Global and Targeted Metabolomics for Revealing Metabolomic Alteration in Niemann-Pick Disease Type C Model Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. LC/MS/MS Equipment
2.3. LC/MS/MS Condition for Global Metabolomics
2.4. Cell Culture Conditions
2.5. Global Metabolomics Procedure for Cell Samples and Selection of Metabolites for Targeted Metabolomics
2.6. Standard Solutions for Targeted Metabolomics
2.7. Optimization of LC/MS/MS Conditions for Targeted Metabolomics
2.8. Calibration Curves for Targeted Metabolomics
2.9. Cell Sample Preparation for Targeted Metabolomics
2.10. Quantification of Metabolites by Targeted Metabolomics
2.11. Variation Analysis of Metabolic Pathways
3. Results and Discussion
3.1. Global Metabolome Analysis of NPC Model Cells
3.2. Targeted Metabolomics for Quantification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vanier, M.T.; Millat, G. Niemann-Pick Disease Type C. Clin. Genet. 2003, 64, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Patterson, M.C.; Clayton, P.; Gissen, P.; Anheim, M.; Bauer, P.; Bonnot, O.; Dardis, A.; Dionisi-Vici, C.; Klünemann, H.-H.; Latour, P.; et al. Recommendations for the Detection and Diagnosis of Niemann-Pick Disease Type C. Neurol. Clin. Pract. 2017, 7, 499–511. [Google Scholar] [CrossRef]
- Patterson, M.C.; Hendriksz, C.J.; Walterfang, M.; Sedel, F.; Vanier, M.T.; Wijburg, F. Recommendations for the Diagnosis and Management of Niemann-Pick Disease Type C: An Update. Mol. Genet. Metab. 2012, 106, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Carstea, E.D.; Polymeropoulos, M.H.; Parker, C.C.; Detera-Wadleigh, S.D.; O’Neill, R.R.; Patterson, M.C.; Goldin, E.; Xiao, H.; Straub, R.E.; Vanier, M.T.; et al. Linkage of Niemann-Pick Disease Type C to Human Chromosome 18. Proc. Natl. Acad. Sci. USA 1993, 90, 2002–2004. [Google Scholar] [CrossRef]
- Naureckiene, S.; Sleat, D.E.; Lackland, H.; Fensom, A.; Vanier, M.T.; Wattiaux, R.; Jadot, M.; Lobel, P. Identification of HE1 as the Second Gene of Niemann-Pick C Disease. Science 2000, 290, 2298–2301. [Google Scholar] [CrossRef] [PubMed]
- Geberhiwot, T.; Moro, A.; Dardis, A.; Ramaswami, U.; Sirrs, S.; Marfa, M.P.; Vanier, M.T.; Walterfang, M.; Bolton, S.; Dawson, C.; et al. Consensus Clinical Management Guidelines for Niemann-Pick Disease Type C. Orphanet J. Rare Dis. 2018, 13, 50. [Google Scholar] [CrossRef]
- Sitarska, D.; Tylki-Szymańska, A.; Ługowska, A. Treatment Trials in Niemann-Pick Type C Disease. Metab. Brain Dis. 2021, 36, 2215. [Google Scholar] [CrossRef]
- Colombo, A.; Dinkel, L.; Müller, S.A.; Sebastian Monasor, L.; Schifferer, M.; Cantuti-Castelvetri, L.; König, J.; Vidatic, L.; Bremova-Ertl, T.; Lieberman, A.P.; et al. Loss of NPC1 Enhances Phagocytic Uptake and Impairs Lipid Trafficking in Microglia. Nat. Commun. 2021, 12, 1158. [Google Scholar] [CrossRef]
- Mengel, E.; Klünemann, H.H.; Lourenço, C.M.; Hendriksz, C.J.; Sedel, F.; Walterfang, M.; Kolb, S.A. Niemann-Pick Disease Type C Symptomatology: An Expert-Based Clinical Description. Orphanet J. Rare Dis. 2013, 8, 166. [Google Scholar] [CrossRef]
- Vanier, M.T. Niemann-Pick Disease Type C. Orphanet J. Rare Dis. 2010, 5, 16. [Google Scholar] [CrossRef]
- Wraith, J.E.; Baumgartner, M.R.; Bembi, B.; Covanis, A.; Levade, T.; Mengel, E.; Pineda, M.; Sedel, F.; Topçu, M.; Vanier, M.T.; et al. Recommendations on the Diagnosis and Management of Niemann-Pick Disease Type C. Mol. Genet. Metab. 2009, 98, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, R.; Mahale, R.R.; Sindhu, D.M.; Stezin, A.; Kamble, N.; Holla, V.V.; Netravathi, M.; Yadav, R.; Pal, P.K. Spectrum of Movement Disorders in Niemann-Pick Disease Type C. Tremor Other Hyperkinetic Mov. 2022, 12, 28. [Google Scholar] [CrossRef]
- Bolton, S.C.; Soran, V.; Marfa, M.P.; Imrie, J.; Gissen, P.; Jahnova, H.; Sharma, R.; Jones, S.; Santra, S.; Crushell, E.; et al. Clinical Disease Characteristics of Patients with Niemann-Pick Disease Type C: Findings from the International Niemann-Pick Disease Registry (INPDR). Orphanet J. Rare Dis. 2022, 17, 51. [Google Scholar] [CrossRef]
