Abstract
Overnutrition with a high-fat or high-sugar diet is widely considered to be the risk factor for various metabolic, chronic, or malignant diseases that are accompanied by alterations in gut microbiota, metabolites, and downstream pathways. In this study, we investigated supplementation with soybean fermentation broth containing saponin (SFBS, also called SAPOZYME) in male C57BL/6 mice fed a high-fat-fructose diet or normal chaw. In addition to the lessening of weight gain, the influence of SFBS on reducing hyperlipidemia and hyperglycemia associated with a high-fat-fructose diet was estimated using the results of related biological tests. The results of gut microbial profiling indicated that the high-fat-fructose diet mediated increases in opportunistic pathogens. In contrast, SFBS supplementation reprogrammed the high-fat-fructose diet-related microbial community with a relatively high abundance of potential probiotics, including Akkermansia and Lactobacillus genera. The metagenomic functions of differential microbial composition in a mouse model and enrolled participants were assessed using the PICRUSt2 algorithm coupled with the MetaCyc and the KEGG Orthology databases. SFBS supplementation exerted a similar influence on an increase in the level of 4-aminobutanoate (also called GABA) through the L-glutamate degradation pathway in the mouse model and the enrolled healthy population. These results suggest the beneficial influence of SFBS supplementation on metabolic disorders associated with a high-fat-fructose diet, and SFBS may function as a nutritional supplement for people with diverse requirements.
1. Introduction
Dietary intake has been widely considered to be a crucial factor in manipulating the constituents of the gut microbial community that are relevant to the healthy status of the host [1]. Recently, probiotic or prebiotic supplementation in symbiotic fermentation has been increasingly applied to manipulate the gut microenvironment in animal models or clinical practice with high-risk conditions [2]. Lactobacillus plantarum supplementation has been documented to strengthen the mucosal integrity or immune response towards opportunistic pathogens within the human gut with the production of short-chain fatty acids [3]. Indeed, the diverse biological activities of Lactobacillus plantarum could alter the gut ecosystem of individuals. The majority of prebiotics are classified as carbohydrate-based ingredients that are generated through the utilization of probiotics, of which their presence is beneficial to gut health [4]. Inulin, which belongs to the classification known as fructans, is a common prebiotic that can be isolated in the roots of different plants. The benefits of inulin have been demonstrated to control blood sugar with the absorption of calcium and magnesium [5] and to facilitate the propagation of probiotics in the human gut [6]. Nevertheless, the metabolism and production of most carbohydrate-based ingredients with dietary intake are varied and affected by the microbial community in each individual.
In this study, the beneficial influence of commercial SFBS was evaluated using a mouse model and a human trial. In summary, consumption of SFBS diminished the impact of a high-fat-fructose (HFF) diet on increases in fasting blood sugar, total triglycerides, and total cholesterol in experimental mice without renal and liver toxicity. Furthermore, SFBS supplementation reprogrammed the HFF-manipulated gut microenvironment to a healthy status within mice or the human gut.
2. Materials and Methods
2.1. Production of SFBS
SFBS (SAPONZYME) is a concentrated symbiotic fermentation broth provided by Sagittarius Life Science Co., Ltd. In brief, soybean powder was applied as the fermentation substrate with Bifidobacterium, Streptococcus, Lactobacillus genera, and Saccharomyces. The fermented products were incubated at 37 °C for 100 h and sterilized to eliminate microorganisms. The compounds in the metabolic profile of SFBS were identified by using non-targeted liquid chromatography mass spectrometry (LC-MS).
2.2. Ethics Statement
The animal experiment procedures executed in this study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Taipei Medical University (approval no. LAC-2021-0511). All the experiments were conducted on animals in accordance with the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council) to minimize the suffering of experimental animals. The protocol regarding the recruitment of healthy participants and fecal sample collection was reviewed and approved by the Institutional Review Board of Taipei Medical University (approval no. N202103179). Healthy participants were enrolled in the Health Examination Center at Taipei Municipal WanFang Hospital. A questionnaire was used to evaluate the lifestyle of all participants.
2.3. Animal Model
C57BL/6 male mice (five weeks old) were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and housed in the animal facility at the Agricultural Technology Research Institute (Hsinchu, Taiwan) at room temperature (22 ± 1 °C), 60% relative humidity, and a 12 h light-dark cycle (light on 8:00 AM). Before the experiments, the mice were acclimatized for a week to the environment and the diet prior to the experiment. Water and a standard laboratory rodent diet 5001 (Scott’s Distribution, Hudson, NH) or a high-fat-fructose (HFF) diet (Research Diet D12451, 24% energy from fat and 41% energy from fructose; New Brunswick, NJ, USA) were provided ad libitum. The mice were randomly assigned to 1 of the following 4 treatments for 8 weeks (n = 5): (1) standard diet, (2) standard diet supplemented with SFBS 700 mg/kg/day, (3) HFF diet, (4) HFF diet supplemented with SFBS 700 mg/kg/day. Feces and peripheral blood were collected every 4 weeks. All the mice were euthanized by 95% CO asphyxiation, and the related organs or tissues were collected immediately.
2.4. Analysis of Biochemical Variables
The fasting blood samples were collected at the onset of the experiment and every 4 weeks throughout the process. The biochemical variables, including blood sugar, total cholesterol (TC), triglycerides (TG), aspartate aminotransferase (AST), blood urea nitrogen (BUN), and creatinine (CRE), were examined using a dry chemistry analyzer (Catalyst One, IDEXX Laboratories, Westbrook, ME, USA).
