Gut Microbiota–Gut Metabolites and Clostridioides difficile Infection: Approaching Sustainable Solutions for Therapy
Abstract
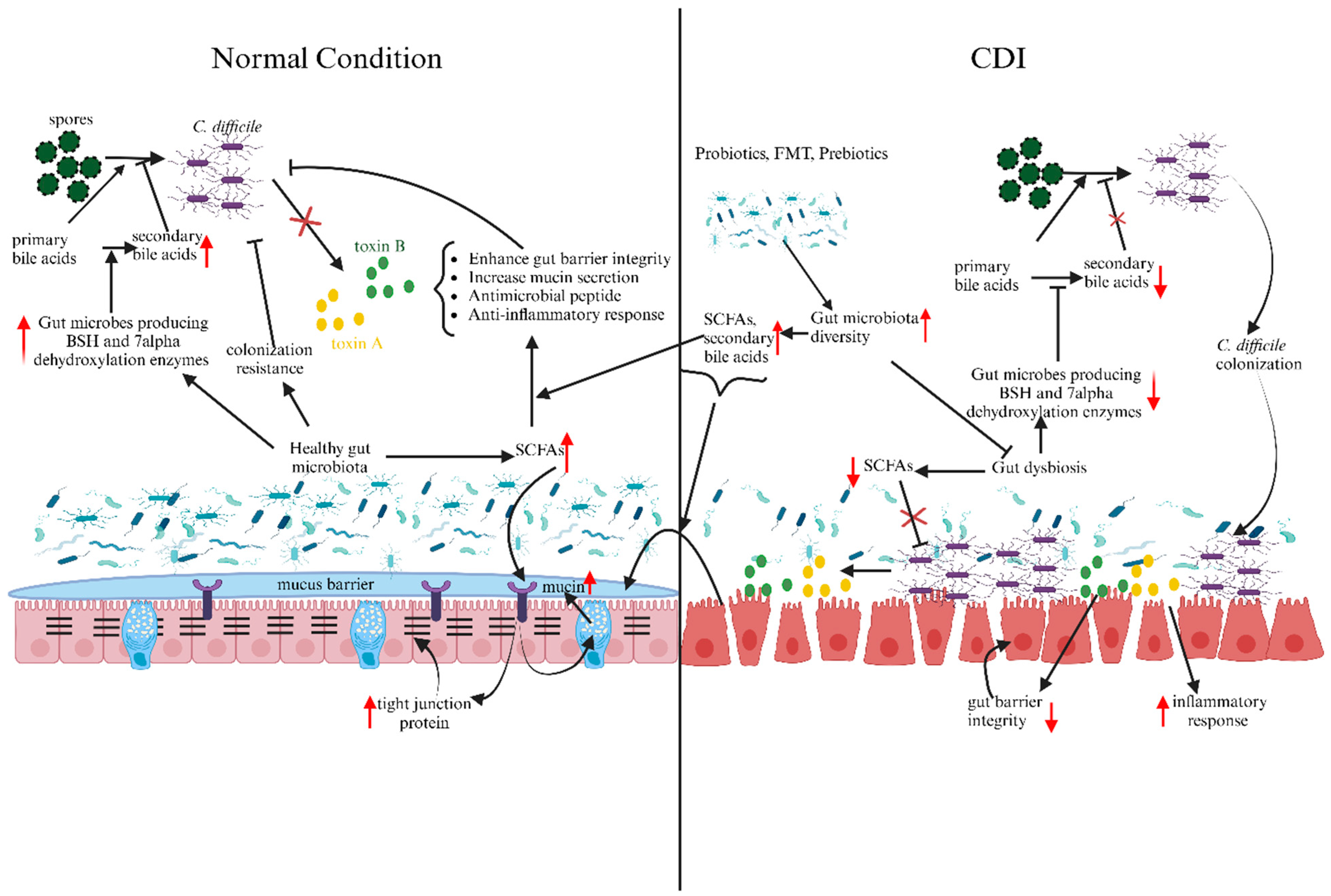
1. Introduction
2. Gut Microbiota and CDI
3. Gut Metabolites and CDI
3.1. Bile Acids
3.2. Short-Chain Fatty Acids (SCFAs)
4. Gut Microbiota Modulators
4.1. Antibiotics
4.2. Proton Pump Inhibitors (PPIs)
4.3. Probiotics
4.4. Prebiotics
5. Therapeutical Strategies against CDI
5.1. Antibiotic Therapy
5.2. Fecal Microbiota Transplantation (FMT)
5.3. Phage Therapy
5.4. Probiotics as a Potential Therapy against CDI
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.; Palazuelos-Munoz, S.; Balsells, E.M.; Nair, H.; Chit, A.; Kyaw, M.H. Cost of Hospital Management of Clostridium difficile Infection in United States—A Meta-Analysis and Modelling Study. BMC Infect. Dis. 2016, 16, 447. [Google Scholar] [CrossRef] [PubMed]
- Paschos, P.; Ioakim, K.; Malandris, K.; Koukoufiki, A.; Nayfeh, T.; Akriviadis, E.; Tsapas, A.; Bekiari, E. Add-on Interventions for the Prevention of Recurrent Clostridioides difficile Infection: A Systematic Review and Network Meta-Analysis. Anaerobe 2021, 71, 102441. [Google Scholar] [CrossRef]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.L.; Oliver, K.B. Antibiotic Resistance Threats in the United States: Stepping Back from the Brink. AFP 2014, 89, 938–941. [Google Scholar]
- Eze, P.; Balsells, E.; Kyaw, M.H.; Nair, H. Risk Factors for Clostridium difficile Infections—An Overview of the Evidence Base and Challenges in Data Synthesis. J. Glob. Health 2017, 7, 010417. [Google Scholar] [CrossRef] [PubMed]
- Lamendella, R.; Wright, J.R.; Hackman, J.; McLimans, C.; Toole, D.R.; Bernard Rubio, W.; Drucker, R.; Wong, H.T.; Sabey, K.; Hegarty, J.P.; et al. Antibiotic Treatments for Clostridium difficile Infection Are Associated with Distinct Bacterial and Fungal Community Structures. mSphere 2018, 3, e00572-17. [Google Scholar] [CrossRef]
- Imhann, F.; Bonder, M.J.; Vila, A.V.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.M.; et al. Proton Pump Inhibitors Affect the Gut Microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef]
- Teng, C.; Reveles, K.R.; Obodozie-Ofoegbu, O.O.; Frei, C.R. Clostridium difficile Infection Risk with Important Antibiotic Classes: An Analysis of the FDA Adverse Event Reporting System. Int. J. Med. Sci. 2019, 16, 630–635. [Google Scholar] [CrossRef]
- Heiss, C.N.; Olofsson, L.E. Gut Microbiota-Dependent Modulation of Energy Metabolism. J. Innate. Immun. 2018, 10, 163–171. [Google Scholar] [CrossRef]
- Jose, S.; Mukherjee, A.; Horrigan, O.; Setchell, K.D.R.; Zhang, W.; Moreno-Fernandez, M.E.; Andersen, H.; Sharma, D.; Haslam, D.B.; Divanovic, S.; et al. Obeticholic Acid Ameliorates Severity of Clostridioides difficile Infection in High Fat Diet-Induced Obese Mice. Mucosal. Immunol. 2021, 14, 500–510. [Google Scholar] [CrossRef]
- Fischer, M.; Sipe, B.; Cheng, Y.-W.; Phelps, E.; Rogers, N.; Sagi, S.; Bohm, M.; Xu, H.; Kassam, Z. Fecal Microbiota Transplant in Severe and Severe-Complicated Clostridium difficile: A Promising Treatment Approach. Gut Microbes 2017, 8, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Alrabaa, S.; Jariwala, R.; Zeitler, K.; Montero, J. Fecal Microbiota Transplantation Outcomes in Immunocompetent and Immunocompromised Patients: A Single-Center Experience. Transpl. Infect. Dis. 2017, 19, e12726. [Google Scholar] [CrossRef] [PubMed]
- Nzabarushimana, E.; Tang, H. Functional Profile of Host Microbiome Indicates Clostridioides difficile Infection. Gut Microbes 2022, 14, 2135963. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.T.; Vernon, J.J.; Loo, V.G.; Kong, L.Y.; Péchiné, S.; Wilcox, M.H.; Kuijper, E.J. Understanding Clostridium difficile Colonization. Clin. Microbiol. Rev. 2018, 31, e00021-17. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.M.; Sorg, J.A. Gut Associated Metabolites and Their Roles in Clostridioides difficile Pathogenesis. Gut Microbes 2022, 14, 2094672. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Jenior, M.L.; Keenan, O.; Hart, J.L.; Specker, J.; Abbas, A.; Rangel, P.C.; Di, C.; Green, J.; Bustin, K.A.; et al. Enterococci Enhance Clostridioides difficile Pathogenesis. Nature 2022, 611, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Fischer, M. Expert Opinion on Fecal Microbiota Transplantation for the Treatment of Clostridioides difficile Infection and Beyond. Expert Opin. Biol. Ther. 2020, 20, 73–81. [Google Scholar] [CrossRef]
