Detection and Validation of Organic Metabolites in Urine for Clear Cell Renal Cell Carcinoma Diagnosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Chemicals and Materials
2.3. Extraction and Chemical Analysis of VOCs from Urine Samples
2.4. Data Processing and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panwoon, C.; Seubwai, W.; Thanee, M.; Sangkhamanon, S. Identification of Novel Biomarkers to Distinguish Clear Cell and Non-Clear Cell Renal Cell Carcinoma Using Bioinformatics and Machine Learning. PLoS ONE 2024, 19, e0305252. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Tu, S.; Li, L.; Li, G.; Zhang, Y. Diagnostic, Predictive and Prognostic Molecular Biomarkers in Clear Cell Renal Cell Carcinoma: A Retrospective Study. Cancer Rep. 2024, 7, e2116. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2023; American Cancer Society: Atlanta, GA, USA, 2023. [Google Scholar]
- Diaz De Leon, A.; Pedrosa, I. Imaging and Screening of Kidney Cancer. Radiol. Clin. N. Am. 2017, 55, 1235–1250. [Google Scholar] [CrossRef]
- Posada Calderon, L.P.; Eismann, L.; Reese, S.W.; Reznik, E.; Hakimi, A.A. Advances in Imaging-Based Biomarkers in Renal Cell Carcinoma: A Critical Analysis of the Current Literature. Cancers 2023, 15, 354. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Henriques, V.; Cimadamore, A.; Santoni, M.; Cheng, L.; Gevaert, T.; Blanca, A.; Massari, F.; Scarpelli, M.; Montironi, R. The Identification of Immunological Biomarkers in Kidney Cancers. Front. Oncol. 2018, 8, 456. [Google Scholar] [CrossRef]
- Yong, C.; Stewart, G.D.; Frezza, C. Oncometabolites in Renal Cancer. Nat. Rev. Nephrol. 2020, 16, 156–172. [Google Scholar] [CrossRef]
- Kaushik, A.K.; Tarangelo, A.; Boroughs, L.K.; Ragavan, M.; Zhang, Y.; Wu, C.-Y.; Li, X.; Ahumada, K.; Chiang, J.-C.; Tcheuyap, V.T.; et al. In Vivo Characterization of Glutamine Metabolism Identifies Therapeutic Targets in Clear Cell Renal Cell Carcinoma. Sci. Adv. 2022, 8, eabp8293. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A Promising Cancer Biomarker. Clin. Transl. Med. 2018, 7, 21. [Google Scholar] [CrossRef]
- Rodrigues, D.; Monteiro, M.; Jerónimo, C.; Henrique, R.; Belo, L.; Bastos, M.d.L.; Guedes de Pinho, P.; Carvalho, M. Renal Cell Carcinoma: A Critical Analysis of Metabolomic Biomarkers Emerging from Current Model Systems. Transl. Res. 2017, 180, 1–11. [Google Scholar] [CrossRef]
- Abooshahab, R.; Hooshmand, K.; Razavi, S.A.; Gholami, M.; Sanoie, M.; Hedayati, M. Plasma Metabolic Profiling of Human Thyroid Nodules by Gas Chromatography-Mass Spectrometry (GC-MS)-Based Untargeted Metabolomics. Front. Cell Dev. Biol. 2020, 8, 385. [Google Scholar] [CrossRef] [PubMed]
- Ather, M.H.; Masood, N.; Siddiqui, T. Current Management of Advanced and Metastatic Renal Cell Carcinoma. Urol. J. 2010, 7, 1–9. [Google Scholar] [PubMed]
- Claps, F.; Mir, M.C. Novel Expanding Renal Cell Carcinoma Biomarkers. Société Int. D’urologie J. 2021, 2, 32–42. [Google Scholar] [CrossRef]
- Kennelley, G.E.; Amaye-Obu, T.; Foster, B.A.; Tang, L.; Paragh, G.; Huss, W.J. Mechanistic Review of Sulforaphane as a Chemoprotective Agent in Bladder Cancer. Am. J. Clin. Exp. Urol. 2023, 11, 103–120. [Google Scholar]
- Amaro, F.; Pinto, J.; Rocha, S.; Araújo, A.M.; Miranda-Gonçalves, V.; Jerónimo, C.; Henrique, R.; Bastos, M.D.L.; Carvalho, M.; Guedes De Pinho, P. Volatilomics Reveals Potential Biomarkers for Identification of Renal Cell Carcinoma: An In Vitro Approach. Metabolites 2020, 10, 174. [Google Scholar] [CrossRef]
- Di Meo, N.A.; Lasorsa, F.; Rutigliano, M.; Loizzo, D.; Ferro, M.; Stella, A.; Bizzoca, C.; Vincenti, L.; Pandolfo, S.D.; Autorino, R.; et al. Renal Cell Carcinoma as a Metabolic Disease: An Update on Main Pathways, Potential Biomarkers, and Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 14360. [Google Scholar] [CrossRef]