- Spiegel, R.; Raas-Rothschild, A.; Reish, O.; Regev, M.; Meiner, V.; Bargal, R.; Sury, V.; Meir, K.; Nadjari, M.; Hermann, G.; et al. The Clinical Spectrum of Fetal Niemann-Pick Type C. Am. J. Med. Genet. A 2009, 149A, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Pineda, M.; Walterfang, M.; Patterson, M.C. Miglustat in Niemann-Pick Disease Type C Patients: A Review. Orphanet J. Rare Dis. 2018, 13, 140. [Google Scholar] [CrossRef]
- Héron, B.; Valayannopoulos, V.; Baruteau, J.; Chabrol, B.; Ogier, H.; Latour, P.; Dobbelaere, D.; Eyer, D.; Labarthe, F.; Maurey, H.; et al. Miglustat Therapy in the French Cohort of Paediatric Patients with Niemann-Pick Disease Type C. Orphanet J. Rare Dis. 2012, 7, 36. [Google Scholar] [CrossRef]
- Pineda, M.; Wraith, J.E.; Mengel, E.; Sedel, F.; Hwu, W.L.; Rohrbach, M.; Bembi, B.; Walterfang, M.; Korenke, G.C.; Marquardt, T.; et al. Miglustat in Patients with Niemann-Pick Disease Type C (NP-C): A Multicenter Observational Retrospective Cohort Study. Mol. Genet. Metab. 2009, 98, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Mano, N. Identification and Evaluation of Biomarkers for Niemann-Pick Disease Type C Based on Chemical Analysis Techniques. Chromatography 2020, 41, 19–29. [Google Scholar] [CrossRef]
- Maekawa, M.; Mano, N. Searching, Structural Determination, and Diagnostic Performance Evaluation of Biomarker Molecules for Niemann-Pick Disease Type C Using Liquid Chromatography/Tandem Mass Spectrometry. Mass Spectrom. 2022, 11, A0111. [Google Scholar] [CrossRef]
- Maekawa, M.; Iwahori, A.; Mano, N. Biomarker Analysis of Niemann-Pick Disease Type C Using Chromatography and Mass Spectrometry. J. Pharm. Biomed. Anal. 2020, 191, 113622. [Google Scholar] [CrossRef]
- Bremova-Ertl, T.; Claassen, J.; Foltan, T.; Gascon-Bayarri, J.; Gissen, P.; Hahn, A.; Hassan, A.; Hennig, A.; Jones, S.A.; Kolnikova, M.; et al. Efficacy and Safety of N-Acetyl-l-Leucine in Niemann–Pick Disease Type C. J. Neurol. 2022, 269, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.; Shraishi, K.; Wada, K.; Ishitsuka, Y.; Doi, H.; Maeda, M.; Mizoguchi, T.; Eto, J.; Mochinaga, S.; Arima, H.; et al. Effects of Intracerebroventricular Administration of 2-Hydroxypropyl-β-Cyclodextrin in a Patient with Niemann–Pick Type C Disease. Mol. Genet. Metab. Rep. 2014, 1, 391–400. [Google Scholar] [CrossRef]
- Rodriguez-Gil, J.L.; Watkins-Chow, D.E.; Baxter, L.L.; Elliot, G.; Harper, U.L.; Wincovitch, S.M.; Wedel, J.C.; Incao, A.A.; Huebecker, M.; Boehm, F.J.; et al. Genetic Background Modifies Phenotypic Severity and Longevity in a Mouse Model of Niemann-Pick Disease Type C1. DMM Dis. Models Mech. 2020, 13, dmm042614. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nanba, E.; Ninomiya, H.; Higaki, K.; Taniguchi, M.; Zhang, H.; Akaboshi, S.; Watanabe, Y.; Takeshima, T.; Inui, K.; et al. NPC1 Gene Mutations in Japanese Patients with Niemann-Pick Disease Type C. Hum. Genet. 1999, 105, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kawazoe, T.; Yamamoto, T.; Narita, A.; Ohno, K.; Adachi, K.; Nanba, E.; Noguchi, A.; Takahashi, T.; Maekawa, M.; Eto, Y.; et al. Phenotypic Variability of Niemann-Pick Disease Type C Including a Case with Clinically Pure Schizophrenia: A Case Report. BMC Neurol. 2018, 18, 117. [Google Scholar] [CrossRef]
- Costanzo, M.C.; Nicotera, A.G.; Vinci, M.; Vitello, A.; Fiumara, A.; Calì, F.; Musumeci, S.A. Novel Compound Heterozygous Mutation in NPC1 Gene Cause Niemann–Pick Disease Type C with Juvenile Onset. J. Genet. 2020, 99, 30. [Google Scholar] [CrossRef]
- López de Frutos, L.; Cebolla, J.J.; Aldámiz-Echevarría, L.; de la Vega, Á.; Stanescu, S.; Lahoz, C.; Irún, P.; Giraldo, P. New Variants in Spanish Niemann-Pick Type c Disease Patients. Mol. Biol. Rep. 2020, 47, 2085–2095. [Google Scholar] [CrossRef]