- Gaisawat, M.B.; MacPherson, C.W.; Tremblay, J.; Piano, A.; Iskandar, M.M.; Tompkins, T.A.; Kubow, S. Probiotic Supplementation in a Clostridium difficile-Infected Gastrointestinal Model Is Associated with Restoring Metabolic Function of Microbiota. Microorganisms 2020, 8, 60. [Google Scholar] [CrossRef]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-Induced Inflammatory Bowel Disease Mice Model. EBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Bruno, G.; Zaccari, P.; Rocco, G.; Scalese, G.; Panetta, C.; Porowska, B.; Pontone, S.; Severi, C. Proton Pump Inhibitors and Dysbiosis: Current Knowledge and Aspects to Be Clarified. World J. Gastroenterol. 2019, 25, 2706–2719. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.; Abe, F.; Osawa, R. Age-Related Changes in Gut Microbiota Composition from Newborn to Centenarian: A Cross-Sectional Study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Thanissery, R.; Winston, J.A.; Theriot, C.M. Inhibition of Spore Germination, Growth, and Toxin Activity of Clinically Relevant C. difficile Strains by Gut Microbiota Derived Secondary Bile Acids. Anaerobe 2017, 45, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere 2016, 1, e00045-15. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, K.; Khanna, S. Gut Microbiome and Clostridioides difficile Infection: A Closer Look at the Microscopic Interface. Therap. Adv. Gastroenterol. 2021, 14, 1–9. [Google Scholar] [CrossRef]
- Bien, J.; Palagani, V.; Bozko, P. The Intestinal Microbiota Dysbiosis and Clostridium difficile Infection: Is There a Relationship with Inflammatory Bowel Disease? Therap. Adv. Gastroenterol. 2013, 6, 53–68. [Google Scholar] [CrossRef]
- Ke, S.; Pollock, N.R.; Wang, X.-W.; Chen, X.; Daugherty, K.; Lin, Q.; Xu, H.; Garey, K.W.; Gonzales-Luna, A.J.; Kelly, C.P.; et al. Integrating Gut Microbiome and Host Immune Markers to Understand the Pathogenesis of Clostridioides difficile Infection. Gut Microbes 2021, 13, 1935186. [Google Scholar] [CrossRef]
- Vázquez-Cuesta, S.; Villar, L.; García, N.L.; Fernández, A.I.; Olmedo, M.; Alcalá, L.; Marín, M.; Muñoz, P.; Bouza, E.; Reigadas, E. Characterization of the Gut Microbiome of Patients with Clostridioides difficile Infection, Patients with Non–C. difficile Diarrhea, and C. difficile–Colonized Patients. Front. Cell. Infect. Microbiol. 2023, 13, 1130701. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The Gut Bacterial Community Potentiates Clostridioides difficile Infection Severity. mBio 2022, 13, e01183-22. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Ge, X.; Xu, H.; Tan, B.; Tian, B.; Shi, Y.; Dai, Y.; Li, Y.; Hu, S.; Qian, J. Gut Microbiome and Mycobiome in Inflammatory Bowel Disease Patients with Clostridioides difficile Infection. Front. Cell. Infect. Microbiol. 2023, 13, 1129043. [Google Scholar] [CrossRef] [PubMed]
- Schäffler, H.; Breitrück, A. Clostridium difficile—From Colonization to Infection. Front. Microbiol. 2018, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.M.; Sinani, H.; Schloss, P.D. Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. mBio 2015, 6, e00974. [Google Scholar] [CrossRef]
- Finn, E.; Andersson, F.L.; Madin-Warburton, M. Burden of Clostridioides difficile Infection (CDI)—A Systematic Review of the Epidemiology of Primary and Recurrent CDI. BMC Infect. Dis. 2021, 21, 456. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Bajinka, O.; Jarju, P.O.; Tan, Y.; Taal, A.M.; Ozdemir, G. The Varying Effects of Antibiotics on Gut Microbiota. AMB Express 2021, 11, 116. [Google Scholar] [CrossRef]
- Leong, C.; Zelenitsky, S. Treatment Strategies for Recurrent Clostridium difficile Infection. Can. J. Hosp. Pharm. 2013, 66, 361–368. [Google Scholar] [CrossRef]
- Bagdasarian, N.; Rao, K.; Malani, P.N. Diagnosis and Treatment of Clostridium difficile in Adults: A Systematic Review. JAMA 2015, 313, 398–408. [Google Scholar] [CrossRef]