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative Diagnostic Methods for Early Prostate Cancer Detection through Urine Analysis: A Review. Cancers 2018, 10, 123. [Google Scholar] [CrossRef]
- Feil, C.; Staib, F.; Berger, M.R.; Stein, T.; Schmidtmann, I.; Forster, A.; Schimanski, C.C. Sniffer Dogs Can Identify Lung Cancer Patients from Breath and Urine Samples. BMC Cancer 2021, 21, 917. [Google Scholar] [CrossRef]
- Fischer-Tenhagen, C.; Johnen, D.; Nehls, I.; Becker, R. A Proof of Concept: Are Detection Dogs a Useful Tool to Verify Potential Biomarkers for Lung Cancer? Front. Vet. Sci. 2018, 5, 52. [Google Scholar] [CrossRef]
- Cornu, J.-N.; Cancel-Tassin, G.; Ondet, V.; Girardet, C.; Cussenot, O. Olfactory Detection of Prostate Cancer by Dogs Sniffing Urine: A Step Forward in Early Diagnosis. Eur. Urol. 2011, 59, 197–201. [Google Scholar] [CrossRef]
- Boedeker, E.; Friedel, G.; Walles, T. Sniffer Dogs as Part of a Bimodal Bionic Research Approach to Develop a Lung Cancer Screening. Interact. Cardiovasc. Thorac. Surg. 2012, 14, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Willis, C.M.; Britton, L.E.; Harris, R.; Wallace, J.; Guest, C.M. Volatile Organic Compounds as Biomarkers of Bladder Cancer: Sensitivity and Specificity Using Trained Sniffer Dogs. Cancer Biomark. 2011, 8, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Costello Bde, L.; Miekisch, W.; Schubert, J.; Buszewski, B.; Pleil, J.; Ratcliffe, N.; Risby, T. The Human Volatilome: Volatile Organic Compounds (VOCs) in Exhaled Breath, Skin Emanations, Urine, Feces and Saliva. J. Breath Res. 2014, 8, 034001. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Smith, D. Volatile Biomarkers: Non-Invasive Diagnosis in Physiology and Medicine; Elsevier: Boston, MA, USA, 2013; ISBN 0-444-62620-4. [Google Scholar]
- Costantini, M.; Filianoti, A.; Anceschi, U.; Bove, A.M.; Brassetti, A.; Ferriero, M.; Mastroianni, R.; Misuraca, L.; Tuderti, G.; Ciliberto, G.; et al. Human Urinary Volatilome Analysis in Renal Cancer by Electronic Nose. Biosensors 2023, 13, 427. [Google Scholar] [CrossRef]
- Monteiro, M.; Moreira, N.; Pinto, J.; Pires-Luis, A.S.; Henrique, R.; Jeronimo, C.; Bastos, M.L.; Gil, A.M.; Carvalho, M.; Guedes de Pinho, P. GC-MS Metabolomics-Based Approach for the Identification of a Potential VOC-Biomarker Panel in the Urine of Renal Cell Carcinoma Patients. J. Cell. Mol. Med. 2017, 21, 2092–2105. [Google Scholar] [CrossRef]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [CrossRef]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging Deeper into Volatile Organic Compounds Associated with Cancer. Biol. Methods Protoc. 2019, 4, bpz014. [Google Scholar] [CrossRef]
- Vuong, L.; Kotecha, R.R.; Voss, M.H.; Hakimi, A.A. Tumor Microenvironment Dynamics in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2019, 9, 1349–1357. [Google Scholar] [CrossRef]
- Weiss, R.H. Metabolomics and Metabolic Reprogramming in Kidney Cancer. Semin. Nephrol. 2018, 38, 175–182. [Google Scholar] [CrossRef]
- Ganti, S.; Weiss, R.H. Urine Metabolomics for Kidney Cancer Detection and Biomarker Discovery. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 551–557. [Google Scholar] [CrossRef]
- Atrih, A.; Mudaliar, M.A.V.; Zakikhani, P.; Lamont, D.J.; Huang, J.T.-J.; Bray, S.E.; Barton, G.; Fleming, S.; Nabi, G. Quantitative Proteomics in Resected Renal Cancer Tissue for Biomarker Discovery and Profiling. Br. J. Cancer 2014, 110, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- McClain, K.M.; Sampson, J.N.; Petrick, J.L.; Mazzilli, K.M.; Gerszten, R.E.; Clish, C.B.; Purdue, M.P.; Lipworth, L.; Moore, S.C. Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Metabolites 2022, 12, 1189. [Google Scholar] [CrossRef] [PubMed]