- Monton, M.R.N.; Soga, T. Metabolome Analysis by Capillary Electrophoresis-Mass Spectrometry. J. Chromatogr. A 2007, 1168, 237–246. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The Human Serum Metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the Metabolome: Current Analytical Technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Stincone, P.; Naimi, A.; Saviola, A.J.; Reher, R.; Petras, D. Decoding the Molecular Interplay in the Central Dogma: An Overview of Mass Spectrometry-based Methods to Investigate Protein-metabolite Interactions. Proteomics 2023, 24, e2200533. [Google Scholar] [CrossRef] [PubMed]
- Costa dos Santos, G.; Renovato-Martins, M.; de Brito, N.M. The Remodel of the “Central Dogma”: A Metabolomics Interaction Perspective. Metabolomics 2021, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.A.; Church, H.J.; Wu, H.Y. Cholestane-3β, 5α, 6β-Triol: Further Insights into the Performance of This Oxysterol in Diagnosis of Niemann-Pick Disease Type C. Mol. Genet. Metab. 2020, 130, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Ory, D.S.; Ottinger, E.A.; Farhat, N.Y.; King, K.A.; Jiang, X.; Weissfeld, L.; Berry-Kravis, E.; Davidson, C.D.; Bianconi, S.; Keener, L.A.; et al. Intrathecal 2-Hydroxypropyl-β-Cyclodextrin Decreases Neurological Disease Progression in Niemann-Pick Disease, Type C1: A Non-Randomised, Open-Label, Phase 1–2 Trial. Lancet 2017, 390, 1758–1768. [Google Scholar] [CrossRef]
- Maekawa, M.; Jinnoh, I.; Narita, A.; Iida, T.; Saigusa, D.; Iwahori, A.; Nittono, H.; Okuyama, T.; Eto, Y.; Ohno, K.; et al. Investigation of Diagnostic Performance of Five Urinary Cholesterol Metabolites for Niemann-Pick Disease Type C. J. Lipid Res. 2019, 60, 2074–2081. [Google Scholar] [CrossRef]
- Maekawa, M.; Jinnoh, I.; Matsumoto, Y.; Narita, A.; Mashima, R.; Takahashi, H.; Iwahori, A.; Saigusa, D.; Fujii, K.; Abe, A.; et al. Structural Determination of Lysosphingomyelin-509 and Discovery of Novel Class Lipids from Patients with Niemann-Pick Disease Type C. Int. J. Mol. Sci. 2019, 20, 5018. [Google Scholar] [CrossRef]
- Porter, F.D.; Scherrer, D.E.; Lanier, M.H.; Langmade, S.J.; Molugu, V.; Gale, S.E.; Olzeski, D.; Sidhu, R.; Dietzen, D.J.; Fu, R.; et al. Cholesterol Oxidation Products Are Sensitive and Specific Blood-Based Biomarkers for Niemann-Pick C1 Disease. Sci. Transl. Med. 2010, 2, 56ra81. [Google Scholar] [CrossRef]
- Fu, R.; Yanjanin, N.M.; Bianconi, S.; Pavan, W.J.; Porter, F.D. Oxidative Stress in Niemann-Pick Disease, Type C. Mol. Genet. Metab. 2010, 101, 214–218. [Google Scholar] [CrossRef]
- Jiang, X.; Sidhu, R.; Mydock-McGrane, L.; Hsu, F.-F.; Covey, D.F.; Scherrer, D.E.; Earley, B.; Gale, S.E.; Farhat, N.Y.; Porter, F.D.; et al. Development of a Bile Acid-Based Newborn Screen for Niemann-Pick Disease Type C. Sci. Transl. Med. 2016, 8, 337ra63. [Google Scholar] [CrossRef]
- Mazzacuva, F.; Mills, P.; Mills, K.; Camuzeaux, S.; Gissen, P.; Nicoli, E.-R.; Wassif, C.; te Vruchte, D.; Porter, F.D.; Maekawa, M.; et al. Identification of Novel Bile Acids as Biomarkers for the Early Diagnosis of Niemann-Pick C Disease. FEBS Lett. 2016, 590, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, R.; Kell, P.; Dietzen, D.J.; Farhat, N.Y.; Do, A.N.D.; Porter, F.D.; Berry-Kravis, E.; Reunert, J.; Marquardt, T.; Giugliani, R.; et al. Application of a Glycinated Bile Acid Biomarker for Diagnosis and Assessment of Response to Treatment in Niemann-Pick Disease Type C1. Mol. Genet. Metab. 2020, 131, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Narita, A.; Jinnoh, I.; Iida, T.; Marquardt, T.; Mengel, E.; Eto, Y.; Clayton, P.T.; Yamaguchi, H.; Mano, N. Diagnostic Performance Evaluation of Sulfate-Conjugated Cholesterol Metabolites as Urinary Biomarkers of Niemann–Pick Disease Type C. Clin. Chim. Acta 2019, 494, 58–63. [Google Scholar] [CrossRef]
