite Analysis During Leaf Development
3.3. Screening of DAMs and KEGG Annotation
3.4. Overview of Transcriptome Sequencing
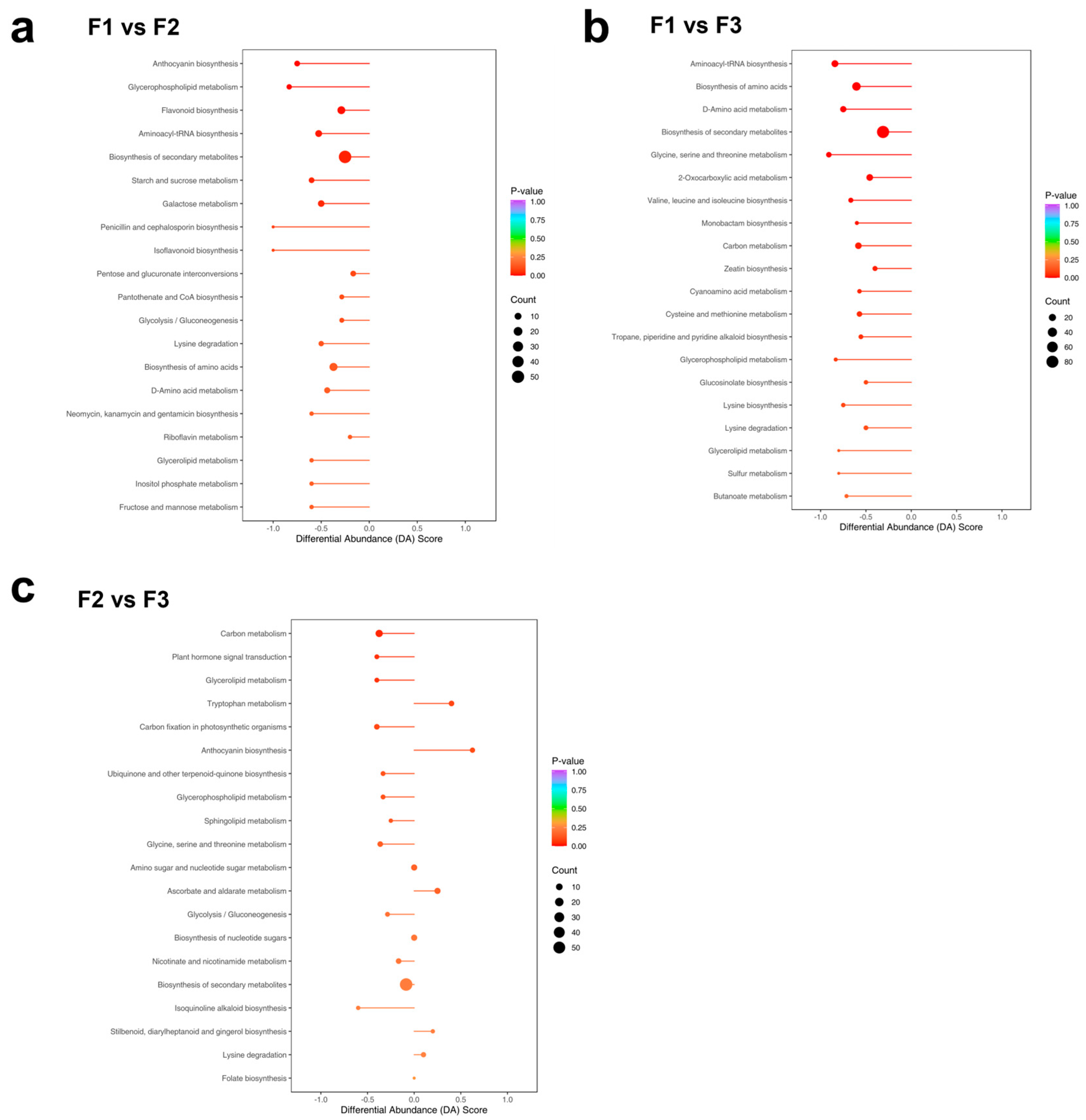
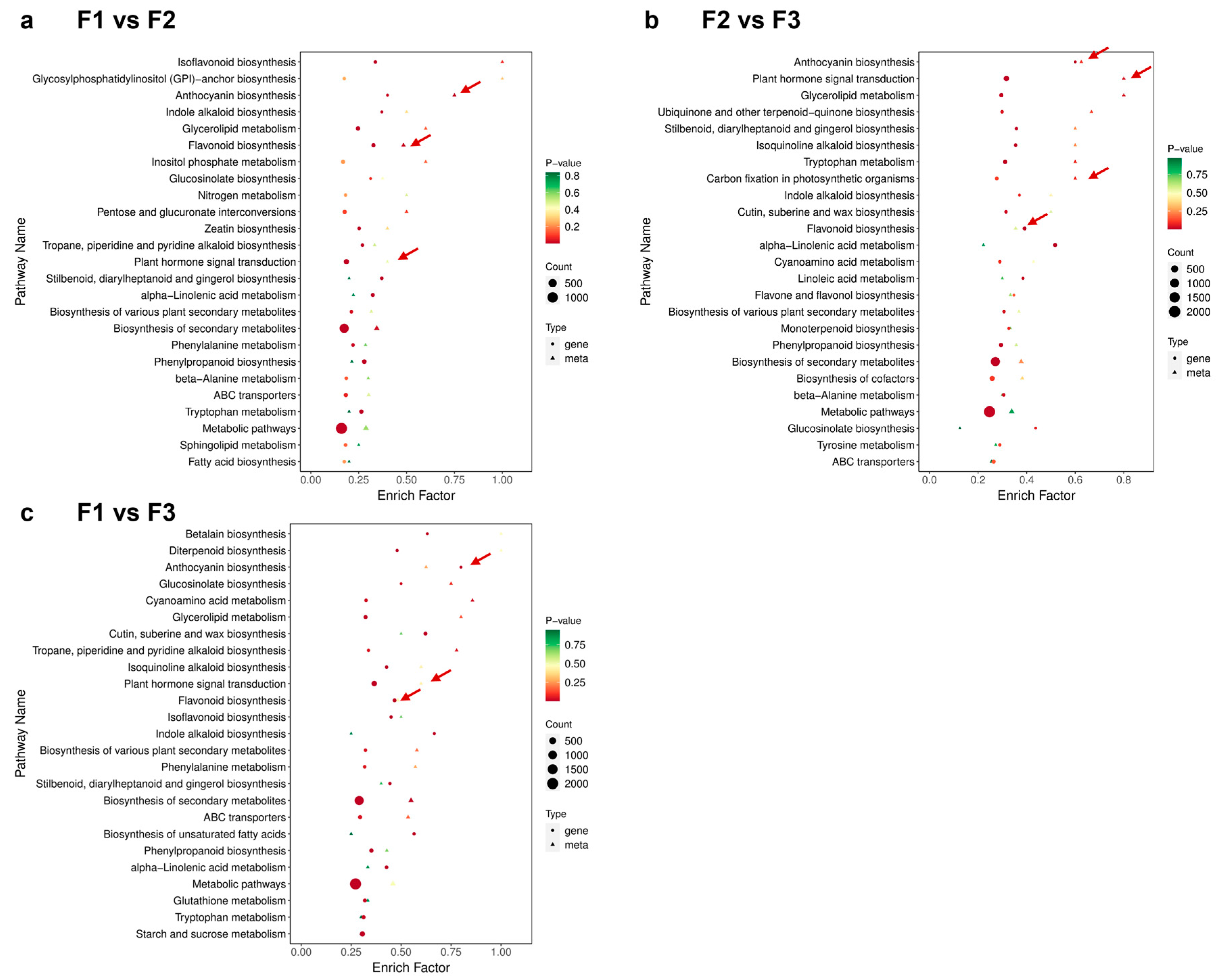
3.5. KEGG Pathway Analysis of DEGs
3.6. Combined Transcriptomic and Metabolomic Analysis
3.7. Analysis of Gene Expression Through qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, P.; Yang, A.; Yao, X.; Huang, H. Isolation and characterization of nine polymorphic microsatellite loci in the endangered shrub Disanthus cercidifolius var. longipes (Hamamelidaceae). Mol. Ecol. Resour. 2009, 9, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wang, J.; Zhang, H. Characterization and phylogenetic analysis of the complete chloroplast genome sequence of Disanthus cercidifolius subsp. longipes (Hamamelidaceae), a rare and endangered wild plant species in China. Mitochondrial DNA B Resour. 2020, 5, 1206–1207. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Xiang, G.; Lv, H.; Zhu, L.; Peng, J.; Li, G.; Mou, C. Transcriptomic and metabolic analysis unveils the mechanism behind leaf color development in Disanthus cercidifolius var. longipes. Front. Mol. Biosci. 2024, 11, 1343123. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Duan, F.Y.; Sun, X.Z.; Song, X.L.; Zhou, W.B. Leaf photosynthetic and anatomical insights into mechanisms of acclimation in rice in response to long-term fluctuating light. Plant Cell Environ. 2021, 44, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Hubbart, S.; Ajigboye, O.O.; Horton, P.; Murchie, E.H. The photoprotective protein PsbS exerts control over CO2 assimilation rate in fluctuating light in rice. Plant J. 2012, 71, 402–412. [Google Scholar] [CrossRef]
- Xiao, Y.; He, P.; Hu, W.; Li, X. Temporal and spatial dynamics of reproductive modules of endangered plant Disanthus cercidifolius var. longipes natural populations. Ying Yong Sheng Tai Xue Bao 2005, 16, 1200–1204. [Google Scholar] [CrossRef]
- Yu, Y.; Fan, Q.; Shen, R.; Guo, W.; Jin, J.; Cui, D.; Liao, W. Genetic variability and population structure of Disanthus cercidifolius subsp. longipes (Hamamelidaceae) based on AFLP analysis. PLoS ONE 2014, 9, e107769. [Google Scholar] [CrossRef]