- Arendowski, A.; Ossolinski, K.; Niziol, J.; Ruman, T. Screening of Urinary Renal Cancer Metabolic Biomarkers with Gold Nanoparticles-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Sci. 2020, 36, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Falegan, O.; Ball, M.; Shaykhutdinov, R.; Pieroraio, P.; Farshidfar, F.; Vogel, H.; Allaf, M.; Hyndman, M. Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma. Metabolites 2017, 7, 6. [Google Scholar] [CrossRef]
- Žalimas, A.; Kubiliūtė, R.; Žukauskaitė, K.; Sabaliauskaitė, R.; Trakymas, M.; Letautienė, S.; Kaubrienė, E.M.; Ušinskienė, J.; Ulys, A.; Jarmalaitė, S. Urine Molecular Biomarkers for Detection and Follow-Up of Small Renal Masses. Int. J. Mol. Sci. 2022, 23, 16110. [Google Scholar] [CrossRef]
- Peter, M.R.; Zhao, F.; Jeyapala, R.; Kamdar, S.; Xu, W.; Hawkins, C.; Evans, A.J.; Fleshner, N.E.; Finelli, A.; Bapat, B. Investigating Urinary Circular RNA Biomarkers for Improved Detection of Renal Cell Carcinoma. Front. Oncol. 2022, 11, 814228. [Google Scholar] [CrossRef]
- Gao, Q.; Su, X.; Annabi, M.H.; Schreiter, B.R.; Prince, T.; Ackerman, A.; Morgas, S.; Mata, V.; Williams, H.; Lee, W.-Y. Application of Urinary Volatile Organic Compounds (VOCs) for the Diagnosis of Prostate Cancer. Clin. Genitourin. Cancer 2019, 17, 183–190. [Google Scholar] [CrossRef]
- Noriega Landa, E.; Quaye, G.E.; Su, X.; Badmos, S.; Holbrook, K.L.; Polascik, T.J.; Adams, E.S.; Deivasigamani, S.; Gao, Q.; Annabi, M.H.; et al. Urinary Fatty Acid Biomarkers for Prostate Cancer Detection. PLoS ONE 2024, 19, e0297615. [Google Scholar] [CrossRef]
- Badmos, S. Urinary Volatile Organic Compounds in Prostate Cancer Biopsy Pathologic Risk Stratification Using Logistic Regression and Multivariate Analysis Models. Am. J. Cancer Res. 2024, 14, 192–209. [Google Scholar] [CrossRef]
- Kleinbaum, F. Logistic Regression: A Self-Learning Text, 3rd ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Schober, P.; Vetter, T.R. Linear Regression in Medical Research. Anesth. Analg. 2021, 132, 108–109. [Google Scholar] [CrossRef]
- Da Fonseca, M.d.J.M.; Juvanhol, L.L.; Rotenberg, L.; Nobre, A.A.; Griep, R.H.; De Mello Alves, M.G.; Cardoso, L.d.O.; Giatti, L.; Nunes, M.A.; Aquino, E.M.L.; et al. Using Gamma and Quantile Regressions to Explore the Association between Job Strain and Adiposity in the ELSA-Brasil Study: Does Gender Matter? Int. J. Environ. Res. Public Health 2017, 14, 1404. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, C.; Pi, X.; Guo, L.; Wang, Y.; Li, M.; Feng, Y.; Lin, Z.; Hou, W.; Li, E. Urinary Volatile Organic Compounds as Potential Biomarkers for Renal Cell Carcinoma. Biomed. Rep. 2016, 5, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.J.; Mellnick, V.M.; Luo, J.; Siegel, M.J.; Figenshau, R.S.; Bhayani, S.; Kharasch, E.D. Evaluation of Urine Aquaporin-1 and Perilipin-2 Concentrations as Biomarkers to Screen for Renal Cell Carcinoma: A Prospective Cohort Study. Jama Oncol. 2015, 1, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Mijuskovic, M.; Stanojevic, I.; Milovic, N.; Cerovic, S.; Petrovic, D.; Jovanovic, D.; Aleksic, P.; Kovacevic, B.; Andjelic, T.; Terzic, B.; et al. Urinary KIM-1 and AQP-1 in Patients with Clear Renal Cell Carcinoma: Potential Noninvasive Biomarkers. Vojnosanit. Pregl. 2016, 73, 266–272. [Google Scholar] [CrossRef]
- Monteiro, M.; Carvalho, M.; Henrique, R.; Jeronimo, C.; Moreira, N.; de Lourdes Bastos, M.; de Pinho, P.G. Analysis of Volatile Human Urinary Metabolome by Solid-Phase Microextraction in Combination with Gas Chromatography-Mass Spectrometry for Biomarker Discovery: Application in a Pilot Study to Discriminate Patients with Renal Cell Carcinoma. Eur. J. Cancer 2014, 50, 1993–2002. [Google Scholar] [CrossRef]