- Gerding, D.N.; File, T.M.; McDonald, L.C. Diagnosis and Treatment of Clostridium difficile Infection (CDI). Infect. Dis. Clin. Pract. 2016, 24, 3–10. [Google Scholar] [CrossRef][Green Version]
- Vázquez-Cuesta, S.; Lozano García, N.; Fernández, A.I.; Olmedo, M.; Kestler, M.; Alcalá, L.; Marín, M.; Bermejo, J.; Díaz, F.F.-A.; Muñoz, P.; et al. Microbiome Profile and Calprotectin Levels as Markers of Risk of Recurrent Clostridioides difficile Infection. Front. Cell. Infect. Microbiol. 2023, 13, 1237500. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Montassier, E.; Schmidt, B.; Patel, R.; Knights, D.; Pardi, D.S.; Kashyap, P.C. Gut Microbiome Predictors of Treatment Response and Recurrence in Primary Clostridium difficile Infection. Aliment. Pharmacol. Ther. 2016, 44, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, L.; Ke, S.; Kelly, C.P.; Pollock, N.R.; Villafuerte Gálvez, J.A.; Daugherty, K.; Xu, H.; Yao, J.; Chen, Y.; et al. Analysis of Intestinal Mycobiota of Patients with Clostridioides difficile Infection among a Prospective Inpatient Cohort. Microbiol. Spectr. 2022, 10, e0136222. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.-K.; Yun, B.-S.; Matsuzaki, K.; Furukawa, M.; Min, H.-K.; Bajaj, J.S.; et al. Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics That Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 2019, 26, 27–34.e4. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, T.J.; Ashaolu, J.O.; Adeyeye, S.A.O. Fermentation of Prebiotics by Human Colonic Microbiota in Vitro and Short-Chain Fatty Acids Production: A Critical Review. J. Appl. Microbiol. 2021, 130, 677–687. [Google Scholar] [CrossRef]
- Hayashi, A.; Nagao-Kitamoto, H.; Kitamoto, S.; Kim, C.H.; Kamada, N. The Butyrate-Producing Bacterium Clostridium Butyricum Suppresses Clostridioides difficile Infection via Neutrophil- and Antimicrobial Cytokine–Dependent but GPR43/109a-Independent Mechanisms. J. Immunol. 2021, 206, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.-M.; Flemer, B.; Joyce, S.A.; Zulquernain, A.; Sheehan, D.; Shanahan, F.; O’Toole, P.W. Changes in Microbiota Composition, Bile and Fatty Acid Metabolism, in Successful Faecal Microbiota Transplantation for Clostridioides difficile Infection. BMC Gastroenterol. 2018, 18, 131. [Google Scholar] [CrossRef]
- Ouyang, Z.; Niu, X.; Wang, W.; Zhao, J. The Role of Short-Chain Fatty Acids in Clostridioides difficile Infection: A Review. Anaerobe 2022, 75, 102585. [Google Scholar] [CrossRef]
- Carter, G.P.; Rood, J.I.; Lyras, D. The Role of Toxin A and Toxin B in Clostridium difficile-Associated Disease. Gut Microbes 2010, 1, 58–64. [Google Scholar] [CrossRef]
- Rotondo-Trivette, S.; Wang, B.; Gayer, C.; Parsana, R.; Luan, Y.; Sun, F.; Michail, S. Decreased Secondary Faecal Bile Acids in Children with Ulcerative Colitis and Clostridioides difficile Infection. Aliment. Pharmacol. Ther. 2021, 54, 792–804. [Google Scholar] [CrossRef]
- Winston, J.A.; Theriot, C.M. Diversification of Host Bile Acids by Members of the Gut Microbiota. Gut Microbes 2019, 11, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Łukawska, A.; Mulak, A. Impact of Primary and Secondary Bile Acids on Clostridioides difficile Infection. Pol. J. Microbiol. 2022, 71, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; Allegretti, J.R. The Contribution of Bile Acid Metabolism to the Pathogenesis of Clostridioides difficile Infection. Therap. Adv. Gastroenterol. 2021, 14, 17562848211017725. [Google Scholar] [CrossRef]
- Sorg, J.A.; Sonenshein, A.L. Bile Salts and Glycine as Cogerminants for Clostridium difficile Spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef]
- Francis, M.B.; Allen, C.A.; Shrestha, R.; Sorg, J.A. Bile Acid Recognition by the Clostridium difficile Germinant Receptor, CspC, Is Important for Establishing Infection. PLOS Pathog. 2013, 9, e1003356. [Google Scholar] [CrossRef] [PubMed]