- Hu, C.; Rao, J.; Song, Y.; Chan, S.A.; Tohge, T.; Cui, B.; Lin, H.; Fernie, A.R.; Zhang, D.; Shi, J. Dissection of flag leaf metabolic shifts and their relationship with those occurring simultaneously in developing seed by application of non-targeted metabolomics. PLoS ONE 2020, 15, e0227577. [Google Scholar] [CrossRef]
- Sticher, L.; Mauch-Mani, B.; Métraux, J.P. Systemic acquired resistance. J. Annu. Rev. Phytopathol. 1997, 35, 235–270. [Google Scholar] [CrossRef]
- Hu, F.; Zhang, Y.; Guo, J. Effects of drought stress on photosynthetic physiological characteristics, leaf microstructure, and related gene expression of yellow horn. Plant Signal. Behav. 2023, 18, 2215025. [Google Scholar] [CrossRef]
- Gutiérrez-Boem, F.H.; Thomas, G.W. Leaf area development in soybean as affected by phosphorus nutrition and water deficit. J. Plant Nutr. 2001, 24, 1711–1729. [Google Scholar] [CrossRef]
- Itagaki, K.; Shibuya, T. Differences in early hyphal development of Podosphaera xanthii on Cucumis sativus leaves acclimatized to high or low relative humidity. Botany 2018, 96, 67–71. [Google Scholar] [CrossRef]
- Liao, F. Physiological habits of Disanthus cercidifolius var. longipes and its application in gardens. North Hortic. 2010, 8, 69–71. [Google Scholar]
- Xie, G.; Zhao, J.; Li, R.; Li, H.; Guo, P.; Chen, Y.; Huang, A. Optimization of the ISSR-PCR reaction system of the endangered plant Disanthus cercidifolius var. longipes. J. Guangzhou Univ. 2010, 9, 45–50. [Google Scholar]
- Gao, P.X.; Li, M.Q.; Zhou, S.X.; Liu, J.; Niu, Y.L.; Du, J.; Ding, J.M. Distribution and current status of the endangered shrub Disanthus cercidifolius var. longipes. Plant Sci. 2013, 31, 34–41. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, G.; Liu, Z. Effects of tree size and organ age on variations in carbon, nitrogen, and phosphorus stoichiometry in Pinus koraiensis. J. Res. 2024, 35, 52. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, J.; Dai, X.; Chen, X.; Wang, Q. Flavonoid contents and free radical scavenging activity of extracts from leaves, stems, rachis and roots of Dryopteris erythrosora. Iran. J. Pharm. Res. 2012, 11, 991–997. [Google Scholar] [PubMed] [PubMed Central]
- Chen, G.L.; Hu, K.; Zhong, N.J.; Guo, J.; Gong, Y.S.; Deng, X.T.; Huang, Y.S.; Chu, D.K.; Gao, Y.Q. Antioxidant capacities and total polyphenol content of nine commercially available tea juices measured by an in vitro digestion model. Eur. Food Res. Technol. 2013, 236, 303–310. [Google Scholar] [CrossRef]