- Buszewska-Forajta, M.; Raczak-Gutknecht, J.; Artymowicz, M.; Wesołowski, W.; Buczkowski, K.; Iżycka-Świeszewska, E.; Markuszewski, M.J. The Potential Role of Fatty Acids in Prostate Cancer Determined by GC–MS Analysis of Formalin-Fixed Paraffin-Embedded Tissue Samples. J. Pharm. Biomed. Anal. 2021, 196, 113907. [Google Scholar] [CrossRef]
- Karmokar, P.F.; Moniri, N.H. Free-Fatty Acid Receptor-4 (FFA4/GPR120) Differentially Regulates Migration, Invasion, Proliferation and Tumor Growth of Papillary Renal Cell Carcinoma Cells. Biochem. Pharmacol. 2023, 213, 115590. [Google Scholar] [CrossRef]
- Lv, W.; Yang, T. Identification of Possible Biomarkers for Breast Cancer from Free Fatty Acid Profiles Determined by GC-MS and Multivariate Statistical Analysis. Clin. Biochem. 2012, 45, 127–133. [Google Scholar] [CrossRef]
- Andreoli, R.; Manini, P.; Corradi, M.; Mutti, A.; Niessen, W.M. Determination of Patterns of Biologically Relevant Aldehydes in Exhaled Breath Condensate of Healthy Subjects by Liquid Chromatography/Atmospheric Chemical Ionization Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 637–645. [Google Scholar] [CrossRef]
- Riccio, G.; Baroni, S.; Urbani, A.; Greco, V. Mapping of Urinary Volatile Organic Compounds by a Rapid Analytical Method Using Gas Chromatography Coupled to Ion Mobility Spectrometry (GC–IMS). Metabolites 2022, 12, 1072. [Google Scholar] [CrossRef] [PubMed]
- Wakayama, T.; Ito, Y.; Sakai, K.; Miyake, M.; Shibata, E.; Ohno, H.; Kamijima, M. Comprehensive Review of 2-ethyl-1-hexanol as an Indoor Air Pollutant. J. Occup. Health 2019, 61, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Lee, H.M.; Kim, J. Oncology Therapeutics Targeting the Metabolism of Amino Acids. Cells 2020, 9, 1904. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Pacheco, A.; Salinero-Bachiller, M.; Iribar, M.C.; López-Luque, A.; Miján-Ortiz, J.L.; Peinado, J.M. Furan and P-Xylene as Candidate Biomarkers for Prostate Cancer. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 243.e21–243.e27. [Google Scholar] [CrossRef]
- Mansour, A.; Chatila, R.; Bejjani, N.; Dagher, C.; Faour, W.H. A Novel Xylene-Free Deparaffinization Method for the Extraction of Proteins from Human Derived Formalin-Fixed Paraffin Embedded (FFPE) Archival Tissue Blocks. MethodsX 2014, 1, 90–95. [Google Scholar] [CrossRef]
- Shala, N.K.; Stenehjem, J.S.; Babigumira, R.; Liu, F.-C.; Berge, L.A.M.; Silverman, D.T.; Friesen, M.C.; Rothman, N.; Lan, Q.; Hosgood, H.D.; et al. Exposure to Benzene and Other Hydrocarbons and Risk of Bladder Cancer among Male Offshore Petroleum Workers. Br. J. Cancer 2023, 129, 838–851. [Google Scholar] [CrossRef]
- Rana, I.; Dahlberg, S.; Steinmaus, C.; Zhang, L. Benzene Exposure and Non-Hodgkin Lymphoma: A Systematic Review and Meta-Analysis of Human Studies. Lancet Planet. Health 2021, 5, e633–e643. [Google Scholar] [CrossRef]
- Lee, K.H.; Bartsch, H.; Nair, J.; Yoo, D.H.; Hong, Y.C.; Cho, S.H.; Kang, D. Effect of Short-Term Fasting on Urinary Excretion of Primary Lipid Peroxidation Products and on Markers of Oxidative DNA Damage in Healthy Women. Carcinogenesis 2006, 27, 1398–1403. [Google Scholar] [CrossRef]
- Ahmad, S.; Ullah, F.; Zeb, A.; Ayaz, M.; Ullah, F.; Sadiq, A. Evaluation of Rumex Hastatus D. Don for Cytotoxic Potential against HeLa and NIH/3T3 Cell Lines: Chemical Characterization of Chloroform Fraction and Identification of Bioactive Compounds. BMC Complement. Altern. Med. 2016, 16, 308. [Google Scholar] [CrossRef]
- Cala, M.; Aldana, J.; Sánchez, J.; Guio, J.; Meesters, R.J.W. Urinary Metabolite and Lipid Alterations in Colombian Hispanic Women with Breast Cancer: A Pilot Study. J. Pharm. Biomed. Anal. 2018, 152, 234–241. [Google Scholar] [CrossRef]