2.5. Isolation of Total Genomic DNA in the Fecal Samples
The fecal samples were preserved using Fecal Collection tubes (Zymo Research, Irvine, CA, USA) to diminish the growth of bacteria. Total genomic DNA was extracted by using a stool DNA extraction kit (Geneaid, Taipei, Taiwan) in accordance with the manufacturer’s instructions. The quantity of purified DNA samples was estimated by using a fluorometric assay (GeneCopoeia, Rockville, MD, USA).
2.6. Profiling of Gut Microenvironment
The gut microbial community was classified using an Oxford Nanopore sequencing platform (ONT, Oxford, UK). In brief, 10 ng total gDNA of each sample was applied for barcoding of 16S ribosomal RNA (or 16S rRNA) gene sequences using the SQK-16S024 kit (ONT) in accordance with the manufacturer’s protocol. Two ng of barcoded DNA for each sample was pooled, ligated with the adapter, and sequenced on a MinION MK1C sequencer and flow cells (FLO-MIN106D R9.4.1, MinION instrument, ONT). The mean read number was 100,000 per sample to meet a sufficient reading depth. The raw reads were quality checked and clustered to operational taxonomic units (OTUs) with 22,596 16S rRNA reference sequences curated from the Bacterial 16S Ribosomal RNA RefSeq Targeted Loci Project (Accession No. PRJNA33175, NCBI) with 97% similarity by using the CLC genomics workbench (v23.0.4) composed of the Minimap2 program and the Microbial Genomics Module (v23.0.1, Qiagen, CLC bio, Aarhus, Denmark). An OTU table was applied to construct the phylogenetic tree, using the MUSCLE 2.0 and Maximum Likelihood Phylogeny tools (CLC Genomics Workbench), which was subjected to estimate the alpha diversity metrics and the inter-sample dissimilarity (beta diversity). The differential abundance of identified OTUs among all groups was assessed using the linear discriminant analysis (LDA) effect size (LEfSe) method via the website algorithm (https://huttenhower.sph.harvard.edu/galaxy/root, accessed on 1 August 2023) with the default setting. The inter-sample difference of identified taxa was statistically convincing with a p-value < 0.05 and an LDA score (log10) >3 or <−3. Potential relevance between the gut microbial community and the metabolic pathway was assessed by using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) platform adopted to the KEGG Ortholog database (KO, https://www.genome.jp/kegg/ko.html, accessed on 1 August 2023), the Enzyme Commission (EC), and the MetaCycdatabase (https://metacyc.org/, accessed on 1 August 2023).
2.7. Metabolites of SFBS Are Profiled Using an Untargeted LC-MS/MS Analysis
The extraction of fermented metabolites within SFBS was commissioned to the BIOTOOLS Co., Ltd. (Taipei, Taiwan). In brief, 200 μL SFBS was mixed with 600 μL 100% methanol. The mixture was homogenized with sonication and incubated at −20 °C for 1 h. The mixture was centrifugated at 12,000 rpm for 15 min, and the supernatant was transferred to a sample vial. Then, 10 μL extract of each sample was injected into a vanquish-focused ultra-performance liquid chromatography (UPLC) system coupled with an Orbitrap Elite Mass Spectrometry system (Thermo Fisher Scientific, San Jose, CA, USA). The parameters were set as follows: The 2.1 × 100 mm Acquity BEH 1.7 μm C18 column (Waters) was applied at 40 °C. The binary mobile phase was composed of deionized water containing 0.1% formic acid as solvent A and LC-MS grade acetonitrile with 0.1% formic acid as solvent B. The flow rate was set at 0.25 mL/min with a linear gradient elution for 15 min. The percentage of solvent B was linearly increased from 0% to 100%, kept constant for 3 min, and decreased to 0% in the last 1 min. A blank injection was carried out after the sample injection to prevent carryover effects, and a QC injection was executed every five samples to normalize the peak area. MS full scan was conducted in profile mode at 60,000 resolutions, followed by data-dependent MS2 scans at 15,000 resolutions, and the scan range was set from 70 to 1000 m/z. The scanned data were converted to the mzXML format using the ProteoWizard software, which was subjected to annotation. The converted results were assessed using an in-house XCMS program for peak detection, extraction, alignment, and integration (BIOTOOLS). An in-house MS2 database (BiotreeDB, BIOTOOLS) was used for the annotation of scanned fragments.
2.8. Statistical Analysis
The statistical significance of the analytic results was shown in the mean ± standard error (SEM). Continuous variables of the analytic results were estimated by using a one-way analysis of variance (ANOVA) combined with Tukey’s multiple comparison post hoc test. The difference was identified as statistically significant with a p-value less than 0.05. * p < 0.05, ** p < 0.01, and *** p < 0.005.
3. Results
3.1. The Metabolite Profile of SFBS Is Assessed Using the LC-MS/MS Approach
Prior to the in vivo or in vitro functional analysis, the compound profile of SFBS was classified by using the UPLC-MS/MS approach. Under the positive-charge mode, the relatively high abundances of 12 metabolites, such as soyasaponin and dopamine, were identified in SFBS as compared to those of fundamental broth (Table 1). The relatively high abundances of 13 metabolites, including ornithine or epigallocatechin, were classified in SFBS under the negative-charge mode (Table 2). The metabolite profile of SFBS suggests its potential benefit to health.

Table 1.
Statistics of identified metabolites in positive-charge mode.