- Wang, C.F.; Fan, L.; Tian, M.; Du, S.S.; Deng, Z.W.; Feng, J.B.; Wang, Y.Y.; Su, X. Cytotoxicity of benzophenanthridme alkaloids from the roots of Zanthoxylum nitidum (Roxb.) DC. var. fastuosum How ex Huang. Nat. Prod. Res. 2015, 29, 1380–1383. [Google Scholar] [CrossRef]
- Choi, B.; Koh, E. Changes in the antioxidant capacity and phenolic compounds of maesil extract during one-year fermentation. Food Sci. Biotechnol. 2016, 26, 89–95. [Google Scholar] [CrossRef]
- Chen, Y.; An, H.M.; Huang, Y.W.; Liu, J.S.; Liu, Z.H.; Li, S.; Huang, J.A. Analysis of Non-Volatile Compounds in Jasmine Tea and Jasmine Based on Metabolomics and Sensory Evaluation. Foods 2023, 12, 3708. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhao, L.; Liu, Y.; Zhang, A.; Xiao, S.; Dai, X.; Yuan, R.; Zhou, Z.; Cao, Q. Metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway for the accumulation of anthocyanins and other flavonoids in sweet potato root skin and leaf vein base. J. Agric. Food Chem. 2022, 70, 2574–2588. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Li, R.; Ren, L.; Gao, X.; Zhang, Y.; Ma, Z.; Ma, D.; Luo, Y. A comparative metabolomics study of favonoids in sweet potato with different flesh colors (lpomoea batatas (L.)Lam). Food Chem. 2018, 260, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Li, K.T.; Hao, S.X.; Zhang, J.; Song, T.T.; Tian, J.; Yao, Y.C. The use of RNA sequencing and correlation network analysis to study potential regulators of crabapple leaf color transformation. Plant Cell Physiol. 2018, 59, 1027–1042. [Google Scholar] [CrossRef]
- Towler, M.J.; Weathers, P.J. Variations in key artemisinic and other metabolites throughout plant development in Artemisia annua L. for potential therapeutic use. Ind. Crops Prod. 2015, 67, 185–191. [Google Scholar] [CrossRef]
- Harvaux, M.; Kloppstech, K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 2001, 213, 953–966. [Google Scholar] [CrossRef]
- Douris, N.; Melman, T.; Pecherer, J.M.; Pissios, P.; Flier, J.S.; Cantley, L.C.; Maratos-Flier, E. Adaptive changes in amino acid metabolism permit normal longevity in mice consuming a low-carbohydrate ketogenic diet. Biochim. Biophys. Acta-Mol. Basis Dis. 2015, 1852, 2056–2065. [Google Scholar] [CrossRef]