- Wolrab, D.; Jirásko, R.; Cífková, E.; Höring, M.; Mei, D.; Chocholoušková, M.; Peterka, O.; Idkowiak, J.; Hrnčiarová, T.; Kuchař, L.; et al. Lipidomic Profiling of Human Serum Enables Detection of Pancreatic Cancer. Nat. Commun. 2022, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T.; Chan, K.G.; Pusparajah, P.; Lee, W.L.; Chuah, L.H.; Khan, T.M.; Lee, L.H.; Goh, B.H. Targeting Membrane Lipid a Potential Cancer Cure? Front. Pharmacol. 2017, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Secor, J.D.; Fligor, S.C.; Tsikis, S.T.; Yu, L.J.; Puder, M. Free Fatty Acid Receptors as Mediators and Therapeutic Targets in Liver Disease. Front. Physiol. 2021, 12, 656441. [Google Scholar] [CrossRef] [PubMed]
- Ganti, S.; Taylor, S.L.; Kim, K.; Hoppel, C.L.; Guo, L.; Yang, J.; Evans, C.; Weiss, R.H. Urinary Acylcarnitines Are Altered in Human Kidney Cancer. Int. J. Cancer 2012, 130, 2791–2800. [Google Scholar] [CrossRef]
- Nagarajan, S.R.; Butler, L.M.; Hoy, A.J. The Diversity and Breadth of Cancer Cell Fatty Acid Metabolism. Cancer Metab. 2021, 9, 2. [Google Scholar] [CrossRef]
- Sheridan, M.; Ogretmen, B. The Role of Ceramide Metabolism and Signaling in the Regulation of Mitophagy and Cancer Therapy. Cancers 2021, 13, 2475. [Google Scholar] [CrossRef]
- Li, R.-Z.; Wang, X.-R.; Wang, J.; Xie, C.; Wang, X.-X.; Pan, H.-D.; Meng, W.-Y.; Liang, T.-L.; Li, J.-X.; Yan, P.-Y.; et al. The Key Role of Sphingolipid Metabolism in Cancer: New Therapeutic Targets, Diagnostic and Prognostic Values, and Anti-Tumor Immunotherapy Resistance. Front. Oncol. 2022, 12, 941643. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid Metabolism in Cancer Signalling and Therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Moro, K.; Kawaguchi, T.; Tsuchida, J.; Gabriel, E.; Qi, Q.; Yan, L.; Wakai, T.; Takabe, K.; Nagahashi, M. Ceramide Species Are Elevated in Human Breast Cancer and Are Associated with Less Aggressiveness. Oncotarget 2018, 9, 19874–19890. [Google Scholar] [CrossRef]
- Mochalski, P.; Unterkofler, K. Quantification of Selected Volatile Organic Compounds in Human Urine by Gas Chromatography Selective Reagent Ionization Time of Flight Mass Spectrometry (GC-SRI-TOF-MS) Coupled with Head-Space Solid-Phase Microextraction (HS-SPME). Analyst 2016, 141, 4796–4803. [Google Scholar] [CrossRef]
- Loomis, D.; Guyton, K.Z.; Grosse, Y.; El Ghissassi, F.; Bouvard, V.; Benbrahim-Tallaa, L.; Guha, N.; Vilahur, N.; Mattock, H.; Straif, K. Carcinogenicity of Benzene. Lancet Oncol. 2017, 18, 1574–1575. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Li, Q.; Chen, Y.; Yang, Y. Recent Metabolomics Analysis in Tumor Metabolism Reprogramming. Front. Mol. Biosci. 2021, 8, 763902. [Google Scholar] [CrossRef] [PubMed]
- Schaeffeler, E.; Büttner, F.; Reustle, A.; Klumpp, V.; Winter, S.; Rausch, S.; Fisel, P.; Hennenlotter, J.; Kruck, S.; Stenzl, A.; et al. Metabolic and Lipidomic Reprogramming in Renal Cell Carcinoma Subtypes Reflects Regions of Tumor Origin. Eur. Urol. Focus 2019, 5, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.K.; Kim, S. An Insight into GPCR and G-Proteins as Cancer Drivers. Cells 2021, 10, 3288. [Google Scholar] [CrossRef]
- Sriram, K.; Salmerón, C.; Wiley, S.Z.; Insel, P.A. GPCRs in Pancreatic Adenocarcinoma: Contributors to Tumour Biology and Novel Therapeutic Targets. Br. J. Pharmacol. 2020, 177, 2434–2455. [Google Scholar] [CrossRef]
- Predescu, D.V.; Crețoiu, S.M.; Crețoiu, D.; Alexandra Pavelescu, L.; Suciu, N.; Radu, B.M.; Voinea, S.C. G Protein-Coupled Receptors (GPCRs)-Mediated Calcium Signaling in Ovarian Cancer: Focus on GPCRs Activated by Neurotransmitters and Inflammation-Associated Molecules. Int. J. Mol. Sci. 2019, 20, 5568. [Google Scholar] [CrossRef]