Table 2.
Statistics of identified metabolites in negative-charge mode.
3.2. SFBS Supplementation Diminishes the Impact of a High-Fat-Fructose Diet on Biochemical Variables in Serum
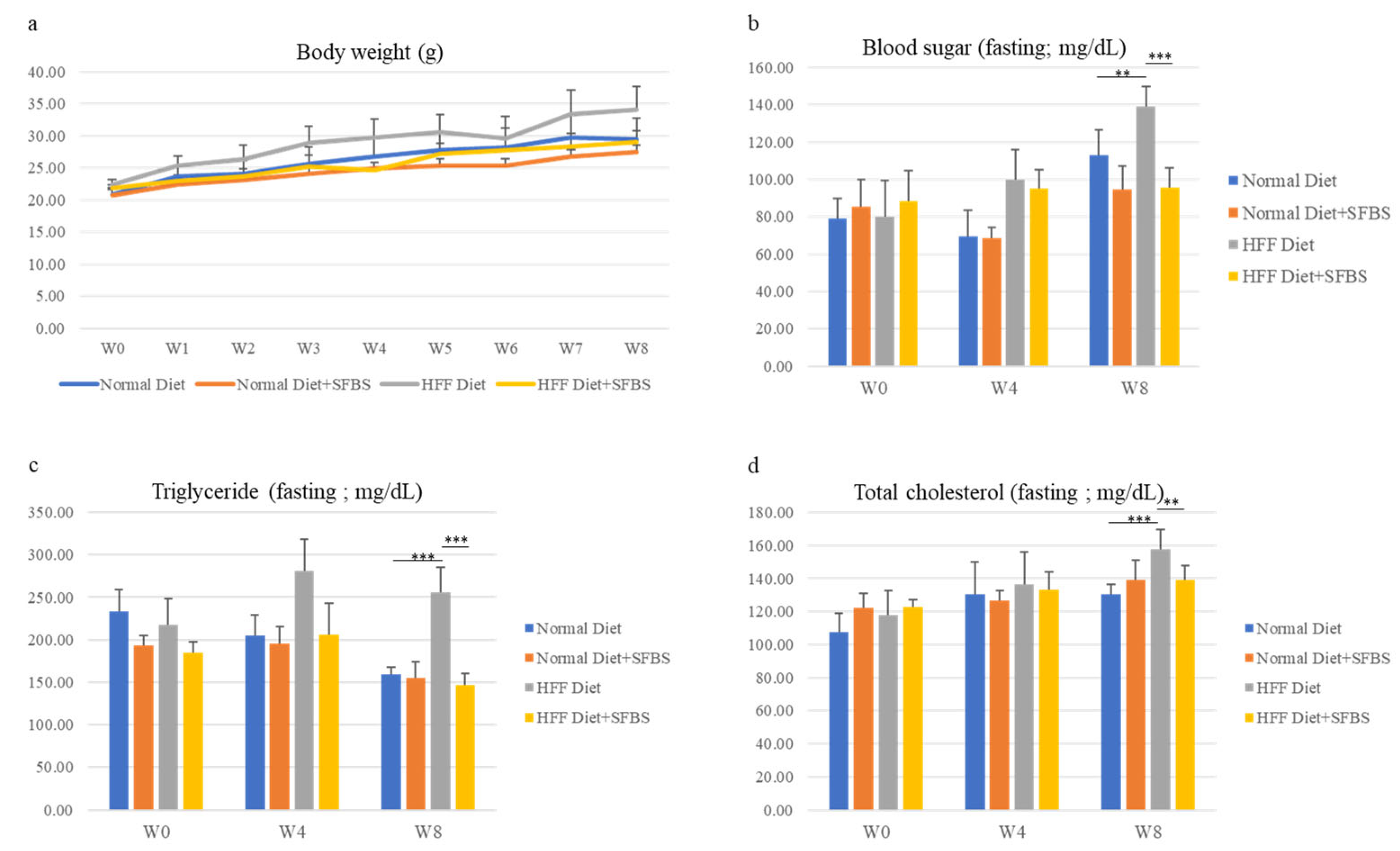
The influence of SFBS supplementation on healthy status was first evaluated using the mouse model with vital signs and biochemical variables. As shown in Figure 1, the steady gain in body weight of the four groups was monitored throughout the experiment, and SFBS supplementation (Figure 1a, yellow line) lessened the influence of the HFF diet (Figure 1a, gray line) on increasing body weight of experimental mice. Relatively high levels of biochemical variables, including blood sugar, total triglyceride, and total cholesterol, were noted in the HFF groups (Figure 1b–d, gray bar) as compared to those of mice fed the HFF diet and SFBS, 8 weeks post the onset of the experiment (Figure 1b–d, yellow bar). These results suggest the effect of SFBS supplementation on maintaining the metabolic homeostasis of experimental mice fed the HFF diet.
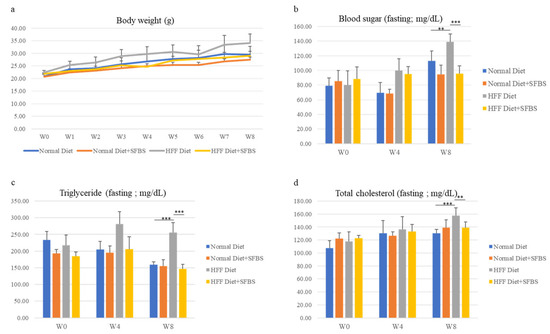
Figure 1.