- Welford, R.W.D.; Garzotti, M.; Marques Lourenço, C.; Mengel, E.; Marquardt, T.; Reunert, J.; Amraoui, Y.; Kolb, S.A.; Morand, O.; Groenen, P. Plasma Lysosphingomyelin Demonstrates Great Potential as a Diagnostic Biomarker for Niemann-Pick Disease Type C in a Retrospective Study. PLoS ONE 2014, 9, e114669. [Google Scholar] [CrossRef]
- Kuchar, L.; Sikora, J.; Gulinello, M.E.; Poupetova, H.; Lugowska, A.; Malinova, V.; Jahnova, H.; Asfaw, B.; Ledvinova, J. Quantitation of Plasmatic Lysosphingomyelin and Lysosphingomyelin-509 for Differential Screening of Niemann-Pick A/B and C Diseases. Anal. Biochem. 2017, 525, 73–77. [Google Scholar] [CrossRef]
- Mashima, R.; Maekawa, M.; Narita, A.; Okuyama, T.; Mano, N. Elevation of Plasma Lysosphingomyelin-509 and Urinary Bile Acid Metabolite in Niemann-Pick Disease Type C-Affected Individuals. Mol. Genet. Metab. Rep. 2018, 15, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Iwamoto, T.; Hossain, M.A.; Akiyama, K.; Igarashi, J.; Miyajima, T.; Eto, Y. A Combination of 7-Ketocholesterol, Lysosphingomyelin and Bile Acid-408 to Diagnose Niemann-Pick Disease Type C Using LC-MS/MS. PLoS ONE 2020, 15, e0238624. [Google Scholar] [CrossRef]
- Sidhu, R.; Mondjinou, Y.; Qian, M.; Song, H.; Kumar, A.B.; Hong, X.; Hsu, F.-F.; Dietzen, D.J.; Yanjanin, N.M.; Porter, F.D.; et al. N-Acyl-O-Phosphocholineserines: Structures of a Novel Class of Lipids That Are Biomarkers for Niemann-Pick C1 Disease. J. Lipid Res. 2019, 60, 1410–1424. [Google Scholar] [CrossRef] [PubMed]
- Iwahori, A.; Maekawa, M.; Narita, A.; Kato, A.; Sato, T.; Ogura, J.; Sato, Y.; Kikuchi, M.; Noguchi, A.; Higaki, K.; et al. Development of a Diagnostic Screening Strategy for Niemann–Pick Diseases Based on Simultaneous Liquid Chromatography-Tandem Mass Spectrometry Analyses of N-Palmitoyl-O-Phosphocholine-Serine and Sphingosylphosphorylcholine. Biol. Pharm. Bull. 2020, 43, 1398–1406. [Google Scholar] [CrossRef]
- Sidhu, R.; Kell, P.; Dietzen, D.J.; Farhat, N.Y.; Do, A.N.D.; Porter, F.D.; Berry-Kravis, E.; Vite, C.H.; Reunert, J.; Marquardt, T.; et al. Application of N-Palmitoyl-O-Phosphocholineserine for Diagnosis and Assessment of Response to Treatment in Niemann-Pick Type C Disease. Mol. Genet. Metab. 2020, 129, 292–302. [Google Scholar] [CrossRef]
- Giese, A.-K.; Mascher, H.; Grittner, U.; Eichler, S.; Kramp, G.; Lukas, J.; te Vruchte, D.; Al Eisa, N.; Cortina-Borja, M.; Porter, F.D.; et al. A Novel, Highly Sensitive and Specific Biomarker for Niemann-Pick Type C1 Disease. Orphanet J. Rare Dis. 2015, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H.; Jones, D.D. An Update of the Enzymology and Regulation of Sphingomyelin Metabolism. Biochim. Biophys. Acta BBA/Lipids Lipid Metab. 1990, 1044, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Grassi, S.; Chiricozzi, E.; Mauri, L.; Sonnino, S.; Prinetti, A. Sphingolipids and Neuronal Degeneration in Lysosomal Storage Disorders. J. Neurochem. 2019, 148, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Hishinuma, E.; Matsukawa, N.; Shirasago, Y.; Watanabe, M.; Sato, T.; Sato, Y.; Kumondai, M.; Kikuchi, M.; Koshiba, S.; et al. Global Proteomics for Identifying the Alteration Pathway of Niemann–Pick Disease Type C Using Hepatic Cell Models. Int. J. Mol. Sci. 2023, 24, 15642. [Google Scholar] [CrossRef] [PubMed]
- Iwahori, A.; Maekawa, M.; Kanemitsu, Y.; Matsumoto, Y.; Tomioka, Y.; Narita, A.; Okuyama, T.; Eto, Y.; Saigusa, D.; Mano, N. Global Metabolomics Analysis of Serum from Patients with Niemann-Pick Disease Type C. Med. Mass Spectrom. 2020, 4, 76–86. [Google Scholar] [CrossRef]
- Saigusa, D.; Okamura, Y.; Motoike, I.N.; Katoh, Y.; Kurosawa, Y.; Saijyo, R.; Koshiba, S.; Yasuda, J.; Motohashi, H.; Sugawara, J.; et al. Establishment of Protocols for Global Metabolomics by LC-MS for Biomarker Discovery. PLoS ONE 2016, 11, e0160555. [Google Scholar] [CrossRef]