- Wexler, A.G.; Guiberson, E.R.; Beavers, W.N.; Shupe, J.A.; Washington, M.K.; Lacy, D.B.; Caprioli, R.M.; Spraggins, J.M.; Skaar, E.P. Clostridioides difficile Infection Induces a Rapid Influx of Bile Acids into the Gut during Colonization of the Host. Cell Rep. 2021, 36, 109683. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.A.K.; Mullish, B.H.; Pechlivanis, A.; Liu, Z.; Brignardello, J.; Kao, D.; Holmes, E.; Li, J.V.; Clarke, T.B.; Thursz, M.R.; et al. Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota. Gastroenterology 2018, 155, 1495–1507.e15. [Google Scholar] [CrossRef]
- Wang, S.; Xiang, L.; Li, F.; Deng, W.; Lv, P.; Chen, Y. Butyrate Protects against Clostridium difficile Infection by Regulating Bile Acid Metabolism. Microbiol. Spectr. 2023, 11, e04479-22. [Google Scholar] [CrossRef]
- Winston, J.A.; Rivera, A.J.; Cai, J.; Thanissery, R.; Montgomery, S.A.; Patterson, A.D.; Theriot, C.M. Ursodeoxycholic Acid (UDCA) Mitigates the Host Inflammatory Response during Clostridioides difficile Infection by Altering Gut Bile Acids. Infect. Immun. 2020, 88, e00045-20. [Google Scholar] [CrossRef]
- Theriot, C.M.; Petri, W.A. Cell Preview: Role of Microbiota-Derived Bile Acids in Enteric Infections. Cell 2020, 181, 1452–1454. [Google Scholar] [CrossRef]
- Tam, J.; Hamza, T.; Ma, B.; Chen, K.; Beilhartz, G.L.; Ravel, J.; Feng, H.; Melnyk, R.A. Host-Targeted Niclosamide Inhibits C. difficile Virulence and Prevents Disease in Mice without Disrupting the Gut Microbiota. Nat Commun 2018, 9, 5233. [Google Scholar] [CrossRef] [PubMed]
- Bourgin, M.; Kriaa, A.; Mkaouar, H.; Mariaule, V.; Jablaoui, A.; Maguin, E.; Rhimi, M. Bile Salt Hydrolases: At the Crossroads of Microbiota and Human Health. Microorganisms 2021, 9, 1122. [Google Scholar] [CrossRef] [PubMed]
- Stofan, M.; Guo, G.L. Bile Acids and FXR: Novel Targets for Liver Diseases. Front. Med. 2020, 7, 544. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.F. The Enterohepatic Circulation of Bile Acids in Mammals: Form and Functions. Front. Biosci. (Landmark Ed.) 2009, 14, 2584–2598. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial Transformations of Human Bile Acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Reed, A.D.; Nethery, M.A.; Stewart, A.; Barrangou, R.; Theriot, C.M. Strain-Dependent Inhibition of Clostridioides difficile by Commensal Clostridia Carrying the Bile Acid-Inducible (Bai) Operon. J. Bacteriol. 2020, 202, e00039-20. [Google Scholar] [CrossRef]
- Tam, J.; Icho, S.; Utama, E.; Orrell, K.E.; Gómez-Biagi, R.F.; Theriot, C.M.; Kroh, H.K.; Rutherford, S.A.; Lacy, D.B.; Melnyk, R.A. Intestinal Bile Acids Directly Modulate the Structure and Function of C. difficile TcdB Toxin. Proc. Natl. Acad. Sci. USA 2020, 117, 6792–6800. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Kalavagunta, P.K.; Liao, J.; Jin, L.; Shang, J.; et al. Taxonomic Profiling and Populational Patterns of Bacterial Bile Salt Hydrolase (BSH) Genes Based on Worldwide Human Gut Microbiome. Microbiome 2019, 7, 9. [Google Scholar] [CrossRef]
- Vital, M.; Rud, T.; Rath, S.; Pieper, D.H.; Schlüter, D. Diversity of Bacteria Exhibiting Bile Acid-Inducible 7α-Dehydroxylation Genes in the Human Gut. Comput. Struct. Biotechnol. J. 2019, 17, 1016–1019. [Google Scholar] [CrossRef]
- Hazleton, K.Z.; Martin, C.G.; Orlicky, D.J.; Arnolds, K.L.; Nusbacher, N.M.; Moreno-Huizar, N.; Armstrong, M.; Reisdorph, N.; Lozupone, C.A. Dietary Fat Promotes Antibiotic-Induced Clostridioides difficile Mortality in Mice. npj Biofilms Microbiomes 2022, 8, 1–13. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Valdes, A.M.; Menni, C. The Role of Short-Chain Fatty Acids in the Interplay between Gut Microbiota and Diet in Cardio-Metabolic Health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef] [PubMed]
- Tsukuda, N.; Yahagi, K.; Hara, T.; Watanabe, Y.; Matsumoto, H.; Mori, H.; Higashi, K.; Tsuji, H.; Matsumoto, S.; Kurokawa, K.; et al. Key Bacterial Taxa and Metabolic Pathways Affecting Gut Short-Chain Fatty Acid Profiles in Early Life. ISME J. 2021, 15, 2574–2590. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating Levels of the Short-Chain Fatty Acid Acetate Mediate the Effect of the Gut Microbiome on Visceral Fat. Front. Microbiol. 2021, 12, 711359. [Google Scholar] [CrossRef]