The body weight (a), sera levels of fasting glucose (b), fasting triglyceride (c), and total cholesterol (d) of each experimental group were monitored at the onset of the experiment (W0), fourth (W4), and eighth week (W8). *** p < 0.001 and ** p < 0.005.
3.3. SFBS Supplementation Reprograms HFF Diet-Manipulated Gut Microenvironment
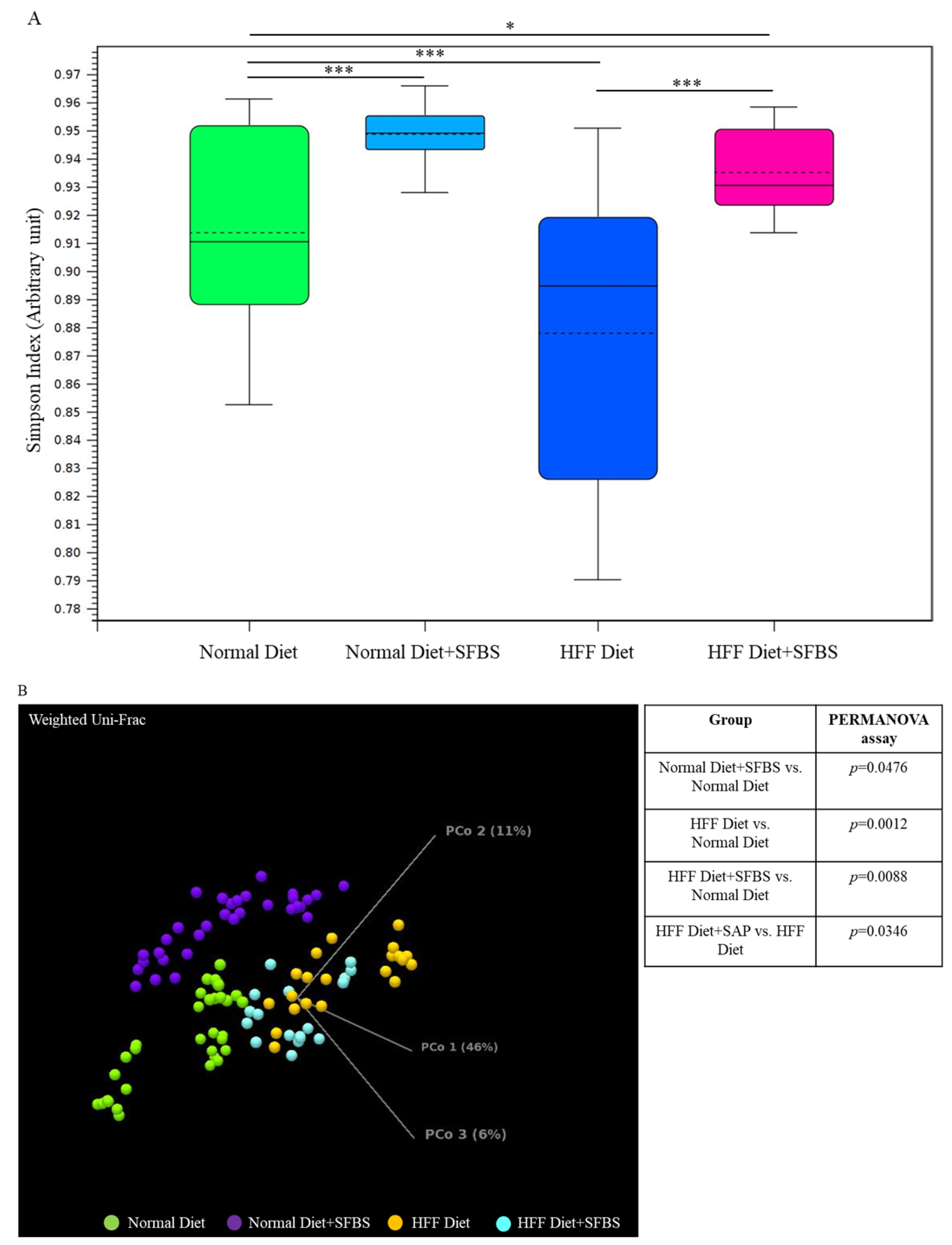
A high-fat or high-glucose diet has been widely considered to be a high-risk factor involved in the occurrence of diverse aging, chronic, and malignant diseases [1,7], which might alter the homeostasis of a gut microbial community. In this study, the classified I table was subjected to the diversity assessment. As shown in Figure 2A, the relatively low Simpson’ s dominance index in the HFF diet group (Figure 2A, dark blue box) indicated poor evenness in the gut microbial community as compared to that of the normal diet group, whereas SFBS supplementation restored the HFF-manipulated evenness in gut microbiota (Figure 2A, pink box). The results of the PERMANOVA assay indicated the dissimilarity between the gut microbiota in the normal diet group and the HFF diet group (Figure 2B, green and orange dots, p = 0.0012). SFBS supplementation diminished the impact of the HFF diet, leading to alterations in the gut microbial community as compared to that of the normal diet group (Figure 2B, green and light blue dot, p = 0.0088).