- Aoyagi, R.; Ikeda, K.; Isobe, Y.; Arita, M. Comprehensive Analyses of Oxidized Phospholipids Using a Measured MS/MS Spectra Library. J. Lipid Res. 2017, 58, 2229–2237. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-Targeted and Targeted Metabolomics Approaches to Diagnosing Lung Cancer and Predicting Patient Prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, B.; Chen, Q.; Su, Y.; Wang, R.; Liu, Z.; Chen, G. Non-Targeted Metabolomic Analysis of the Variations in the Metabolites of Two Genotypes of Glycyrrhiza Uralensis Fisch. under Drought Stress. Ind. Crops Prod. 2022, 176, 114402. [Google Scholar] [CrossRef]
- Gertsman, I.; Gangoiti, J.A.; Barshop, B.A. Validation of a Dual LC-HRMS Platform for Clinical Metabolic Diagnosis in Serum, Bridging Quantitative Analysis and Untargeted Metabolomics. Metabolomics 2014, 10, 312. [Google Scholar] [CrossRef]
- Andrews, G.L.; Simons, B.L.; Young, J.B.; Hawkridge, A.M.; Muddiman, D.C. Performance Characteristics of a New Hybrid Quadrupole Time-of-Flight Tandem Mass Spectrometer (TripleTOF 5600). Anal. Chem. 2011, 83, 5442–5446. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Maekawa, M.; Kanamori, M.; Yamauchi, M.; Abe, A.; Shimoda, Y.; Saito, R.; Endo, H.; Mano, N. Investigation of Cystine as Differential Diagnostic Biomarker between Astrocytomas and Oligodendrogliomas Based on Global- and Targeted Analysis Using Liquid Chromatography/Tandem Mass Spectrometric Analysis. Adv. Biomark. Sci. Technol. 2023, 5, 76–85. [Google Scholar] [CrossRef]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A Lipidome Atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Redestig, H.; Fukushima, A.; Stenlund, H.; Moritz, T.; Arita, M.; Saito, K.; Kusano, M. Compensation for Systematic Cross-Contribution Improves Normalization of Mass Spectrometry Based Metabolomics Data. Anal. Chem. 2009, 81, 7974–7980. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of Bioactive Metabolites Using Activity Metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-Mcintyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for Large-Scale Metabolic Profiling of Serum and Plasma Using Gas Chromatography and Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Wehrens, R.; Hageman, J.A.; van Eeuwijk, F.; Kooke, R.; Flood, P.J.; Wijnker, E.; Keurentjes, J.J.B.; Lommen, A.; van Eekelen, H.D.L.M.; Hall, R.D.; et al. Improved Batch Correction in Untargeted MS-Based Metabolomics. Metabolomics 2016, 12, 88. [Google Scholar] [CrossRef]
- Kamleh, M.A.; Ebbels, T.M.D.; Spagou, K.; Masson, P.; Want, E.J. Optimizing the Use of Quality Control Samples for Signal Drift Correction in Large-Scale Urine Metabolic Profiling Studies. Anal. Chem. 2012, 84, 2670–2677. [Google Scholar] [CrossRef]
- Sato, T.; Kawasaki, Y.; Maekawa, M.; Takasaki, S.; Saigusa, D.; Ota, H.; Shimada, S.; Yamashita, S.; Mitsuzuka, K.; Yamaguchi, H.; et al. Value of Global Metabolomics in Association with Diagnosis and Clinicopathological Factors of Renal Cell Carcinoma. Int. J. Cancer 2019, 145, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards More Transparent and Integrative Metabolomics Analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Zagare, A.; Preciat, G.; Nickels, S.L.; Luo, X.; Monzel, A.S.; Gomez-Giro, G.; Robertson, G.; Jaeger, C.; Sharif, J.; Koseki, H.; et al. Omics Data Integration Suggests a Potential Idiopathic Parkinson’s Disease Signature. Commun. Biol. 2023, 6, 1179. [Google Scholar] [CrossRef] [PubMed]
- Barzegar Behrooz, A.; Latifi-Navid, H.; da Silva Rosa, S.C.; Swiat, M.; Wiechec, E.; Vitorino, C.; Vitorino, R.; Jamalpoor, Z.; Ghavami, S. Integrating Multi-Omics Analysis for Enhanced Diagnosis and Treatment of Glioblastoma: A Comprehensive Data-Driven Approach. Cancers 2023, 15, 3158. [Google Scholar] [CrossRef]
- Jinks, M.; Davies, E.C.; Boughton, B.A.; Lodge, S.; Maker, G.L. 1H NMR Spectroscopic Characterisation of HepG2 Cells as a Model Metabolic System for Toxicology Studies. Toxicol. In Vitro 2024, 99, 105881. [Google Scholar] [CrossRef]