- Hu, G.H.; Ning, H.L.; Wang, H.D.; Wang, J.A.; Zhang, D.Y.; Li, W.B. Effect of photo intensity on quality and yiel d of soybeansI_Flfect of light -intensity on oil content and protein content of soybeans in the whde growth period. Chin. J. Oil Crops 2004, 22, 197–206. [Google Scholar] [CrossRef]
- Jiang, X.L.; Shi, Y.F.; Dai, X.L.; Zhuang, J.H.; Fu, Z.P.; Zhao, X.Q.; Xia, T. Four flavonoid glycosyltransferases present in tea overexpressed in model plants Arabidopsis thaliana and Nicotiana tabacum for functional identification. J. Chromatogr. B-Anal. Technol. Biomed. Life Sci. 2018, 1100, 148–157. [Google Scholar] [CrossRef]
- Navarro-Hoyos, M.; Acuña-Quirós, M.; Quirós-Fallas, M.I.; Vargas-Huertas, F.; Wilhelm-Romero, K.; Vásquez-Castro, F.; Sánchez-Kopper, A. UPLC-HRMS Polyphenolic Characterization, Contents and Antioxidant Activity of Zingiber officinale Roscoe rhizomes from Costa Rica. Processes 2022, 10, 691. [Google Scholar] [CrossRef]
- Wang, Y.S.; Gao, L.P.; Shan, Y.; Liu, Y.J.; Tian, Y.W.; Xia, T. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2012, 141, 7–16. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Y.M.; Zhang, S.Y.; Che, Z.J.; Yuan, W.J.; Yu, D.Y. GWAS reveals two novel loci for photosynthesis-related traits in soybean. Mol. Genet. Genom. 2022, 295, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Nordeen, K.W.; Nordeen, E.J. Synaptic and molecular mechanisms regulating plasticity during early learning. Ann. N. Y. Acad. Sci. 2004, 1016, 416–437. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Ahmad, S.; Cheng, T.; Wang, J.; Pan, H.; Zhao, L.; Zhang, Q. Red to Far-Red Light Ratio Modulates Hormonal and Genetic Control of Axillary bud Outgrowth in Chrysanthemum (Dendranthema grandiflorum “Jinba’). Int. J. Mol. Sci. 2018, 19, 1590. [Google Scholar] [CrossRef]
- Mueller, D.; Leyser, O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011, 107, 1203–1212. [Google Scholar] [CrossRef]
- Shi, J.; Wang, N.; Zhou, H.; Xu, Q.; Yan, G. Transcriptome analyses provide insights into the homeostatic regulation of axillary buds in upland cotton (G. hirsutum L.). BMC Plant Biol. 2020, 20, 228. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, L.; Pang, S.; Jia, Z.; Wang, L.; Li, W.; Jin, B. UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind. Crops Prod. 2020, 151, 112483. [Google Scholar] [CrossRef]