- Ghosh, C.; Luong, G.; Sun, Y. A Snapshot of the PD-1/PD-L1 Pathway. J. Cancer 2021, 12, 2735–2746. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fraile-Martínez, O.; Asúnsolo, Á.; Buján, J.; García-Honduvilla, N.; Coca, S. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J. Oncol. 2020, 2020, 9258396. [Google Scholar] [CrossRef]
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Seyed Forootan, F.; Nasr Esfahani, M.H.; Ghaedi, K. Signaling Pathways Involved in Colorectal Cancer Progression. Cell Biosci. 2019, 9, 97. [Google Scholar] [CrossRef]
- Rasheed, S.A.K.; Subramanyan, L.V.; Lim, W.K.; Udayappan, U.K.; Wang, M.; Casey, P.J. The Emerging Roles of Gα12/13 Proteins on the Hallmarks of Cancer in Solid Tumors. Oncogene 2022, 41, 147–158. [Google Scholar] [CrossRef]
- Lauriola, A.; Davalli, P.; Marverti, G.; Santi, S.; Caporali, A.; D’Arca, D. Targeting the Interplay of Independent Cellular Pathways and Immunity: A Challenge in Cancer Immunotherapy. Cancers 2023, 15, 3009. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Mondul, A.M.; Weinstein, S.J.; Albanes, D. Serum Retinol and Risk of Overall and Site-Specific Cancer in the ATBC Study. Am. J. Epidemiol. 2020, 189, 532–542. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Pandey, K.; Panda, M.; Spinella, M.J.; Rengasamy, K.R.; Biswal, B.K. The Potential of Retinoids for Combination Therapy of Lung Cancer: Updates and Future Directions. Pharmacol. Res. 2019, 147, 104331. [Google Scholar] [CrossRef] [PubMed]
- Duitama, M.; Moreno, Y.; Santander, S.P.; Casas, Z.; Sutachan, J.J.; Torres, Y.P.; Albarracín, S.L. TRP Channels as Molecular Targets to Relieve Cancer Pain. Biomolecules 2021, 12, 1. [Google Scholar] [CrossRef]
- Saldías, M.P.; Maureira, D.; Orellana-Serradell, O.; Silva, I.; Lavanderos, B.; Cruz, P.; Torres, C.; Cáceres, M.; Cerda, O. TRP Channels Interactome as a Novel Therapeutic Target in Breast Cancer. Front. Oncol. 2021, 11, 621614. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.-H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef]
- Beretov, J.; Wasinger, V.C.; Schwartz, P.; Graham, P.H.; Li, Y. A Standardized and Reproducible Urine Preparation Protocol for Cancer Biomarkers Discovery. Biomark. Cancer 2014, 6, BIC-S17991. [Google Scholar] [CrossRef]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard Operating Procedures for Pre-Analytical Handling of Blood and Urine for Metabolomic Studies and Biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef]
- Nam, S.L.; de la Mata, A.P.; Dias, R.P.; Harynuk, J.J. Towards Standardization of Data Normalization Strategies to Improve Urinary Metabolomics Studies by GC×GC-TOFMS. Metabolites 2020, 10, 376. [Google Scholar] [CrossRef]
- Favé, G.; Beckmann, M.; Lloyd, A.J.; Zhou, S.; Harold, G.; Lin, W.; Tailliart, K.; Xie, L.; Draper, J.; Mathers, J.C. Development and Validation of a Standardized Protocol to Monitor Human Dietary Exposure by Metabolite Fingerprinting of Urine Samples. Metabolomics 2011, 7, 469–484. [Google Scholar] [CrossRef]
- Holbrook, K.L.; Badmos, S.; Habib, A.; Landa, E.N.; Quaye, G.E.; Pokojovy, M.; Su, X.; Lee, W.-Y. Investigating the Effects of Storage Conditions on Urinary Volatilomes for Their Reliability in Disease Diagnosis. Am. J. Clin. Exp. Urol. 2023, 11, 481–499. [Google Scholar]
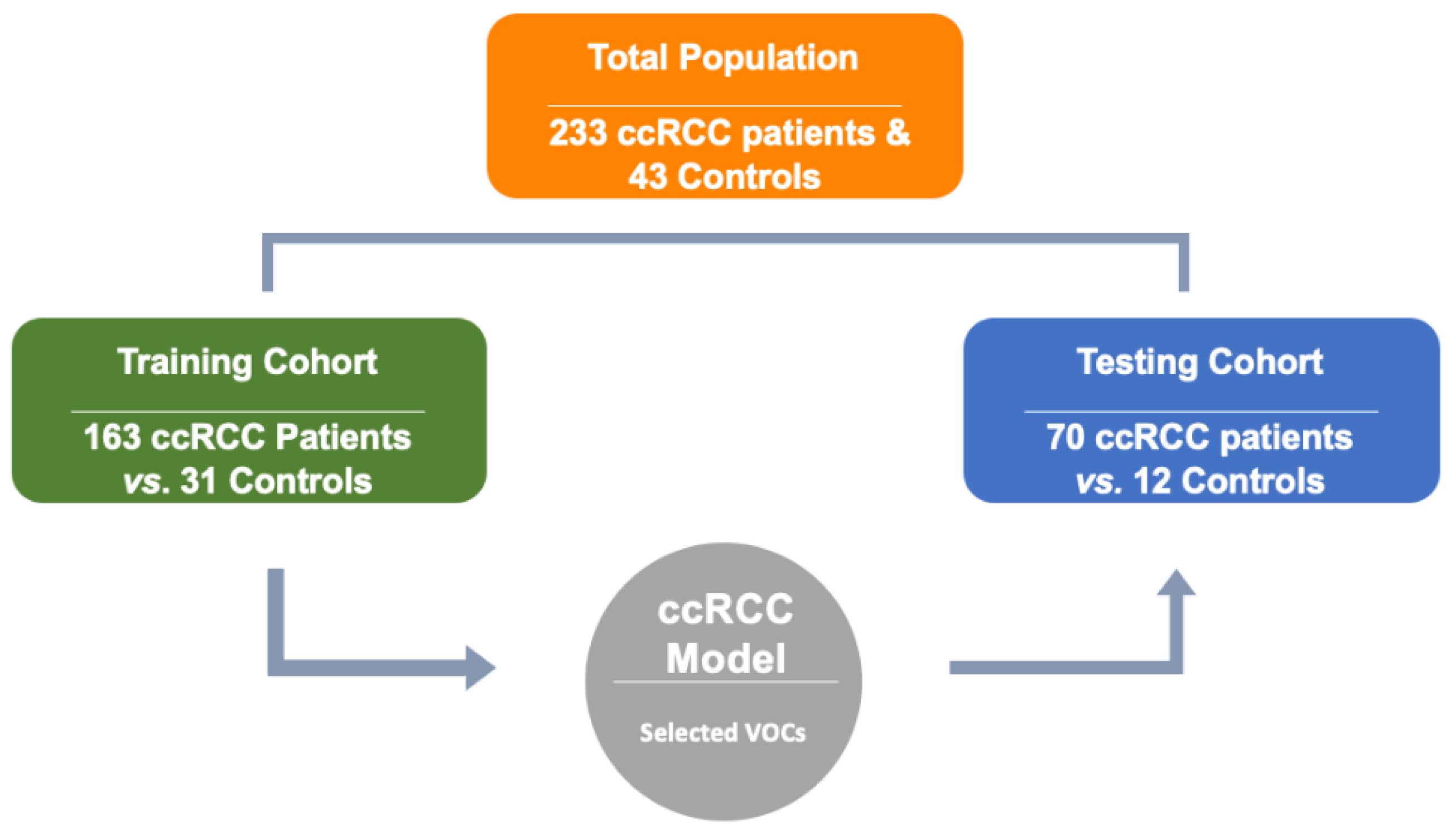
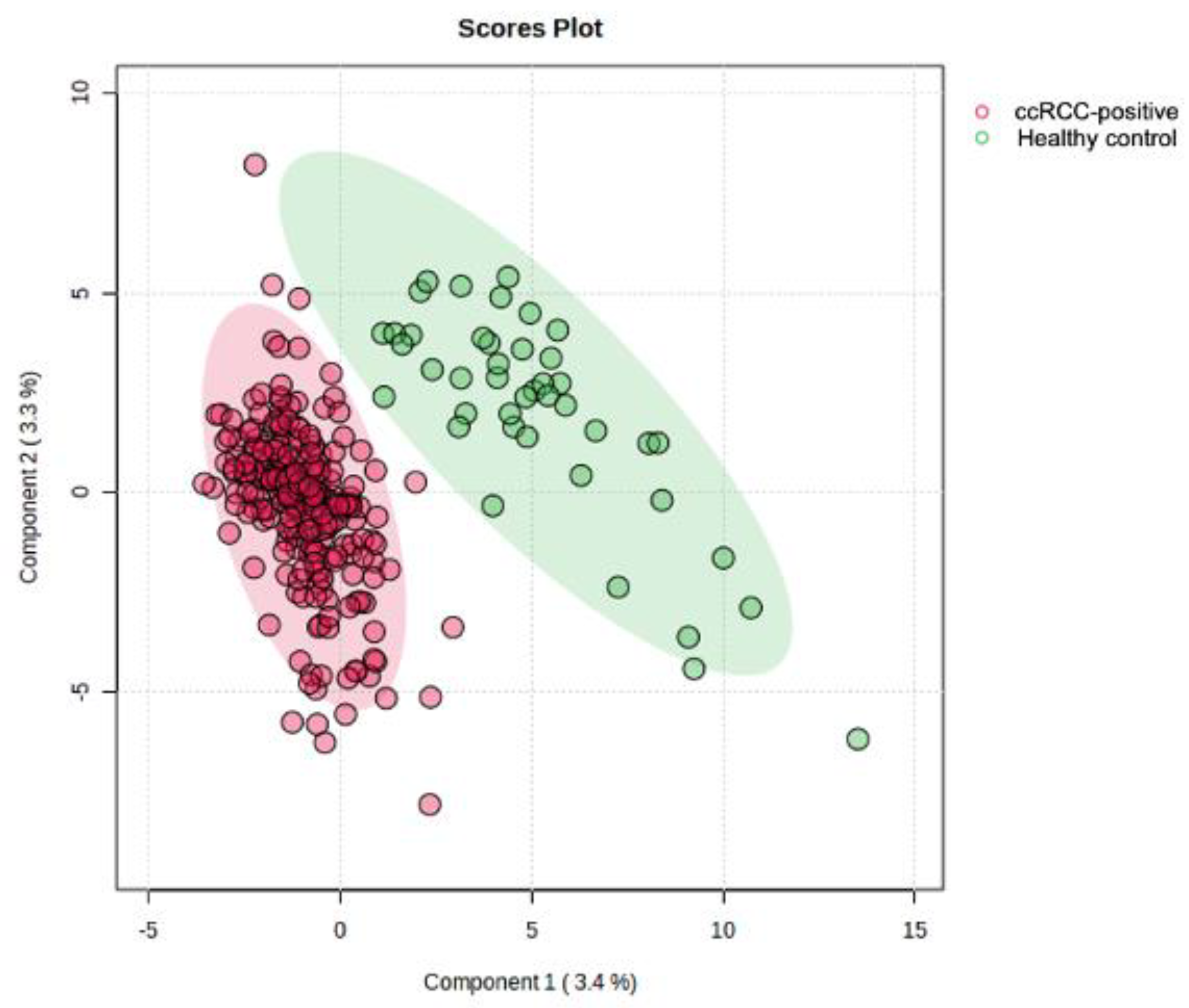
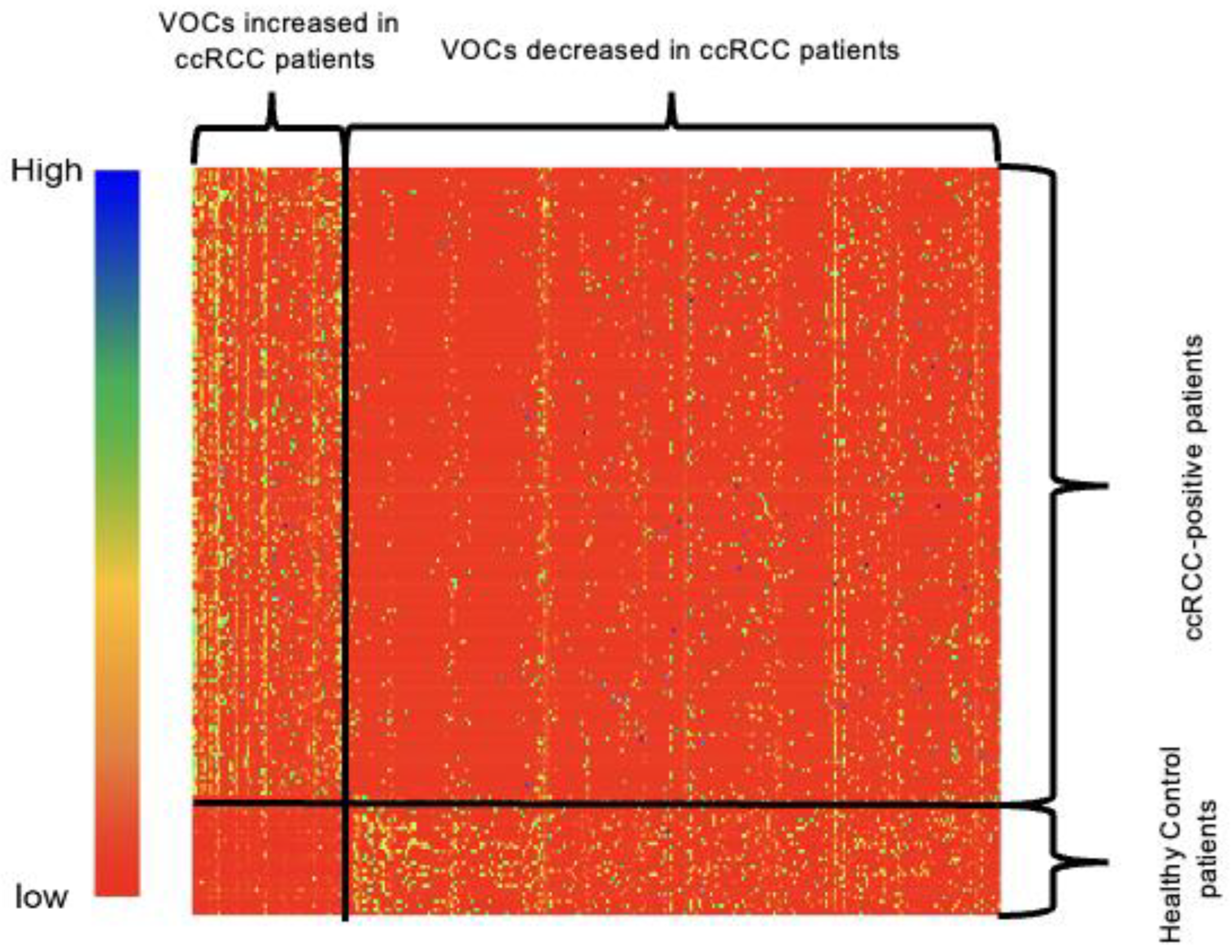
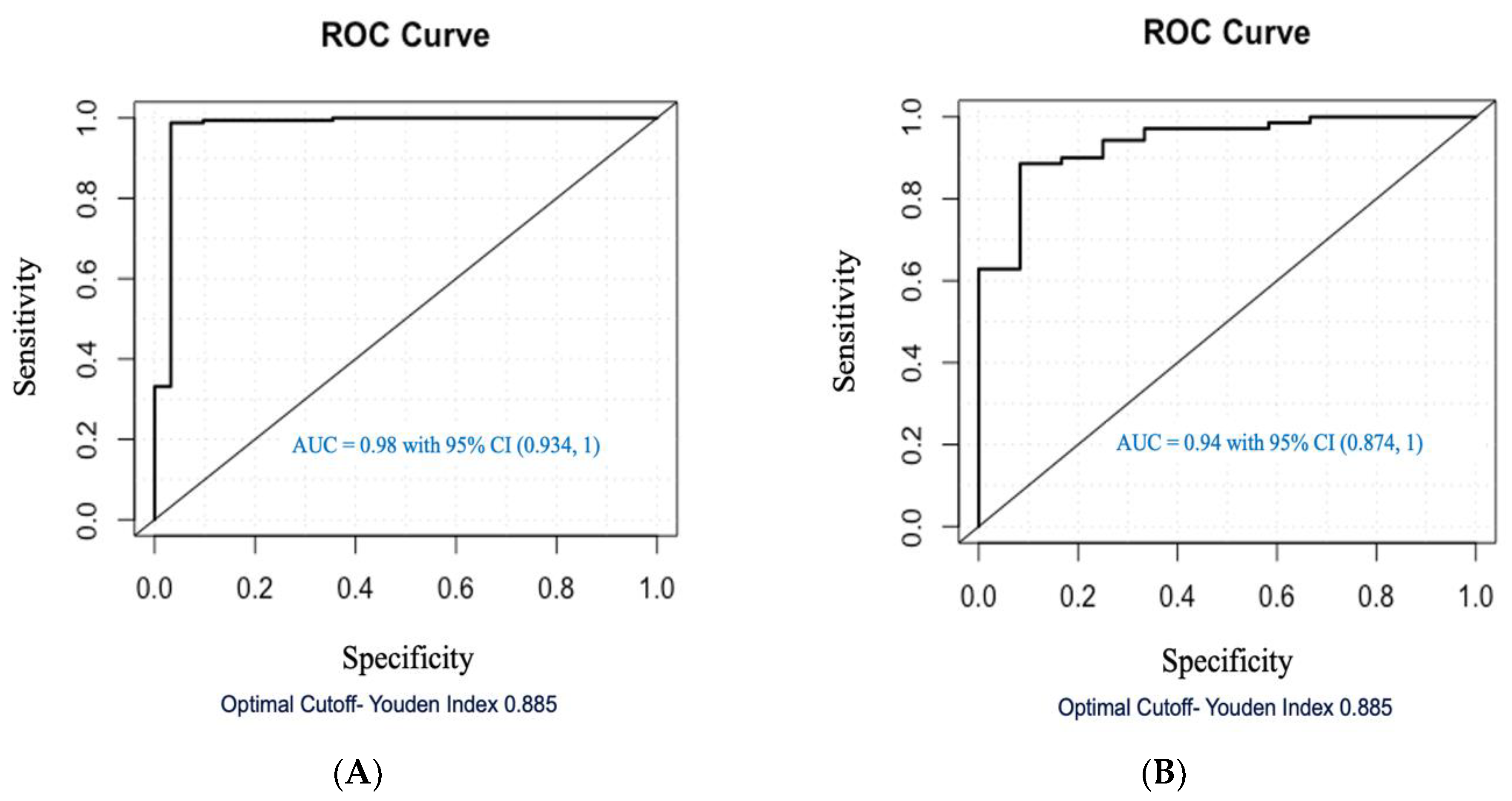
| (A) | Training Cohort (Model Development) | Testing Cohort (Model Validation) | ||
|---|---|---|---|---|
| ccRCC Group | Control Group | ccRCC Group | Control Group | |
| No. | 163 | 31 | 70 | 12 |
| Age | 63 (26–87) | 46 (22–78) | 60 (33–87) | 57 (26–73) |