- Patin, F.; Corcia, P.; Vourc’h, P.; Nadal-Desbarats, L.; Baranek, T.; Goossens, J.F.; Marouillat, S.; Dessein, A.F.; Descat, A.; Madji Hounoum, B.; et al. Omics to Explore Amyotrophic Lateral Sclerosis Evolution: The Central Role of Arginine and Proline Metabolism. Mol. Neurobiol. 2017, 54, 5361–5374. [Google Scholar] [CrossRef]
- Ding, X.; Qin, J.; Huang, F.; Feng, F.; Luo, L. The Combination of Machine Learning and Untargeted Metabolomics Identifies the Lipid Metabolism -Related Gene CH25H as a Potential Biomarker in Asthma. Inflamm. Res. 2023, 72, 1099–1119. [Google Scholar] [CrossRef]
- Xu, T.; Wu, Z.; Yuan, Q.; Zhang, X.; Liu, Y.; Wu, C.; Song, M.; Wu, J.; Jiang, J.; Wang, Z.; et al. Proline Is Increased in Allergic Asthma and Promotes Airway Remodeling. JCI Insight 2023, 8, e167395. [Google Scholar] [CrossRef]
- Berquez, M.; Chen, Z.; Festa, B.P.; Krohn, P.; Keller, S.A.; Parolo, S.; Korzinkin, M.; Gaponova, A.; Laczko, E.; Domenici, E.; et al. Lysosomal Cystine Export Regulates MTORC1 Signaling to Guide Kidney Epithelial Cell Fate Specialization. Nat. Commun. 2023, 14, 3994. [Google Scholar] [CrossRef]
- Liu, B.; Du, H.; Rutkowski, R.; Gartner, A.; Wang, X. LAAT-1 Is the Lysosomal Lysine/Arginine Transporter That Maintains Amino Acid Homeostasis. Science 2012, 337, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Bräuer, A.U.; Kuhla, A.; Holzmann, C.; Wree, A.; Witt, M. Current Challenges in Understanding the Cellular and Molecular Mechanisms in Niemann–Pick Disease Type C1. Int. J. Mol. Sci. 2019, 20, 4392. [Google Scholar] [CrossRef]
- Häberle, J.; Burlina, A.; Chakrapani, A.; Dixon, M.; Karall, D.; Lindner, M.; Mandel, H.; Martinelli, D.; Pintos-Morell, G.; Santer, R.; et al. Suggested Guidelines for the Diagnosis and Management of Urea Cycle Disorders: First Revision. J. Inherit. Metab. Dis. 2019, 42, 1192–1230. [Google Scholar] [CrossRef] [PubMed]
- Kazak, L.; Cohen, P. Creatine Metabolism: Energy Homeostasis, Immunity and Cancer Biology. Nat. Rev. Endocrinol. 2020, 16, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Greenhill, C. Role for Creatine Metabolism in Energy Expenditure. Nat. Rev. Endocrinol. 2017, 13, 624. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Maekawa, M.; Sato, T.; Sato, Y.; Kumondai, M.; Takahashi, H.; Kikuchi, M.; Higaki, K.; Ogura, J.; Mano, N. Metabolic Alteration Analysis of Steroid Hormones in Niemann–Pick Disease Type C Model Cell Using Liquid Chromatography/Tandem Mass Spectrometry. Int. J. Mol. Sci. 2022, 23, 4459. [Google Scholar] [CrossRef]
- Chiang, F.-F.; Chao, T.-H.; Huang, S.-C.; Cheng, C.-H.; Tseng, Y.-Y.; Huang, Y.-C. Cysteine Regulates Oxidative Stress and Glutathione-Related Antioxidative Capacity before and after Colorectal Tumor Resection. Int. J. Mol. Sci. 2022, 23, 9581. [Google Scholar] [CrossRef]
- Paul, B.D.; Sbodio, J.I.; Snyder, S.H. Cysteine Metabolism in Neuronal Redox Homeostasis. Trends Pharmacol. Sci. 2018, 39, 513–524. [Google Scholar] [CrossRef]
- Zampieri, S.; Mellon, S.H.; Butters, T.D.; Nevyjel, M.; Covey, D.F.; Bembi, B.; Dardis, A. Oxidative Stress in NPC1 Deficient Cells: Protective Effect of Allopregnanolone. J. Cell. Mol. Med. 2009, 13, 3786–3796. [Google Scholar] [CrossRef]
- Dunlop, R.A.; Powell, J.; Guillemin, G.J.; Cox, P.A. Mechanisms of L-Serine Neuroprotection in Vitro Include ER Proteostasis Regulation. Neurotox. Res. 2018, 33, 123–132. [Google Scholar] [CrossRef]
- Dunlop, R.A.; Carney, J.M. Mechanisms of L-Serine-Mediated Neuroprotection Include Selective Activation of Lysosomal Cathepsins B and L. Neurotox. Res. 2021, 39, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.S.; Dunlop, R.A.; Powell, J.T.; Banack, S.A.; Cox, P.A. L-Serine: A Naturally-Occurring Amino Acid with Therapeutic Potential. Neurotox. Res. 2018, 33, 213–221. [Google Scholar] [CrossRef]