- Gao, Y.; Ma, L.; Su, J. Host and Microbial-Derived Metabolites for Clostridioides difficile Infection: Contributions, Mechanisms and Potential Applications. Microbiol. Res. 2022, 263, 127113. [Google Scholar] [CrossRef]
- Pensinger, D.A.; Fisher, A.T.; Dobrila, H.A.; Van Treuren, W.; Gardner, J.O.; Higginbottom, S.K.; Carter, M.M.; Schumann, B.; Bertozzi, C.R.; Anikst, V.; et al. Butyrate Differentiates Permissiveness to Clostridioides difficile Infection and Influences Growth of Diverse C. difficile Isolates. Infect. Immun. 2023, 91, e00570-22. [Google Scholar] [CrossRef]
- Fachi, J.L.; de Souza Felipe, J.; Pral, L.P.; da Silva, B.K.; Corrêa, R.O.; de Andrade, M.C.P.; da Fonseca, D.M.; Basso, P.J.; Câmara, N.O.S.; de Sales e Souza, É.L.; et al. Butyrate Protects Mice from Clostridium difficile-Induced Colitis through an HIF-1-Dependent Mechanism. Cell Rep. 2019, 27, 750–761.e7. [Google Scholar] [CrossRef]
- Fachi, J.L.; Sécca, C.; Rodrigues, P.B.; de Mato, F.C.P.; Di Luccia, B.; de Souza Felipe, J.; Pral, L.P.; Rungue, M.; de Melo Rocha, V.; Sato, F.T.; et al. Acetate Coordinates Neutrophil and ILC3 Responses against C. difficile through FFAR2. J. Exp. Med. 2019, 217, e20190489. [Google Scholar] [CrossRef]
- Nam, H.J.; Kang, J.K.; Kim, S.-K.; Ahn, K.J.; Seok, H.; Park, S.J.; Chang, J.S.; Pothoulakis, C.; Lamont, J.T.; Kim, H. Clostridium difficile Toxin A Decreases Acetylation of Tubulin, Leading to Microtubule Depolymerization through Activation of Histone Deacetylase 6, and This Mediates Acute Inflammation*. J. Biol. Chem. 2010, 285, 32888–32896. [Google Scholar] [CrossRef]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound Alterations of Intestinal Microbiota Following a Single Dose of Clindamycin Results in Sustained Susceptibility to Clostridium difficile-Induced Colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, N.A.; Schubert, A.M.; Sinani, H.; Schloss, P.D. Clearance of Clostridioides difficile Colonization Is Associated with Antibiotic-Specific Bacterial Changes. mSphere 2021, 6, e01238-20. [Google Scholar] [CrossRef] [PubMed]
- D’Silva, K.M.; Mehta, R.; Mitchell, M.; Lee, T.C.; Singhal, V.; Wilson, M.G.; McDonald, E.G. Proton Pump Inhibitor Use and Risk for Recurrent Clostridioides difficile Infection: A Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2021, 27, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Tawam, D.; Baladi, M.; Jungsuwadee, P.; Earl, G.; Han, J. The Positive Association between Proton Pump Inhibitors and Clostridium difficile Infection. Innov. Pharm. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Mössner, J. The Indications, Applications, and Risks of Proton Pump Inhibitors: A Review after 25 Years. Dtsch. Ärzteblatt Int. 2016, 113, 477. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H. Gut Microbiota Modulation: A Tool for the Management of Colorectal Cancer. J. Transl. Med. 2022, 20, 178. [Google Scholar] [CrossRef]
- Li, H.-Y.; Zhou, D.-D.; Gan, R.-Y.; Huang, S.-Y.; Zhao, C.-N.; Shang, A.; Xu, X.-Y.; Li, H.-B. Effects and Mechanisms of Probiotics, Prebiotics, Synbiotics, and Postbiotics on Metabolic Diseases Targeting Gut Microbiota: A Narrative Review. Nutrients 2021, 13, 3211. [Google Scholar] [CrossRef]
- Fong, W.; Li, Q.; Yu, J. Gut Microbiota Modulation: A Novel Strategy for Prevention and Treatment of Colorectal Cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Kaewarsar, E.; Chaiyasut, C.; Lailerd, N.; Makhamrueang, N.; Peerajan, S.; Sirilun, S. Effects of Synbiotic Lacticaseibacillus Paracasei, Bifidobacterium Breve, and Prebiotics on the Growth Stimulation of Beneficial Gut Microbiota. Foods 2023, 12, 3847. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Wu, C. Modulation of Gut Microbiota and Immune System by Probiotics, Pre-Biotics, and Post-Biotics. Front. Nutr. 2022, 8, 634897. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, Q.; Gu, S.; Chen, Y.; Lv, L.; Zheng, B.; Wang, Q.; Wang, K.; Wang, S.; Xia, J.; et al. Akkermansia Muciniphila Ameliorates Clostridioides difficile Infection in Mice by Modulating the Intestinal Microbiome and Metabolites. Front. Microbiol. 2022, 13, 841920. [Google Scholar] [CrossRef] [PubMed]