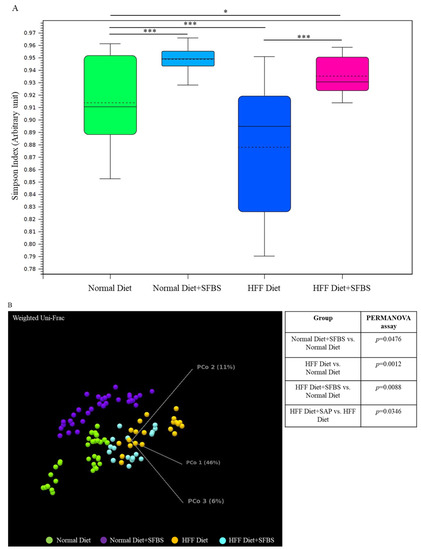
Figure 2.
Dietary intake or SFBS supplementation leads to alterations in the gut microbiota of experimental mice: (A) HFF diet or SFBS supplementation exhibited the opposite effect on changing the evenness (Simpson’s index) of the gut microbial community; (B) HFF diet or SFBS supplementation enhanced the dissimilarity in the IOTU profile compared to the normal diet group, which was estimated by using the Weighted Unifrac principal component analysis (PCoA) and PERMANOVA index. *** p < 0.001 and * p < 0.01.
In summary, intake of the HFF diet resulted in increases in specific OTUs, such as Akkermansia, Romboutsia, and Parabacteroides genera, with a concomitant decrease in Lactobacillus genera in the gut microbial community as compared to those of the normal diet group (Table 3, upper panel). By contrast, the alterations in these OTUs were less robust in the HFF diet group supplemented with SFBS as compared to the normal diet group (Table 3, lower panel).

Table 3.
Statistics of identified OTUs with differential abundance in the mouse model.
The influence of HFF diet-manipulated gut microbiota on the metabolic profile was imputed by deploying the PICRUSt2 platform coupled with the MetaCyc database, the Enzyme Commission (EC) pathway, or the KEGG Orthology database. The analytic results indicated that the intake of the HFF diet potentially led to increases in the generation of 4-aminobutanoate (Table 4, 10.07 folds, also known as gamma-aminobutyric acid) and histamine (Table 4, 6.84 folds) as compared to the normal diet group. Nevertheless, the synchronous SFBS supplementation with the HFF diet resulted in a further high level of 4-aminobutanoate (Table 4, 53.42 folds) with a concomitant decrease in histamine production (Table 4, 4.97 folds). Taking these results together, SFBS supplementation lessened the impact of the HFF diet on disturbing gut homeostasis.

Table 4.
Statistics of the HFF diet-modulated metabolic pathway.
3.4. SFBS Supplementation Reprograms the Gut Microenvironment of Enrolled Participants
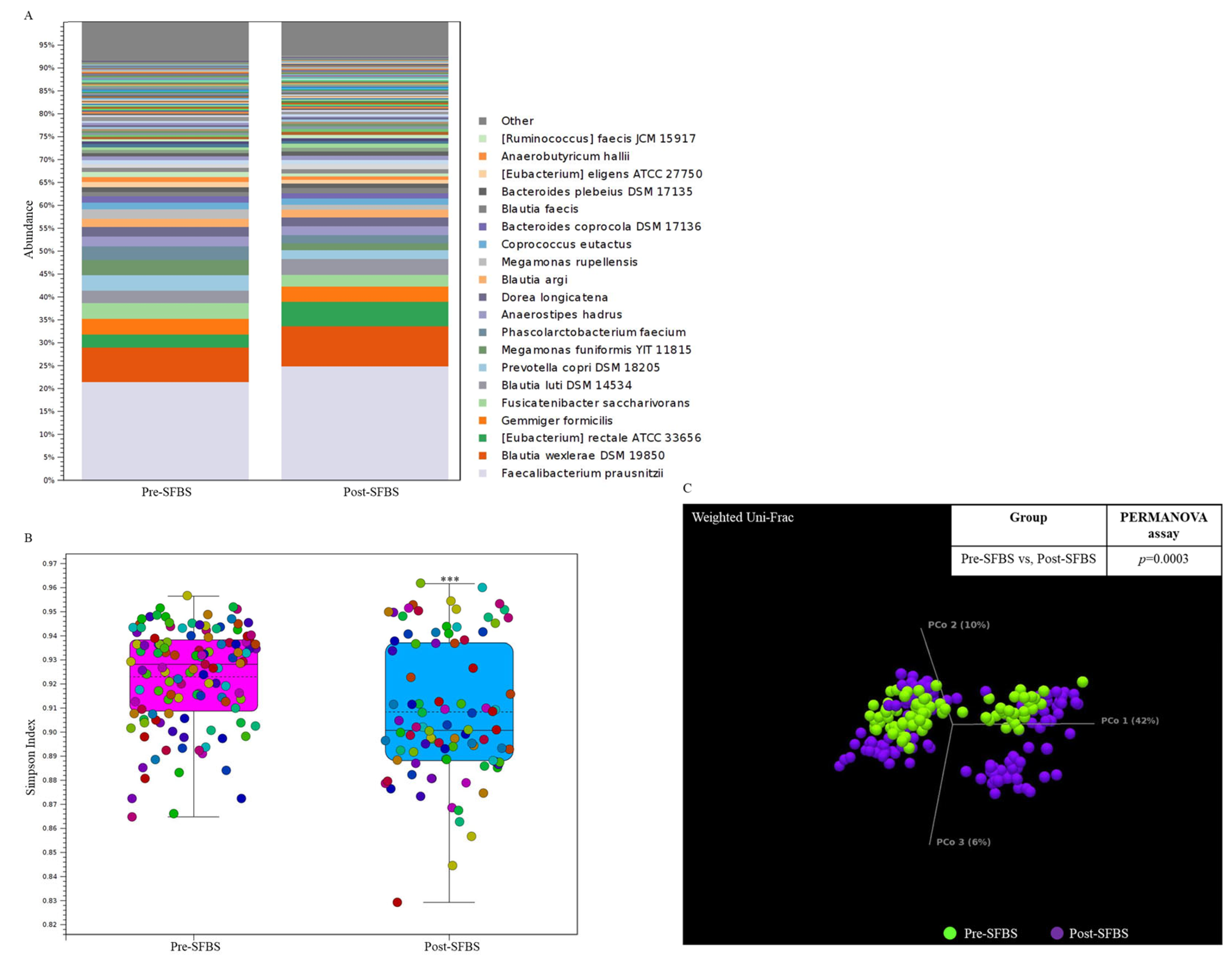
In this study, the influence of SFBS was further validated by conducting a human trial with the recruitment of 10 healthy participants. As shown in Figure 3, SFBS supplementation resulted in an increased abundance of normal flora in the gut (Figure 3A), which was relevant to the less evenness of the OTU profile in the post-SFBS group with the downregulated Simpson’s dominance index (Figure 3B, blue bar). Although SFBS supplementation exhibited no influence on the dissimilarity between these two groups (Figure 3C, p = 0.0003), the relative abundances of colorectal cancer (CRC) or irritable bowel syndrome (IBS)-related OTUs, including Eubacterium rectale, Roseburia intestinalis, and Clostridium saudiense, were diminished in the participants supplemented with SFBS (Table 5). The OTU profile was further subjected to predict the metabolic pathway through the MetaCyc algorithm. The analytic results indicated upregulated activity associated with the production of 4-aminobutanoate, 1-amino-2-propanol O-2-phosphate, and catechol, in the gut microenvironment of recruited participants with SFBS supplementation (Table 6), which were results that were consistent with those of the mouse model fed with SFBS (Table 4). Taken together, SFBS supplementation exerted a consistent influence on manipulating the gut microbial communities and metabolite profiles in distinct species.
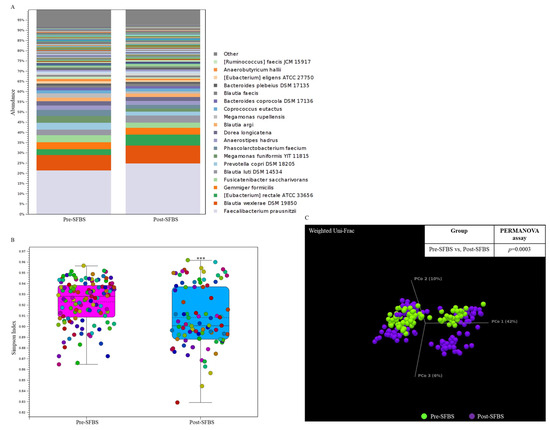
Figure 3.