| Gender | ||||
| M | 103 | 14 | 49 | 5 |
| F | 60 | 17 | 21 | 7 |
| Tumor grade 1 | N/A 2 | N/A 2 | ||
| 1 | 20 (12%) | 10 (14%) | ||
| 2 | 23 (14%) | 9 (13%) | ||
| 3 | 19 (12%) | 11 (16%) | ||
| 4 | 10 (6%) | 6 (8%) | ||
| Unknown | 90 (56%) | 34 (49%) | ||
| (B) | Characteristic | Control N = 43 3 | Positive N = 233 3 | p-Value 4 |
| Age | 50 (29, 60) | 63 (56, 70) | <0.001 | |
| Gender | 0.011 | |||
| M | 19 (44%) | 152 (65%) | ||
| F | 24 (56%) | 81 (35%) | ||
| CAS Number 1 | Chemical Formula | Chemical Name | Dominating Group | p-Value 2 | Occurrence | |
|---|---|---|---|---|---|---|
| Cancer (+) 3 | Control (−) 4 | |||||
| * 000104-76-7 | C8H18O | 1-Hexanol, 2-ethyl- | ccRCC | 3.07 × 10−12 | 140 | 9 |
| * 005637-97-8 | C17H32O | Heptadecanolide | ccRCC | 1.35 × 10−1 | 27 | 2 |
| 1000465-65-6 | C17H24O4 | 2-Ethylhexyl methyl isophthalate | Control | 2.22 × 10−19 | 8 | 21 |
| 015356-70-4 | C10H20O | Cyclohexanol, 5-methyl-2-(1-methylethyl)-, (1.alpha.,2.beta.,5.alpha.)-(.+/-.)- | Control | 5.76 × 10−16 | 14 | 21 |
| 001490-04-6 | C12H22O2 | Cyclohexanol, 5-methyl-2-(1-methylethyl)- | Control | 1.39 × 10−13 | 1 | 11 |
| 007568-58-3 | C18H30O | 1-Propene-1,2,3-tricarboxylic acid, tributyl ester | Control | 1.42 × 10−13 | 2 | 12 |
| 028336-57-4 | C24H24 | Cyclohexane, 1,3,5-triphenyl- | Control | 2.47 × 10−12 | 2 | 11 |
| 016982-00-6 | C15H22 | Benzene, 1-methyl-4-(1,2,2-trimethylcyclopentyl)-, (R)- | Control | 7.43 × 10−10 | 0 | 7 |
| 000491-02-1 | C10H20O | Cyclohexanol, 5-methyl-2-(1-methylethyl)-, (1.alpha.,2.alpha.,5.alpha.)- | Control | 1.68 × 10−8 | 1 | 7 |
| 000075-31-0 | C3H9N | 2-Propanamine | Control | 2.29 × 10−7 | 0 | 5 |
| * 000506-17-2 | C18H34O2 | cis-Vaccenic acid | ccRCC | 1.22 × 10−6 | 38 | 19 |
| * 013151-34-3 | C11H24 | Decane, 3-methyl- | ccRCC | 1.29 × 10−6 | 14 | 12 |
| 002305-05-7 | C10H18O2 | .gamma.-Dodecalactone | Control | 7.63 × 10−6 | 4 | 7 |
| 1000140-05-6 | C15H22 | Cadala-1(10),3,8-triene | Control | 2.12 × 10−5 | 3 | 6 |
| * 004630-07-3 | C15H24 | Naphthalene, 1,2,3,5,6,7,8,8a-octahydro-1,8a-dimethyl-7-(1-methylethenyl)-, [1R-(1.alpha.,7.beta.,8a.alpha.)]- | ccRCC | 3.98 × 10−5 | 13 | 10 |
| 1000427-45-5 | C5H6O2 | 4-Methylamino-2(5H)-furanone | Control | 8.11 × 10−5 | 1 | 4 |
| * 013183-70-5 | C12H22Si2 | 1,4-Bis(trimethylsilyl)benzene | ccRCC | 1.13 × 10−3 | 65 | 18 |
| * 1000383-15-8 | C20H40O3 | Carbonic acid, decyl nonyl ester | ccRCC | 6.09 × 10−3 | 6 | 5 |
| 000095-75-0 | C7H6Cl | Benzene, 1,2-dichloro-4-methyl- | Control | 6.27 × 10−3 | 2 | 3 |
| 000589-08-2 | C9H13N | Benzeneethanamine, N-methyl- | Control | 6.77 × 10−3 | 2 | 3 |
| 1000130-20-8 | C5H7N3O2 | l-Guanidinosuccinimide | Control | 7.03 × 10−3 | 2 | 3 |
| * 028474-90-0 | C38H68O8 | l-(+)-Ascorbic acid 2,6-dihexadecanoate | ccRCC | 1.13 × 10−2 | 7 | 5 |
| * 005951-67-7 | C15H24 | Cyclohexene, 6-ethenyl-6-methyl-1-(1-methylethyl)-3-(1-methylethylidene)-, (S)- | ccRCC | 2.66 × 10−2 | 9 | 5 |
| * 038142-57-3 | C15H22O | 2-Methyl-6-(p-tolyl)hept-2-en-4-ol | ccRCC | 4.57 × 10−2 | 4 | 3 |
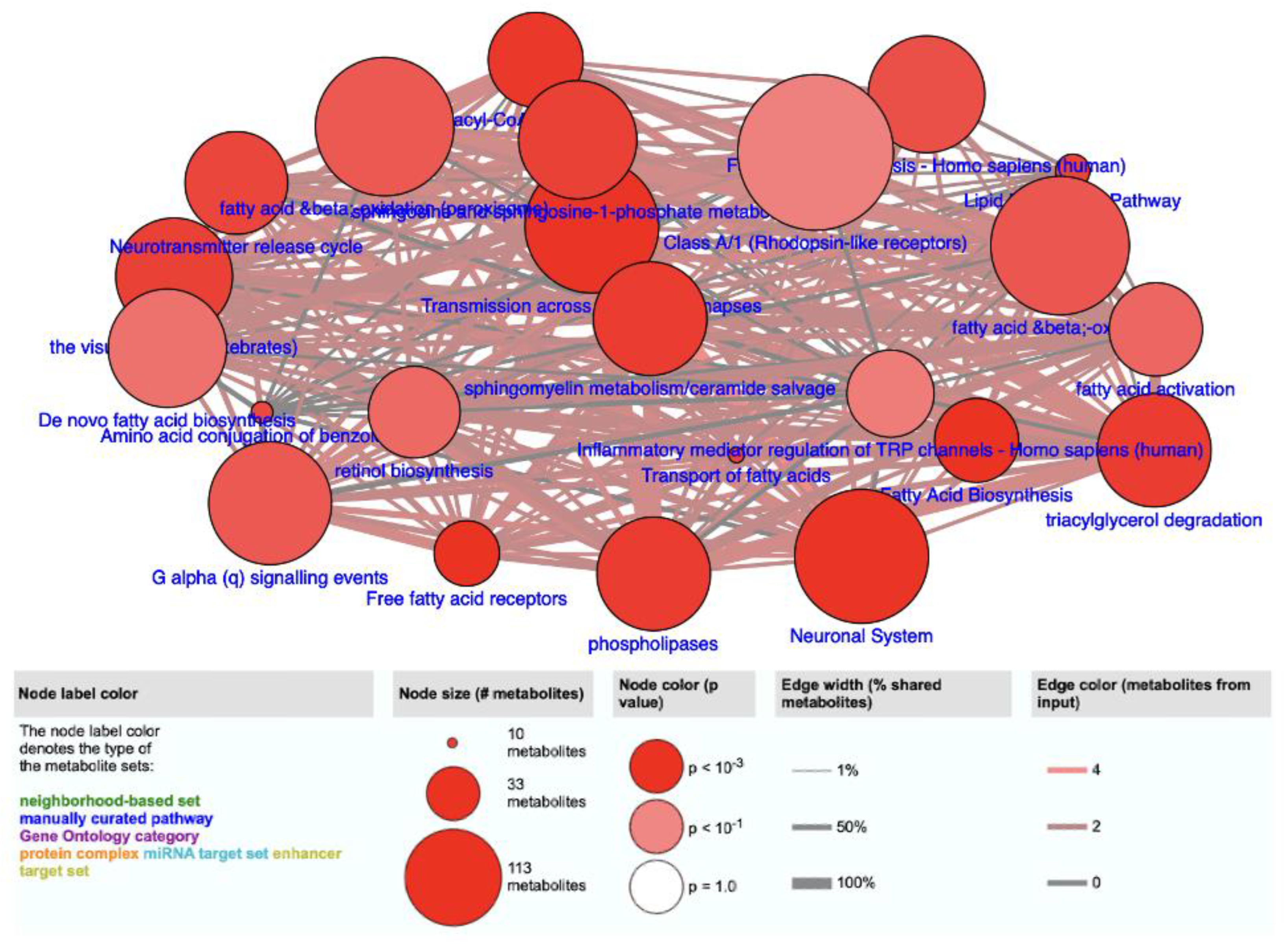
| Pathway Name | Pathway Source * | p-Value |
|---|---|---|
| Free fatty acid receptors | Reactome | 1.16 × 10−4 |
| Fatty acid biosynthesis | SMPDB | 1.16 × 10−4 |
| Transmission across chemical synapses | Reactome | 1.63 × 10−4 |
| Neuronal system | Reactome | 1.63 × 10−4 |
| Acyl-CoA hydrolysis | HumanCyc | 2.29 × 10−4 |
| Phospholipases | HumanCyc | 3.34 × 10−4 |
| Triacylgycerol degradation | HumanCyc | 3.34 × 10−4 |
| Sphingomyelin metabolism/ceramide salvage | HumanCyc | 3.34 × 10−4 |
| The visual cycle I (vertebrates) | HumanCyc | 4.66 × 10−4 |
| Sphingosine and sphingosine-1-phosphate metabolism | HumanCyc | 4.66 × 10−4 |