- Moens, F.; Van den Abbeele, P.; Basit, A.W.; Dodoo, C.; Chatterjee, R.; Smith, B.; Gaisford, S. A Four-Strain Probiotic Exerts Positive Immunomodulatory Effects by Enhancing Colonic Butyrate Production in Vitro. Int. J. Pharm. 2019, 555, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Thananimit, S.; Pahumunto, N.; Teanpaisan, R. Characterization of Short Chain Fatty Acids Produced by Selected Potential Probiotic Lactobacillus Strains. Biomolecules 2022, 12, 1829. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Wolfe, T.J.D.; Eggers, S.; Barker, A.K.; Kates, A.E.; Dill-McFarland, K.A.; Suen, G.; Safdar, N. Oral Probiotic Combination of Lactobacillus and Bifidobacterium Alters the Gastrointestinal Microbiota during Antibiotic Treatment for Clostridium difficile Infection. PLoS ONE 2018, 13, e0204253. [Google Scholar] [CrossRef] [PubMed]
- Éliás, A.J.; Barna, V.; Patoni, C.; Demeter, D.; Veres, D.S.; Bunduc, S.; Erőss, B.; Hegyi, P.; Földvári-Nagy, L.; Lenti, K. Probiotic Supplementation during Antibiotic Treatment Is Unjustified in Maintaining the Gut Microbiome Diversity: A Systematic Review and Meta-Analysis. BMC Med. 2023, 21, 262. [Google Scholar] [CrossRef]
- Allen, S.J.; Wareham, K.; Wang, D.; Bradley, C.; Hutchings, H.; Harris, W.; Dhar, A.; Brown, H.; Foden, A.; Gravenor, M.B.; et al. Lactobacilli and Bifidobacteria in the Prevention of Antibiotic-Associated Diarrhoea and Clostridium difficile Diarrhoea in Older Inpatients (PLACIDE): A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 2013, 382, 1249–1257. [Google Scholar] [CrossRef]
- Heil, E.L.; Harris, A.D.; Brown, C.; Seung, H.; Thom, K.A.; von Rosenvinge, E.; Sorongon, S.; Pineles, L.; Goodman, K.E.; Leekha, S. A Multicenter Evaluation of Probiotic Use for the Primary Prevention of Clostridioides difficile Infection. Clin. Infect. Dis. 2021, 73, 1330–1337. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Al-Sheraji, S.H.; Ismail, A.; Manap, M.Y.; Mustafa, S.; Yusof, R.M.; Hassan, F.A. Prebiotics as Functional Foods: A Review. J. Funct. Foods 2013, 5, 1542–1553. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, J.; Lopez, J.M.; Mills, D.A. Inulin Fermentation by Lactobacilli and Bifidobacteria from Dairy Calves. Appl. Environ. Microbiol. 2020, 87, e01738-20. [Google Scholar] [CrossRef] [PubMed]
- Rubin, I.M.C.; Mollerup, S.; Broholm, C.; Baker, A.; Holm, M.K.A.; Pedersen, M.S.; Pinholt, M.; Westh, H.; Petersen, A.M. Synbiotic Intervention with Lactobacilli, Bifidobacteria, and Inulin in Healthy Volunteers Increases the Abundance of Bifidobacteria but Does Not Alter Microbial Diversity. Appl. Environ. Microbiol. 2022, 88, e01087-22. [Google Scholar] [CrossRef]
- Sivaprakasam, S.; Gurav, A.; Paschall, A.V.; Coe, G.L.; Chaudhary, K.; Cai, Y.; Kolhe, R.; Martin, P.; Browning, D.; Huang, L.; et al. An Essential Role of Ffar2 (Gpr43) in Dietary Fibre-Mediated Promotion of Healthy Composition of Gut Microbiota and Suppression of Intestinal Carcinogenesis. Oncogenesis 2016, 5, e238. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.B.; Costa, C.M.; Bonifácio-Lopes, T.; Silva, S.; Veiga, M.; Monforte, A.R.; Nunes, J.; Vicente, A.A.; Pintado, M. Prebiotic Effects of Olive Pomace Powders in the Gut: In Vitro Evaluation of the Inhibition of Adhesion of Pathogens, Prebiotic and Antioxidant Effects. Food Hydrocoll. 2021, 112, 106312. [Google Scholar] [CrossRef]
- Shen, E.P.; Surawicz, C.M. Current Treatment Options for Severe Clostridium difficile–Associated Disease. Gastroenterol. Hepatol. 2008, 4, 134–139. [Google Scholar]
- Kim, J.; Kim, J.; Kim, B.; Pai, H. Which Is the Preferred Regimen for Non-Severe Clostridioides difficile Infection in Korea, Vancomycin or Metronidazole? Infect. Chemother. 2022, 54, 213–219. [Google Scholar] [CrossRef]
- Skinner, A.M.; Scardina, T.; Kociolek, L.K. Fidaxomicin for the Treatment of Clostridioides difficile in Children. Future Microbiol. 2020, 15, 967–979. [Google Scholar] [CrossRef]
- Credito, K.L.; Appelbaum, P.C. Activity of OPT-80, a Novel Macrocycle, Compared with Those of Eight Other Agents against Selected Anaerobic Species. Antimicrob. Agents Chemother. 2004, 48, 4430–4434. [Google Scholar] [CrossRef][Green Version]