SFBS supplementation reprograms the gut microbial community in recruited participants. (A) The operational taxonomy unit (OTU) profile with long-read sequencing in the enrolled participants was analyzed by using the ONT sequencing platform and presented in a stacked bar chart. (B) Supplementation of SFBS mediated the reduced evenness of gut microbial community with low Simpson’ s index. (C) SFBS supplementation mediated the dissimilarity in OTU profile of the same participants, which is evaluated by using Weighted Unifrac principal component analysis (PCoA) coupled with PERMANOVA index. (***, p < 0.001).

Table 5.
Statistics of identified OTUs with SFBS supplementation in the human trial.

Table 6.
Statistics of SFBS-modulated metabolic pathway in the human trial.
4. Discussion
High-fat-sugar dietary intake has been widely classified as a high-risk diet associated with the occurrence of diverse diseases with elevated levels of serum triglyceride, total cholesterol, and blood sugar [1,7]. Saponin compound, identified in SFBS using an untargeted LC-MS/MS approach in this study, has been demonstrated to exhibit a pharmacological impact on modulating immune response and metabolic homeostasis [8]. The identified compounds, such as eugenol, homocarnosine, genistein, dopamine, ornithine, and distinct short-chain fatty acids, within SFBS have been demonstrated to exert pharmacological effects on diverse physiological events [9,10,11,12,13,14]. These results suggest that the SFBS could function as a nutritional supplement for populations with diverse requirements.
In addition to the biochemical parameters, the relevance between a long-term HFF diet and the gut microbial community has been recently and widely pursued. The differential influence between a high-fat or high-glucose diet on reprogramming gut microbiota has been noted. In summary, increases in the relative abundance of Firmicutes with concomitant decreases in the Bacteroides and Akkermansia have mostly been classified in mouse models with high-fat diets (from 45% to 75%) [15,16]. In contrast, supplementation of a high-glucose or high-fructose diet has resulted in an elevated abundance of Akkermansia muciniphila which has been proposed to exert a protective effect against obesity, diabetes, or other metabolic disorders [17,18,19]. In this study, increases in the relative abundance of Akkermansia, Romboutsia, Parabacteroides, and Clostridium genera with a concomitant decrease in the relative level of Lactobacillus genera were classified in the gut microbial community of the HFF group, which was consistent with other reports [20]. In a previous study, supplementation with the non-edible components of green asparagus, which include xylose, inulin, flavonoids, and saponins, was demonstrated to facilitate the growth of Lactobacillus and Bifidobacteria genera with the execution of an in vitro culture model [21]. Additionally, Synbiotic supplement composed of probiotics, including Lactobacillus and Bifidobacteria genera, and prebiotics has been demonstrated to exhibit a statistically significant effect on decreases in obesity-related biomarkers in conducted clinical trials [22]. These studies have consistently illustrated the relevance between gut microenvironment and host health. In this study, the presence of saponin within SFBS supplementation diminished the impact of an HFF diet on reducing the relative levels of probiotic bacteria in a mouse model. Moreover, SFBS supplementation composed of variable compounds may lessen the influence of an HFF diet on disturbing the composition of gut microbiota via a more complex but solid network, which is beneficial to maintain the hemostasis of the gut microenvironment of a host. Nevertheless, the influence of SFBS supplementation on the gut microbial community associated with distinct dietary types by using animal models or human trials is worthy of further investigation.