- Riedel, W.J.; Klaassen, T.; Schmitt, J.A.J. Tryptophan, Mood, and Cognitive Function. Brain Behav. Immun. 2002, 16, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Ikushiro, H. Study on Serine Palmitoyltransferase, the Rate Limiting Enzyme of Sphingolipid Biosynthesis. Vitamins 2008, 82, 101–114. [Google Scholar] [CrossRef]
- Hojjati, M.R.; Li, Z.; Jiang, X.C. Serine Palmitoyl-CoA Transferase (SPT) Deficiency and Sphingolipid Levels in Mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2005, 1737, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Breiden, B.; Sandhoff, K. Acid Sphingomyelinase, a Lysosomal and Secretory Phospholipase C, Is Key for Cellular Phospholipid Catabolism. Int. J. Mol. Sci. 2021, 22, 9001. [Google Scholar] [CrossRef]
- Vanier, M.T.; Rodriguez-Lafrasse, C.; Rousson, R.; Gazzah, N.; Juge, M.C.; Pentchev, P.G.; Revol, A.; Louisot, P. Type C Niemann-Pick Disease: Spectrum of Phenotypic Variation in Disruption of Intracellular LDL-Derived Cholesterol Processing. Biochim. Biophys. Acta 1991, 1096, 328–337. [Google Scholar] [CrossRef]
- Vanier, M.T. Biochemical Studies in Niemann-Pick Disease. I. Major Sphingolipids of Liver and Spleen. Biochim. Biophys. Acta 1983, 750, 178–184. [Google Scholar] [CrossRef]
- Deodato, F.; Boenzi, S.; Taurisano, R.; Semeraro, M.; Sacchetti, E.; Carrozzo, R.; Dionisi-Vici, C. The Impact of Biomarkers Analysis in the Diagnosis of Niemann-Pick C Disease and Acid Sphingomyelinase Deficiency. Clin. Chim. Acta 2018, 486, 387–394. [Google Scholar] [CrossRef]
- Paul, B.D. Cysteine Metabolism and Hydrogen Sulfide Signaling in Huntington’s Disease. Free Radic. Biol. Med. 2022, 186, 93–98. [Google Scholar] [CrossRef]
- Soria, F.N.; Zabala, A.; Pampliega, O.; Palomino, A.; Miguelez, C.; Ugedo, L.; Sato, H.; Matute, C.; Domercq, M. Cystine/Glutamate Antiporter Blockage Induces Myelin Degeneration. Glia 2016, 64, 1381–1395. [Google Scholar] [CrossRef] [PubMed]
- Stepien, K.M.; Roncaroli, F.; Turton, N.; Hendriksz, C.J.; Roberts, M.; Heaton, R.A.; Hargreaves, I. Mechanisms of Mitochondrial Dysfunction in Lysosomal Storage Disorders: A Review. J. Clin. Med. 2020, 9, 2596. [Google Scholar] [CrossRef] [PubMed]
- Torres, S.; García-Ruiz, C.M.; Fernandez-Checa, J.C. Mitochondrial Cholesterol in Alzheimer’s Disease and Niemann–Pick Type C Disease. Front. Neurol. 2019, 10, 1168. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.; Sillence, D.J. Niemann–Pick Type C Disease: Cellular Pathology and Pharmacotherapy. J. Neurochem. 2020, 153, 674–692. [Google Scholar] [CrossRef]
- Torres, S.; Matías, N.; Baulies, A.; Nuñez, S.; Alarcon-Vila, C.; Martinez, L.; Nuño, N.; Fernandez, A.; Caballeria, J.; Levade, T.; et al. Mitochondrial GSH Replenishment as a Potential Therapeutic Approach for Niemann Pick Type C Disease. Redox Biol. 2017, 11, 60–72. [Google Scholar] [CrossRef]
- Goicoechea, L.; Conde de la Rosa, L.; Torres, S.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial Cholesterol: Metabolism and Impact on Redox Biology and Disease. Redox Biol. 2023, 61, 102643. [Google Scholar] [CrossRef]
- Maekawa, M.; Miyoshi, K.; Narita, A.; Sato, T.; Sato, Y.; Kumondai, M.; Kikuchi, M.; Higaki, K.; Okuyama, T.; Eto, Y.; et al. Development of a Highly Sensitive and Rapid Liquid Chromatography–Tandem Mass Spectrometric Method Using a Basic Mobile Phase Additive to Determine the Characteristics of the Urinary Metabolites for Niemann–Pick Disease Type C. Biol. Pharm. Bull. 2022, 45, 1259–1268. [Google Scholar] [CrossRef]
- Jiang, X.; Sidhu, R.; Porter, F.D.; Yanjanin, N.M.; Speak, A.O.; te Vruchte, D.T.; Platt, F.M.; Fujiwara, H.; Scherrer, D.E.; Zhang, J.; et al. A Sensitive and Specific LC-MS/MS Method for Rapid Diagnosis of Niemann-Pick C1 Disease from Human Plasma. J. Lipid Res. 2011, 52, 1435–1445. [Google Scholar] [CrossRef]
- Jiang, X.; Sidhu, R.; Orsini, J.J.; Farhat, N.Y.; Porter, F.D.; Berry-Kravis, E.; Schaffer, J.E.; Ory, D.S. Diagnosis of Niemann-Pick C1 by Measurement of Bile Acid Biomarkers in Archived Newborn Dried Blood Spots. Mol. Genet. Metab. 2019, 126, 183–187. [Google Scholar] [CrossRef]