- Cornely, O.A.; Crook, D.W.; Esposito, R.; Poirier, A.; Somero, M.S.; Weiss, K.; Sears, P.; Gorbach, S.; OPT-80-004 Clinical Study Group. Fidaxomicin versus Vancomycin for Infection with Clostridium difficile in Europe, Canada, and the USA: A Double-Blind, Non-Inferiority, Randomised Controlled Trial. Lancet Infect. Dis. 2012, 12, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Miller, M.A.; Louie, T.J.; Crook, D.W.; Gorbach, S.L. Treatment of First Recurrence of Clostridium difficile Infection: Fidaxomicin Versus Vancomycin. Clin. Infect. Dis. 2012, 55, S154–S161. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Umscheid, C.A.; Fishman, N.; Lee, B.Y. Is Fidaxomicin Worth the Cost? An Economic Analysis. Clin. Infect. Dis. 2013, 57, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.; Hedenstierna, M.; Ursing, J.; Lidman, C.; Nowak, P. Efficacy of Routine Fecal Microbiota Transplantation for Treatment of Recurrent Clostridium difficile Infection: A Retrospective Cohort Study. Int. J. Microbiol. 2019, 2019, 7395127. [Google Scholar] [CrossRef]
- Goldenberg, S.D.; Merrick, B. The Role of Faecal Microbiota Transplantation: Looking beyond Clostridioides difficile Infection. Ther. Adv. Infect. 2021, 8, 1–16. [Google Scholar] [CrossRef]
- El-Salhy, M.; Valeur, J.; Hausken, T.; Gunnar Hatlebakk, J. Changes in Fecal Short-Chain Fatty Acids Following Fecal Microbiota Transplantation in Patients with Irritable Bowel Syndrome. Neurogastroenterol. Motil. 2021, 33, e13983. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.; Allen, L.; Masirah M Zain, N.; Forbes, B.; Shawcross, D.L.; Goldenberg, S.D. Regulation, Risk and Safety of Faecal Microbiota Transplant. Infect. Prev. Pract. 2020, 2, 100069. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Bloom, P.P.; Torres Soto, M.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.-B.; Hohmann, E.L. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef]
- Wilcox, M.H.; Gerding, D.N.; Poxton, I.R.; Kelly, C.; Nathan, R.; Birch, T.; Cornely, O.A.; Rahav, G.; Bouza, E.; Lee, C.; et al. Bezlotoxumab for Prevention of Recurrent Clostridium difficile Infection. N. Engl. J. Med. 2017, 376, 305–317. [Google Scholar] [CrossRef]
- Venhorst, J.; van der Vossen, J.M.B.M.; Agamennone, V. Battling Enteropathogenic Clostridia: Phage Therapy for Clostridioides difficile and Clostridium Perfringens. Front. Microbiol. 2022, 13, 891790. [Google Scholar] [CrossRef]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepiński, P.; Douce, G.R.; Clokie, M.R.J. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Hudson, S.L.; Arnoczy, G.; Gibson, H.; Thurber, C.; Lee, J.; Kessell, A. Probiotic Use as Prophylaxis for Clostridium difficile-Associated Diarrhea in a Community Hospital. Am. J. Infect. Control 2019, 47, 1028–1029. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.P.; Rao, K.; Young, V.B. Probiotics for Prevention of Clostridium difficile Infection. Curr. Opin. Gastroenterol. 2018, 34, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.T.; Maw, A.; Tmanova, L.L.; Pino, A.; Ancy, K.; Crawford, C.V.; Simon, M.S.; Evans, A.T. Timely Use of Probiotics in Hospitalized Adults Prevents Clostridium difficile Infection: A Systematic Review With Meta-Regression Analysis. Gastroenterology 2017, 152, 1889–1900.e9. [Google Scholar] [CrossRef]
| Study Group | Result | References |
|---|---|---|
| CDI patients, recurrent CDI patients, non-C. difficile diarrhea patients, asymptomatic C. difficile patients, and control | Decrease in alpha diversity, with several genera like Parabacteroides, Faecalicoccus, and Clostridium cluster XVIII as potential biomarkers for colonization. For CDI, potential biomarkers included bacteria genera Batceroides, Proteus, Paraprevotella, and Eggerthella. For the recurrent CDI, Veillonella, Enterococcus, Lactobacillus, Clostridium cluster XIVa, etc., were potential biomarkers. | [31] |
| CDI patients, Asymptomatic carriers, non-CDI diarrhea, and Control | Lower diversity of the gut microbiome. Increased variation of immune markers between the study group. Several bacterial groups were identified as a potential influencer of CDI, like Klebsiella, Streptococcus, and Veillonella. | [30] |