Although there have been considerable efforts to identify or to pursue disease-relevant OTUs or the enterotype to function as the biomarker [23], the gut microbiota-driven metabolic network and generated metabolites have been shown to have a direct effect on manipulating the gut environment of host physiology [24]. To fill the knowledge gap or diminish the ambiguous results of OTU classification, the PICRUSt2 platform coupled with the Enzyme Commission, and the MetaCyc and KEGG Orthology databases have constituted a widely used workflow for predicting the metagenomic or metabolic function of the microbial community [25]. The analytic results have indicated relatively active metabolism pathways with the gut microbiota identified in high-fat or high-sugar mouse models, which have suggested the credibility of PICRUSt2-based predictions [26,27]. Except for energy metabolism, an HFF diet has been estimated to enhance diverse enzymatic activities of the gut microbial community, such as the synthesis of 4-aminobutanoate, histamine, and bacterial toxin in this study, whereas SFBS supplementation has exhibited an opposite effect on manipulating HFF-activated 4-aminobutanoate or histamine synthesis. Moreover, SFBS supplementation has exerted a consistent influence on similar pathway profiles associated with gut microbiota in mouse models and human trials. The targeted LC-MS/MS approach is worthy of conduction to further validate these analytic results.
5. Conclusions
To conclude, synchronous SFBS supplementation generated with multiple probiotics and soybean lessened the increases in weight gain and biochemical parameters of energy metabolism in the HFF diet group. Intervention with SFBS intake diminished the impact of the HFF diet on manipulating the gut microbial community and corresponding enzymatic activity and pathway with animal experiments. Furthermore, SFBS supplementation exhibited a consistent effect on reprogramming the gut microenvironment in experimental mice and enrolled participants without adverse events. These results suggest the benefit and safety of SFBS as a nutritional supplement.
Author Contributions
Conceptualization, J.-C.L.; methodology, J.-C.L.; software, J.-C.L.; validation, J.-C.L.; formal analysis, C.-Y.W., C.-K.H., W.-S.H., Y.-H.L. and M.-C.S.; investigation, C.-Y.W., C.-K.H., W.-S.H., Y.-H.L. and M.-C.S.; resources, J.-C.L.; data curation, C.-Y.W., C.-K.H., W.-S.H., Y.-H.L. and M.-C.S.; writing—original draft preparation, J.-C.L.; writing—review and editing, J.-C.L.; visualization, J.-C.L.; supervision, J.-C.L.; project administration, J.-C.L.; funding acquisition, J.-C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant (110-6603-005-400) from the Ministry of Education, Taiwan, and funding (A-111-008) from Sagittarius Life Science Corporations, Taipei, Taiwan.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Taipei Medical University for studies involving humans (approval no. N202103179). The animal study protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Taipei Medical University (approval no. LAC-2021-0511) for studies involving animals.
Informed Consent Statement
Informed consent was obtained from all subjects enrolled in the study. Written informed consent was obtained from the enrollments to publish this paper” if applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author due to privacy restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yang, J.; Wei, H.; Zhou, Y.; Szeto, C.H.; Li, C.; Lin, Y.; Coker, O.O.; Lau, H.C.H.; Chan, A.W.H.; Sung, J.J.Y.; et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterol. 2022, 162, 135–149. [Google Scholar] [CrossRef]
- Zepeda-Hernández, A.; Garcia-Amezquita, L.E.; Requena, T.; García-Cayuela, T. Probiotics, prebiotics, and synbiotics added to dairy products: Uses and applications to manage type 2 diabetes. Food Res. Int. 2021, 142, 110208. [Google Scholar] [CrossRef] [PubMed]
- Kusumo, P.D.; Maulahela, H.; Utari, A.P.; Surono, I.S.; Soebandrio, A.; Abdullah, M. Probiotic Lactobacillus plantarum IS 10506 supplementation increase SCFA of women with functional constipation. Iran. J. Microbiol. 2019, 11, 389–396. [Google Scholar] [PubMed]
- Rezende, E.S.V.; Lima, G.C.; Naves, M.M.V. Dietary fibers as beneficial microbiota modulators: A proposal classification by prebiotic categories. Nutrition 2021, 89, 111217. [Google Scholar] [CrossRef]
- Menne, E.; Guggenbuhl, N.; Roberfroid, M. Fn-type chicory inulin hydrolysate has a prebiotic effect in humans. J. Nutr. 2000, 130, 1197–1199. [Google Scholar] [CrossRef] [PubMed]