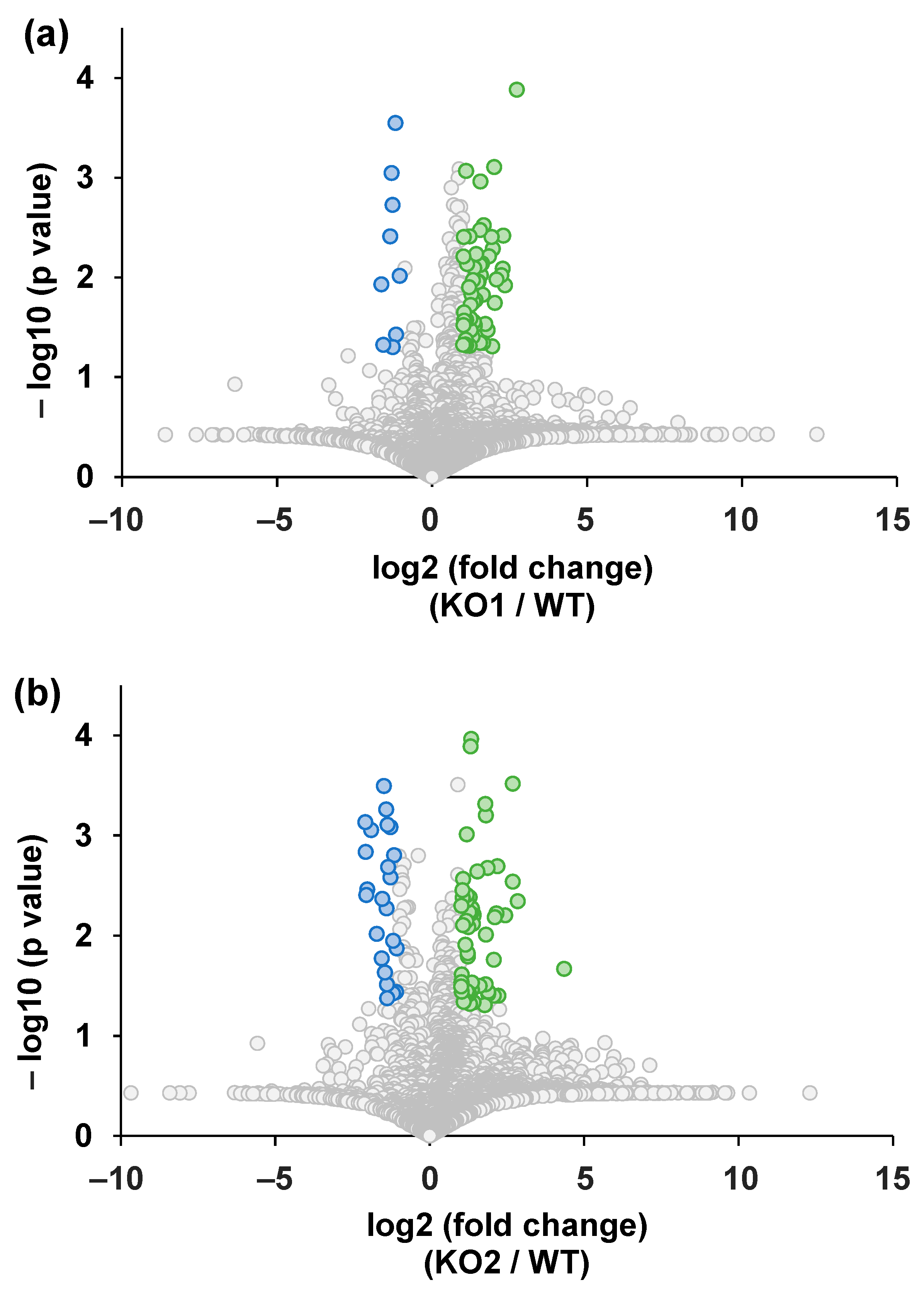
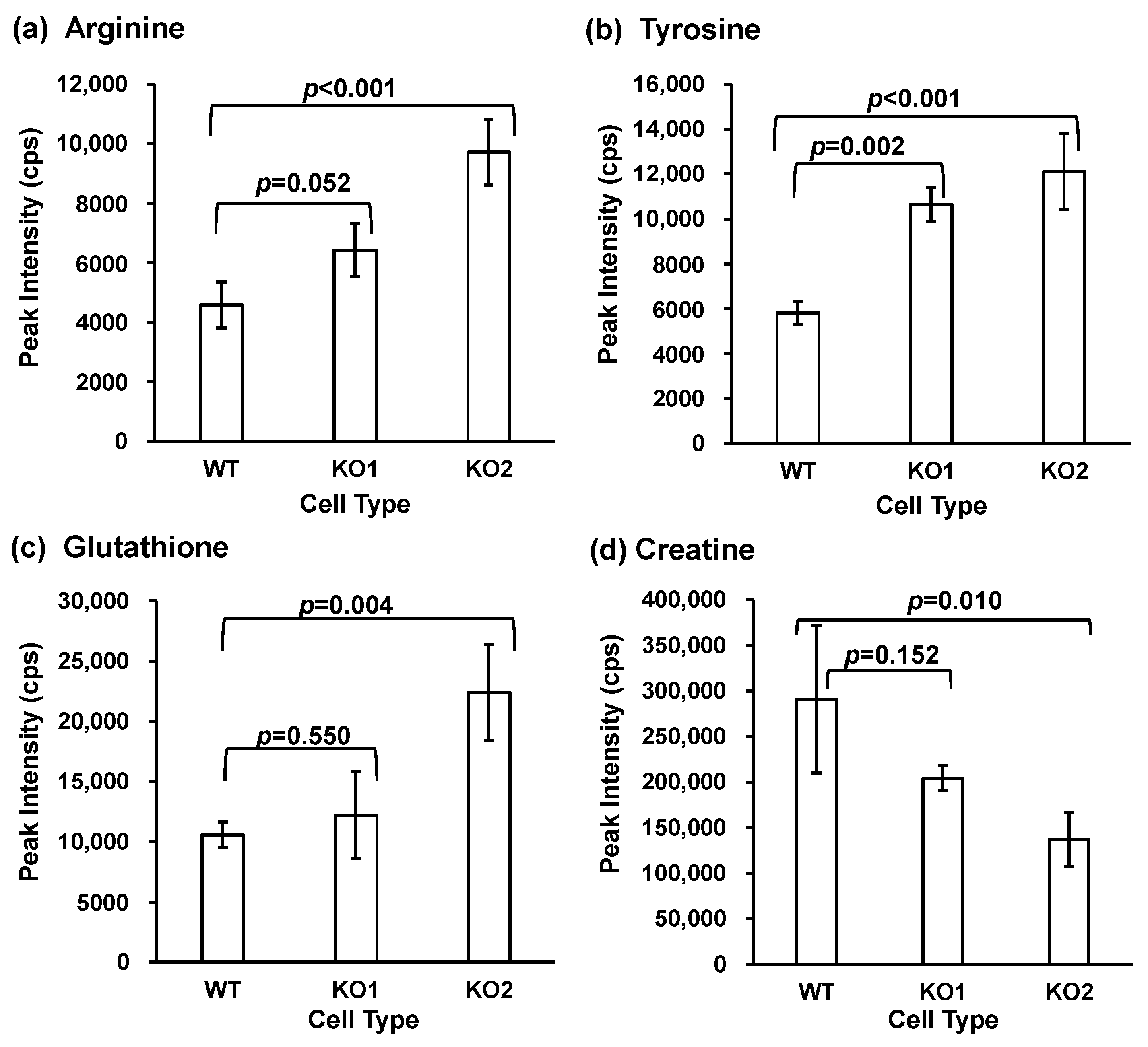
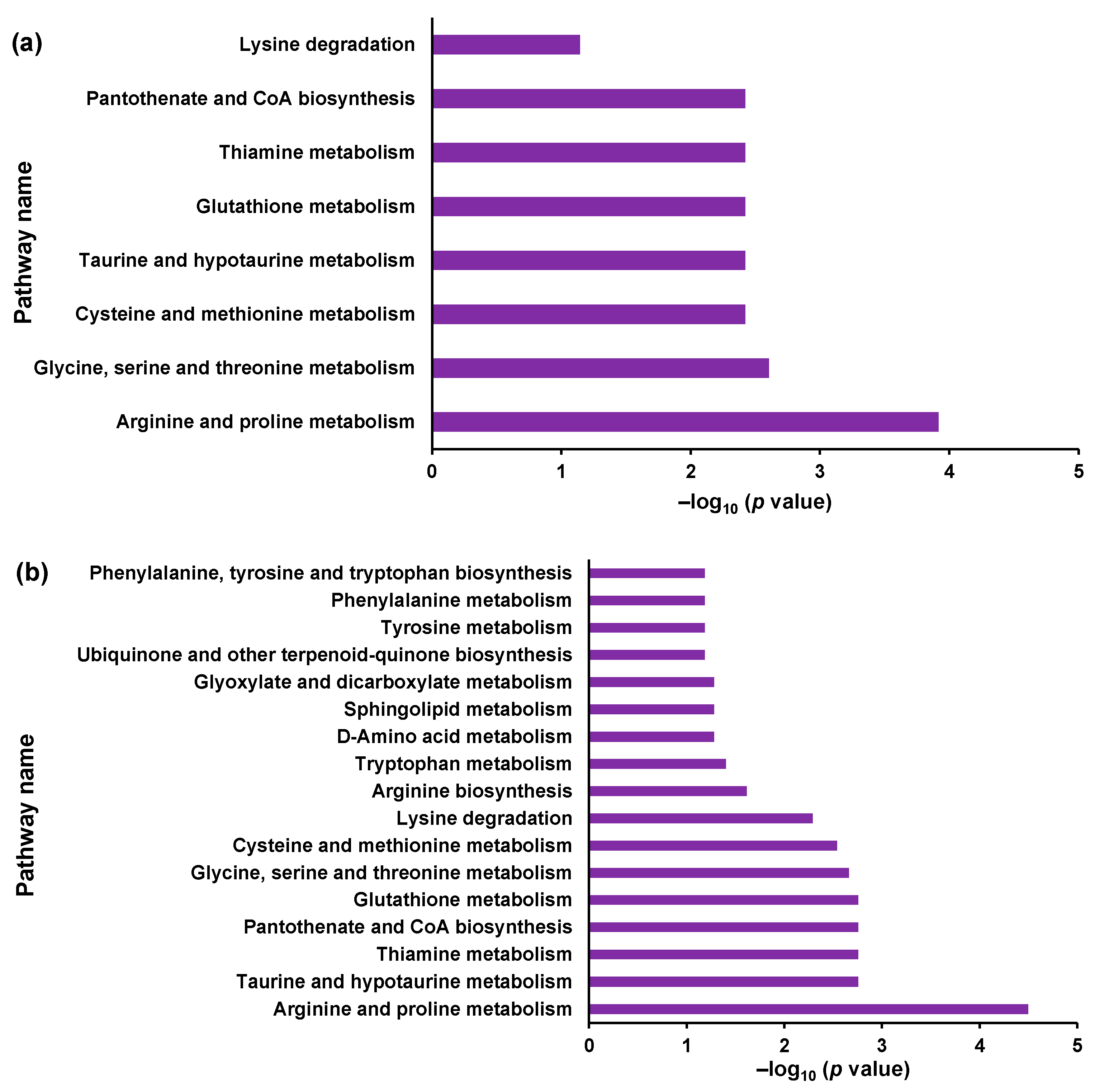
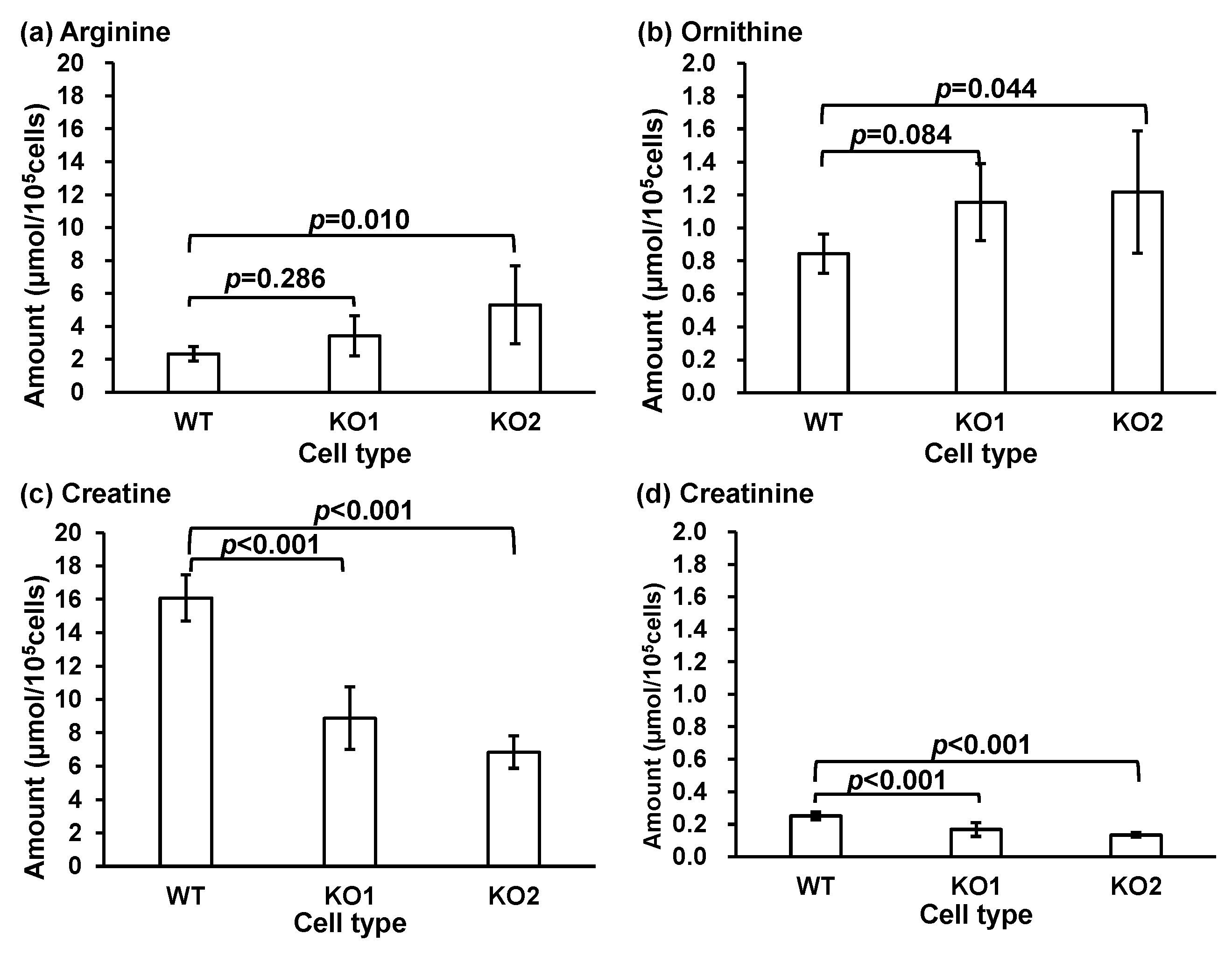
| Analyte | Equation | Correlation Coefficient (R2) | Range (ng/mL) | IS |
|---|---|---|---|---|
| Arginine | y = 0.75221x + 0.00341 | 0.9987 | 50–5000 | Arginine-[13C6,15N4] |
| Carnitine | y = 1.18022x + 0.04072 | 0.9945 | 75–2500 | Arginine-[13C6,15N4] |
| Creatine | y = 0.38279x − 0.00191 | 0.9993 | 50–7500 | Creatine-[2H3] |
| Creatinine | y = 0.68207x + 0.00105 | 0.9962 | 5–250 | Arginine-[13C6,15N4] |
| Cysteine | y = 0.13442x − 0.16923 | 0.9959 | 2500–25,000 | Arginine-[13C6,15N4] |
| Cystine | y = 0.08118x − 0.00016050 | 0.9947 | 10–2500 | Creatine-[2H3] |
| Glutamic acid | y = 0.18196x − 0.42880 | 0.9940 | 5000–50,000 | Creatine-[2H3] |
| Glutamine | y = 0.29085x − 0.03538 | 0.9975 | 500–12,500 | Creatine-[2H3] |
| Glutathione | y = 0.22382x − 0.36368 | 0.9869 | 2500–20,000 | Arginine-[13C6,15N4] |
| Glycocyamine | y = 0.07604x − 0.00212 | 0.9989 | 100–75,000 | Creatine-[2H3] |
| Methionine | y = 0.10443x − 0.00765 | 0.9937 | 100–15,000 | Creatine-[2H3] |
| Ornithine | y = 0.38240x + 0.00333 | 0.9943 | 10–1000 | Arginine-[13C6,15N4] |
| Proline | y = 0.86494x − 0.00627 | 0.9970 | 250–10,000 | Creatine-[2H3] |
| Serine | y = 0.06414x − 0.05882 | 0.9963 | 2500–25,000 | Arginine-[13C6,15N4] |
| Tryptophan | y = 0.36811x − 0.00050155 | 0.9994 | 10–1000 | Creatine-[2H3] |
| Tyrosine | y = 0.58450x − 0.16496 | 0.9972 | 750–20,000 | Creatine-[2H3] |
| Compound | WT Cells (µmol/105 Cells) | KO1 Cells (µmol/105 Cells) | KO2 Cells (µmol/105 Cells) |
|---|---|---|---|
| Arginine | 2.33 ± 0.43 | 3.43 ± 1.22 | 5.32 ± 2.37 * |
| Carnitine | 1.29 ± 0.19 | 0.95 ± 0.31 * | 0.95 ± 0.04 * |
| Creatine | 16.1 ± 1.38 | 8.88 ± 1.87 *** | 6.84 ± 0.97 *** |
| Creatinine | 0.25 ± 0.02 | 0.17 ± 0.04 *** | 0.13 ± 0.01 *** |
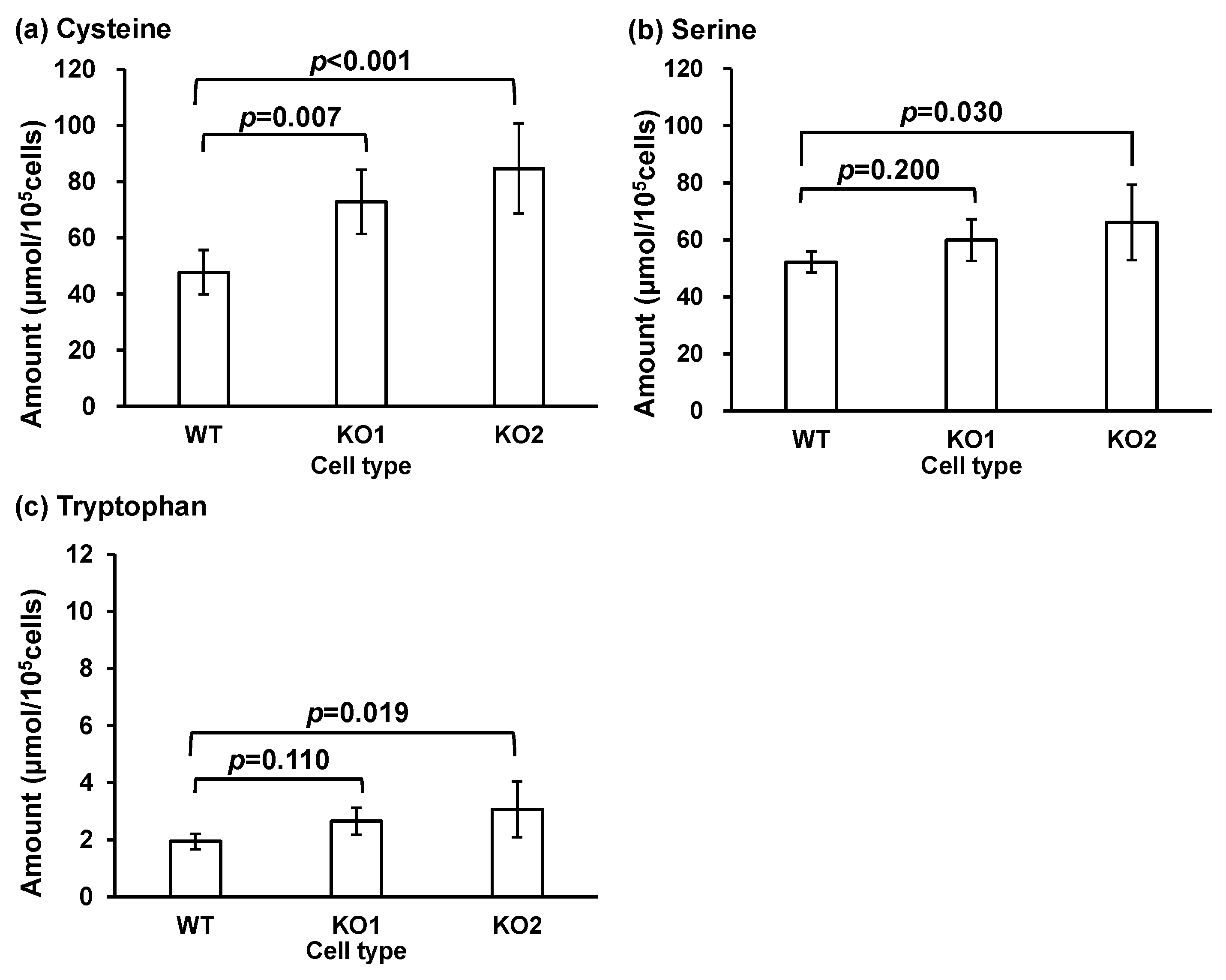
| Cysteine | 47.7 ± 7.89 | 72.8 ± 11.5 ** | 84.7 ± 16.1 *** |
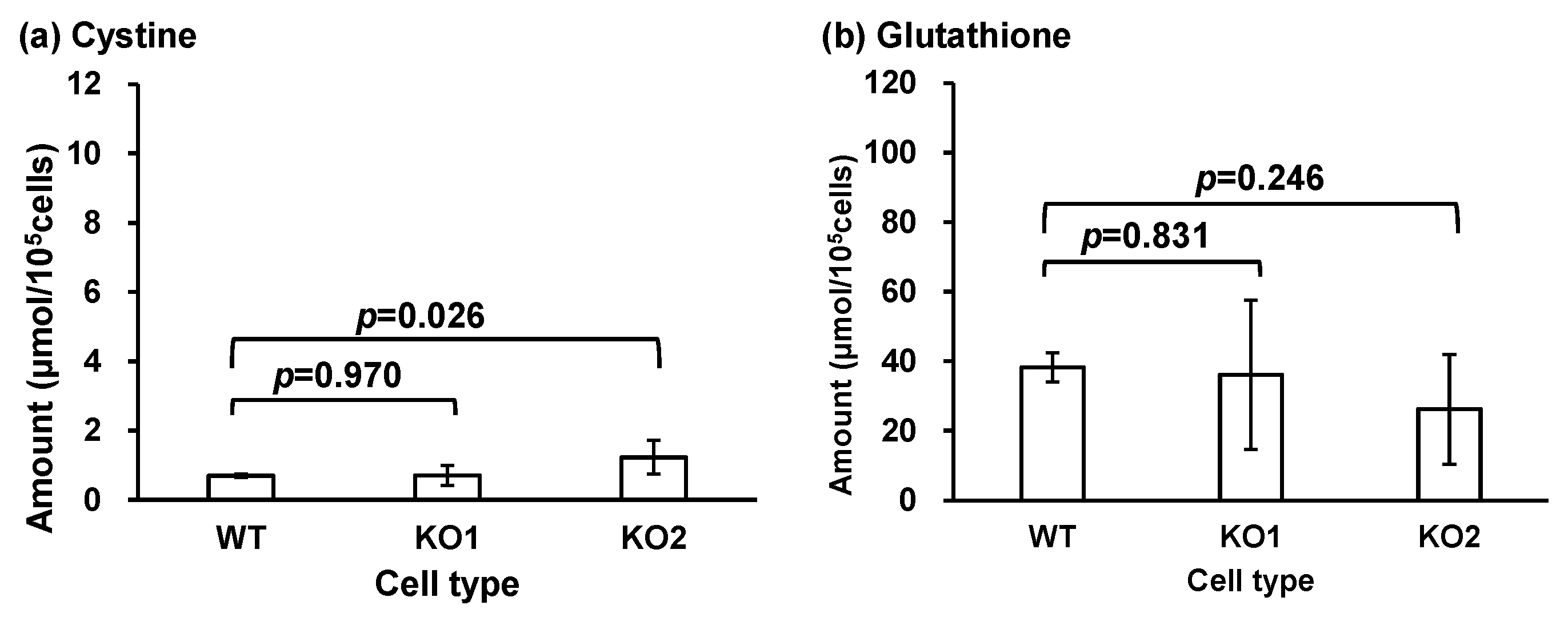
| Cystine | 0.70 ± 0.05 | 0.71 ± 0.29 | 1.23 ± 0.49 * |
| Glutamic acid | 99.4 ± 13.2 | 89.5 ± 17.7 | 95.1 ± 5.51 |
| Glutamine | 7.28 ± 1.01 | 8.25 ± 1.92 | 10.5 ± 3.76 |
| Glutathione | 38.2 ± 4.15 | 36.1 ± 21.4 | 26.2 ± 15.8 |
| Glycocyamine | N.D. | N.D. | N.D. |
| Methionine | 6.63 ± 0.94 | 7.56 ± 1.72 | 8.79 ± 2.95 |
| Ornithine | 0.84 ± 0.12 | 1.16 ± 0.23 | 1.22 ± 0.37 * |
| Proline | 43.3 ± 4.55 | 36.7 ± 5.55 | 38.7 ± 5.72 |
| Serine | 52.2 ± 3.71 | 59.9 ± 7.35 | 66.1 ± 13.2 * |
| Tryptophan | 1.94 ± 0.27 | 2.65 ± 0.48 | 3.06 ± 0.98 * |
| Tyrosine | 8.34 ± 1.09 | 10.3 ± 1.92 | 11.8 ± 3.45 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, M.; Maekawa, M.; Miyoshi, K.; Sato, T.; Sato, Y.; Kumondai, M.; Fukasawa, M.; Mano, N. Global and Targeted Metabolomics for Revealing Metabolomic Alteration in Niemann-Pick Disease Type C Model Cells. Metabolites 2024, 14, 515. https://doi.org/10.3390/metabo14100515
Watanabe M, Maekawa M, Miyoshi K, Sato T, Sato Y, Kumondai M, Fukasawa M, Mano N. Global and Targeted Metabolomics for Revealing Metabolomic Alteration in Niemann-Pick Disease Type C Model Cells. Metabolites. 2024; 14(10):515. https://doi.org/10.3390/metabo14100515
Chicago/Turabian StyleWatanabe, Masahiro, Masamitsu Maekawa, Keitaro Miyoshi, Toshihiro Sato, Yu Sato, Masaki Kumondai, Masayoshi Fukasawa, and Nariyasu Mano. 2024. "Global and Targeted Metabolomics for Revealing Metabolomic Alteration in Niemann-Pick Disease Type C Model Cells" Metabolites 14, no. 10: 515. https://doi.org/10.3390/metabo14100515
APA StyleWatanabe, M., Maekawa, M., Miyoshi, K., Sato, T., Sato, Y., Kumondai, M., Fukasawa, M., & Mano, N. (2024). Global and Targeted Metabolomics for Revealing Metabolomic Alteration in Niemann-Pick Disease Type C Model Cells. Metabolites, 14(10), 515. https://doi.org/10.3390/metabo14100515