| Lipid metabolism pathway | Wikipathways | 5.92 × 10−4 |
| Transport of fatty acids | Reactome | 5.92 × 10−4 |
| Neurotransmitter release cycle | Reactome | 6.28 × 10−4 |
| Amino acid conjugation of benzoic acid | Wikipathways | 7.38 × 10−4 |
| Fatty acid biosynthesis—Homo sapiens (human) | KEGG | 1.08 × 10−3 |
| Fatty acid β-oxidation | HumanCyc | 1.54 × 10−3 |
| G alpha (q) signaling events | Reactome | 1.62 × 10−3 |
| Fatty acid β-oxidation (peroxisome) | HumanCyc | 1.62 × 10−3 |
| Fatty acid activation | HumanCyc | 3.04 × 10−3 |
| Retinol biosynthesis | HumanCyc | 3.35 × 10−3 |
| De novo fatty acid biosynthesis | EHMN | 4.74 × 10−3 |
| Inflammatory mediator regulation of TRP-channels- Homo sapiens (human) | KEGG | 7.25 × 10−3 |
| Class A/1 (Rhodopsin-like receptors) | Reactome | 8.77 × 10−3 |
| Reference | Cohort Size | Analytical Methods | Statistical Methods | AUC-ROC (Sensitivity/Specificity) | Selected VOCs or Biomarkers |
|---|---|---|---|---|---|
| Monteiro et al. [27] | 30 RCC; 37 healthy (RCC urine) | HS-SPME-GC-IT/MS | PCA | ND * | 2-oxopropanal and 2,5,8-trimethyl-1,2,3,4-tetrahydronaphthalene-1-ol |
| Wang et al. [47] | 22 RCC; 25 healthy (RCC urine) | UPLC-MS | Welch Two Sample T-Test, Variable Importance in the Projection (VIP Values), PLS-DA | H vs. RCC: 0.702 (76% and 79%); Pre vs. Post: 0.833 (61% and 88%) | phenol, decanal,1,6-dioxacyclododecane-7,12-dione; 1-brom o-1-(3-methyl-1-pentenylidene)-2,2,3,3-tetramethyl-cyclopropane; nonanal; 3-ethyl-3-methylheptane; isolongifolene-5-ol; 2,5-cyclohexadiene-1,4-dione, 2,6-bis(1,1-dimethylethyl); tetradecane; aniline; 2,6,10,14-tetramethyl-pentadecane; styrene, 4-heptanone; dimethylsilanediol; 2-ethyl-1-hexanol; cyclohexanone; 6-t-butyl-2,2,9,9-tetramethyl-3,5-decadien-7-yne |
| Amaro et al. [16] | RCC cell lines | HS-SPME-GC-MS | PCA and PLS-DA | ND * for entire VOC panel | cyclohexanone; acetaldehyde; cyclohexanol; decanal; decane; dodecane; and 4-methylbenzaldehyde |
| Morrissey et al. [48] | 19 RCC; 80 healthy (RCC urine) | ELISA and Western Blot | One-way ANOVA and Pearson Chi-square Test | 1.0 (100% and 100%); 0.99 (100% and 98%) | AQP-1 and PLIN |
| Mijuskovic et al. [49] | 40 RCC; 40 healthy (RCC urine) | ELISA | Smirnov Test and Mann–Whitney Test | ND * | KIM-1 and AQP-1 |
| Holbrook et al. (this study) | 233 ccRCC; 43 healthy (RCC urine) | SBSE-GC-MS | Linear Regression | 0.94 (86% and 92%) | 24 (Table 2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holbrook, K.L.; Quaye, G.E.; Noriega Landa, E.; Su, X.; Gao, Q.; Williams, H.; Young, R.; Badmos, S.; Habib, A.; Chacon, A.A.; et al. Detection and Validation of Organic Metabolites in Urine for Clear Cell Renal Cell Carcinoma Diagnosis. Metabolites 2024, 14, 546. https://doi.org/10.3390/metabo14100546
Holbrook KL, Quaye GE, Noriega Landa E, Su X, Gao Q, Williams H, Young R, Badmos S, Habib A, Chacon AA, et al. Detection and Validation of Organic Metabolites in Urine for Clear Cell Renal Cell Carcinoma Diagnosis. Metabolites. 2024; 14(10):546. https://doi.org/10.3390/metabo14100546
Chicago/Turabian StyleHolbrook, Kiana L., George E. Quaye, Elizabeth Noriega Landa, Xiaogang Su, Qin Gao, Heinric Williams, Ryan Young, Sabur Badmos, Ahsan Habib, Angelica A. Chacon, and et al. 2024. "Detection and Validation of Organic Metabolites in Urine for Clear Cell Renal Cell Carcinoma Diagnosis" Metabolites 14, no. 10: 546. https://doi.org/10.3390/metabo14100546
APA StyleHolbrook, K. L., Quaye, G. E., Noriega Landa, E., Su, X., Gao, Q., Williams, H., Young, R., Badmos, S., Habib, A., Chacon, A. A., & Lee, W.-Y. (2024). Detection and Validation of Organic Metabolites in Urine for Clear Cell Renal Cell Carcinoma Diagnosis. Metabolites, 14(10), 546. https://doi.org/10.3390/metabo14100546