- Guaragni, A.; Boiago, M.M.; Bottari, N.B.; Morsch, V.M.; Lopes, T.F.; Schafer da Silva, A. Feed supplementation with inulin on broiler performance and meat quality challenged with Clostridium perfringens: Infection and prebiotic impacts. Microb. Pathog. 2020, 139, 103889. [Google Scholar] [CrossRef]
- Miao, M.; Wang, Q.; Wang, X.; Fan, C.; Luan, T.; Yan, L.; Zhang, Y.; Zeng, X.; Dai, Y.; Li, P. The protective effects of inulin-type fructans against high-fat/sucrose diet-induced gestational diabetes mice in association with gut microbiota regulation. Front. Microbiol. 2022, 13, 832151. [Google Scholar] [CrossRef]
- Marrelli, M.; Conforti, F.; Araniti, F.; Statti, G.A. Effects of saponins on lipid metabolism: A review of potential health benefits in the treatment of obesity. Molecules 2016, 21, 1404. [Google Scholar] [CrossRef]
- Hughes, K.; Le, T.B.; Van Der Smissen, P.; Tyteca, D.; Mingeot-Leclercq, M.P.; Quetin-Leclercq, J. The antileishmanial activity of eugenol associated with lipid storage reduction rather than membrane properties alterations. Molecules 2023, 28, 3871. [Google Scholar] [CrossRef]
- Creighton, J.V.; de Souza Gonçalves, L.; Artioli, G.G.; Tan, D.; Elliott-Sale, K.J.; Turner, M.D.; Doig, C.L.; Sale, C. Physiological roles of carnosine in myocardial function and health. Adv. Nutr. 2022, 13, 1914–1929. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.S.; Prasad, S.K.; Shivamallu, C.; Prasad, K.S.; Syed, A.; Reddy, P.; Cull, C.A.; Amachawadi, R.G. Genistein: A potent anti-breast cancer agent. Curr Issues Mol. Biol. 2021, 43, 1502–1517. [Google Scholar] [CrossRef]
- Wert-Carvajal, C.; Reneaux, M.; Tchumatchenko, T.; Clopath, C. Dopamine and serotonin interplay for valence-based spatial learning. Cell Rep. 2022, 39, 110645. [Google Scholar] [CrossRef]
- Marshall, A.; McGrath, J.W.; Graham, R.; McMullan, G. Food for thought-The link between Clostridioides difficile metabolism and pathogenesis. PLoS Pathog. 2023, 19, e1011034. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, F.A.; Keenan, C.M.; Wallace, L.E.; Woods, C.; Cavin, J.B.; Flockton, A.R.; Macklin, W.B.; Belkind-Gerson, J.; Hirota, S.A.; Sharkey, K.A. Intestinal microbiota shapes gut physiology and regulates enteric neurons and glia. Microbiome 2021, 9, 210. [Google Scholar] [CrossRef]
- Yin, W.; Zhang, S.-Q.; Pang, W.-L.; Chen, X.-J.; Wen, J.; Hou, J.; Wang, C.; Song, L.Y.; Qiu, Z.M.; Liang, P.T. Tang-Ping-San decoction remodel intestinal flora and barrier to ameliorate type 2 diabetes mellitus in rodent model. Diabetes Metab. Syndr. Obes. Targets Ther. 2022, 15, 2563–2581. [Google Scholar] [CrossRef] [PubMed]
- Ju, M.; Liu, Y.; Li, M.; Cheng, M.; Zhang, Y.; Deng, G.; Kang, X.; Liu, H. Baicalin improves intestinal microecology and abnormal metabolism induced by high-fat diet. Eur. J. Pharmacol. 2019, 857, 172457. [Google Scholar] [CrossRef]
- Moon, H.D.; Eunjung, L.; Mi-Jin, O.; Yoonsook, K.; Ho-Young, P. High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G.A. High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef]
- Redondo-Cuenca, A.; García-Alonso, A.; Rodríguez-Arcos, R.; Castro, I.; Alba, C.; Rodríguez, J.M.; Goñi, I. Nutritional composition of green asparagus (Asparagus officinalis L.), edible part and by-products, and assessment of their effect on the growth of human gut-associated bacteria. Food Res. Int. 2023, 163, 112284. [Google Scholar] [CrossRef]
- Sergeev, I.N.; Aljutaily, T.; Walton, G.; Huarte, E. Effects of Synbiotic Supplement on Human Gut Microbiota, Body Composition and Weight Loss in Obesity. Nutrients 2020, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A.; et al. Gut microbiota conditions the therapeutic efficacy of trastuzumab in her2-positive breast cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell. 2018, 9, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, H.; Zhao, X.; Zhou, R.; Liu, H.; Sun, Y.; Fan, Y.; Shi, Y.; Qiao, S.; Liu, S.; et al. Multi-omics study reveals that statin therapy is associated with restoration of gut microbiota homeostasis and improvement in outcomes in patients with acute coronary syndrome. Theranostics 2021, 11, 5778–5793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Lu, F.; Mohammed, S.A.D.; Liu, H.; Ding, S.; Liu, S.M. Mechanism of action of shenerjiangzhi formulation on hyperlipidemia induced by consumption of a high-fat diet in rats using network pharmacology and analyses of the gut microbiota. Front. Pharmacol. 2022, 13, 745074. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.X.; Jiang, X.M.; Wang, H.W.; Ge, L.; Lai, Y.T.; Jiang, X.Y.; Chen, F.; Huang, P.P. Probiotic supplements alleviate gestational diabetes mellitus by restoring the diversity of gut microbiota: A study based on 16S rRNA sequencing. J. Microbiol. 2021, 59, 827–839. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).