- Wu, L.; Meng, F.; Su, X.; Chen, N.; Peng, D.; Xing, S. Transcriptomic responses to cold stress in Dendrobium huoshanense. Physiol. Mol. Biol. Plants 2023, 29, 163–1646. [Google Scholar] [CrossRef]
- Ali, A.; Cavallaro, V.; Santoro, P.; Mori, J.; Ferrante, A.; Cocetta, G. Quality and physiological evaluation of tomato subjected to different supplemental lighting systems. Sci. Hortic. 2024, 323, 112469. [Google Scholar] [CrossRef]




| Different Development Stages | F1 | F2 | F3 |
|---|---|---|---|
| Free amino acids (FW mg/g) | 2.32 ± 0.03 a | 0.82 ± 0.00 b | 0.36 ± 0.01 c |
| Soluble protein content (FW mg/g) | 4.65 ± 0.19 a | 2.03 ± 0.05 b | 1.45 ± 0.04 c |
| Soluble sugar content (FW mg/g) | 56.70 ± 2.36 a | 43.58 ± 1.02 c | 40.40 ± 1.92 b |
| Total flavonoids content (FW mg/g) | 21.34 ± 0.99 b | 18.17 ± 0.58 c | 23.21 ± 0.52 a |
| Total phenols content (FW mg/g) | 14.52 ± 0.14 a | 11.04 ± 0.48 b | 13.86 ± 0.41 a |
| Total alkaloids content (FW mg/g) | 14.01 ± 0.55 b | 18.97 ± 0.90 a | 10.70 ± 0.33 c |
| Total triterpenoids content (DW mg/g) | 55.46 ± 2.13 a | 62.19 ± 3.08 b | 55.20 ± 1.29 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, X.; Xiang, G.; Mou, C.; Zhu, L.; Song, J.; Li, G.; Lv, H. Integrated Transcriptomic and Metabolomic Analysis Reveals Possible Molecular Mechanisms of Leaf Growth and Development in Disanthus cercidifolius var. longipes. Metabolites 2024, 14, 654. https://doi.org/10.3390/metabo14120654
Tian X, Xiang G, Mou C, Zhu L, Song J, Li G, Lv H. Integrated Transcriptomic and Metabolomic Analysis Reveals Possible Molecular Mechanisms of Leaf Growth and Development in Disanthus cercidifolius var. longipes. Metabolites. 2024; 14(12):654. https://doi.org/10.3390/metabo14120654
Chicago/Turabian StyleTian, Xiaoming, Guangfeng Xiang, Cun Mou, Lu Zhu, Jing Song, Gaofei Li, and Hao Lv. 2024. "Integrated Transcriptomic and Metabolomic Analysis Reveals Possible Molecular Mechanisms of Leaf Growth and Development in Disanthus cercidifolius var. longipes" Metabolites 14, no. 12: 654. https://doi.org/10.3390/metabo14120654
APA StyleTian, X., Xiang, G., Mou, C., Zhu, L., Song, J., Li, G., & Lv, H. (2024). Integrated Transcriptomic and Metabolomic Analysis Reveals Possible Molecular Mechanisms of Leaf Growth and Development in Disanthus cercidifolius var. longipes. Metabolites, 14(12), 654. https://doi.org/10.3390/metabo14120654