| Mice were separated into groups based on human fecal samples used to inoculate the mice | Mice showing lower clinical scores and comparatively healthy had a higher proportion of Akkermansia, Anaerotignum, Blautia, Enterocloster, etc. Mice showing higher clinical scores had a prevalence of bacterial community from Bacteroides, Enterococcus, and Klebsiella. | [32] |
| Patients having both inflammatory bowel disease (IBD) and CDI, patients with just IBD, healthy control | Variation in the gut microbial (bacterial and fungal) diversity was significantly different between study groups. Bacterial species like Enterococcus faecium, Clostridium inoculum, Ruminococcus gnavus, and fungus Saccharomyces cerevisiae were found in high proportion in IBD-CDI patients. | [33] |
| Patients with primary CDI are further divided into two groups based on recurrent CDI | Calprotectin level combined with the gut microbiome composition provided better insight into the severity of the CDI. In patients with recurrent CDI, calprotectin level was higher, and it was accompanied by an increased proportion of Fusobacterium and a decreased proportion of Ruminococcus, Prevotella, and Collinsella | [41] |
| Gut Metabolite | Result | References |
|---|---|---|
| Bile acids | C. difficile was involved in the increased flux of primary bile acids in the gut, enhancing spore germination and pathogenesis. | [56] |
| Secondary bile acids | In children who have ulcerative colitis and CDI, secondary bile acids like lithocholic acids in the fecal sample were significantly lower, and there was a significant decrease in the genes encoding for enzymes responsible for bile acid transformations. | [50] |
| Secondary bile acids | Antibiotic-associated gut microbiome disruptions led to decreased secondary bile acid production and increased outgrowth of C. difficile in the intestine. Bacteria from Firmicutes phylum, Lachnospiraceae, and Ruminococcaceae were involved in secondary bile acid production and, ultimately, showed resistance to C. difficile. | [26] |
| Secondary bile acids and antibiotics | Along with the production of the secondary bile acids DCA and LCA, Clostridium cinders and Clostridium sordellii were found to produce tryptophan-based antibiotics, which, in concert with secondary bile acids, had an excellent inhibitory effect against C. difficile. | [44] |
| Butyrate (SCFA) | Sodium butyrate was involved in the anti-inflammatory response via activation of GPR109A, which is involved in anti-inflammatory response, and reduces the intestinal permeability and increased production of tight junctions and Mucin 2. | [19] |
| Valerate (SCFA) | Clindamycin treatment was found to reduce the valerate concentration in the gut. Regarding the role of valerate in C. difficile pathogenesis, both in vitro and in vivo studies showed inhibitory effects against C. difficile. | [57] |
| Butyrate (SCFA) | Butryate levels were found to be reduced in patients with CDI. Regarding its protective activity, several mechanisms involved were bile acid metabolism regulation, intestinal barrier strengthening, and gut microbiota modulation. | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurung, B.; Stricklin, M.; Wang, S. Gut Microbiota–Gut Metabolites and Clostridioides difficile Infection: Approaching Sustainable Solutions for Therapy. Metabolites 2024, 14, 74. https://doi.org/10.3390/metabo14010074
Gurung B, Stricklin M, Wang S. Gut Microbiota–Gut Metabolites and Clostridioides difficile Infection: Approaching Sustainable Solutions for Therapy. Metabolites. 2024; 14(1):74. https://doi.org/10.3390/metabo14010074
Chicago/Turabian StyleGurung, Bijay, Maranda Stricklin, and Shaohua Wang. 2024. "Gut Microbiota–Gut Metabolites and Clostridioides difficile Infection: Approaching Sustainable Solutions for Therapy" Metabolites 14, no. 1: 74. https://doi.org/10.3390/metabo14010074
APA StyleGurung, B., Stricklin, M., & Wang, S. (2024). Gut Microbiota–Gut Metabolites and Clostridioides difficile Infection: Approaching Sustainable Solutions for Therapy. Metabolites, 14(1), 74. https://doi.org/10.3390/metabo14010074






