Untargeted Metabolomics and Body Mass in Adolescents: A Cross-Sectional and Longitudinal Analysis
Abstract
1. Introduction
2. Materials and Methods
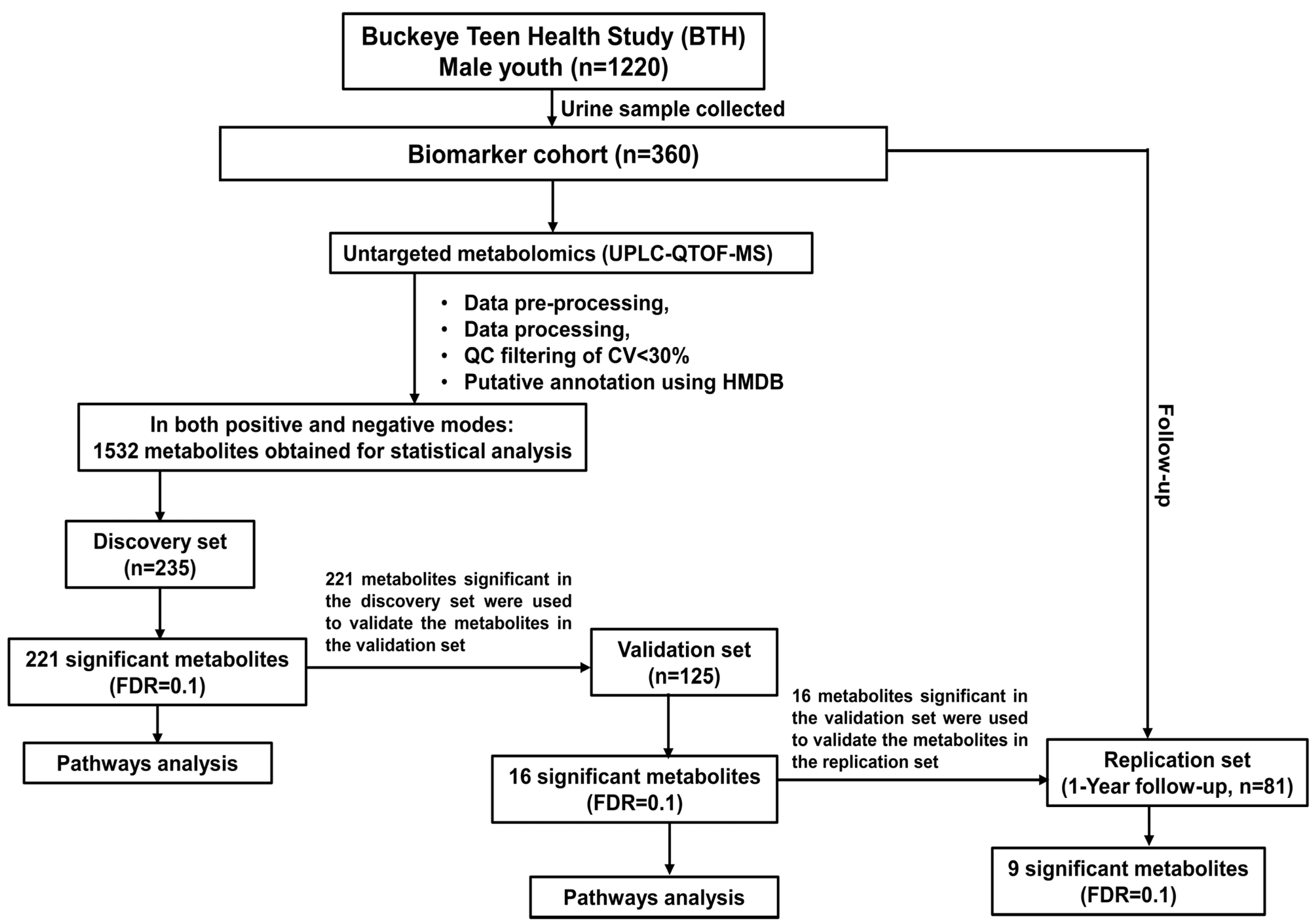
2.1. Study Recruitment and Design
2.2. Urine Sample Collection
2.3. Reagents and Chemicals
2.4. Urine Sample Preparation for Metabolomics
2.5. UPLC–QTOF-MS Analysis
2.6. Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Characteristics of Study Subjects
3.2. Untargeted Metabolic Profiling of Urine by UPLC-QTOF-MS
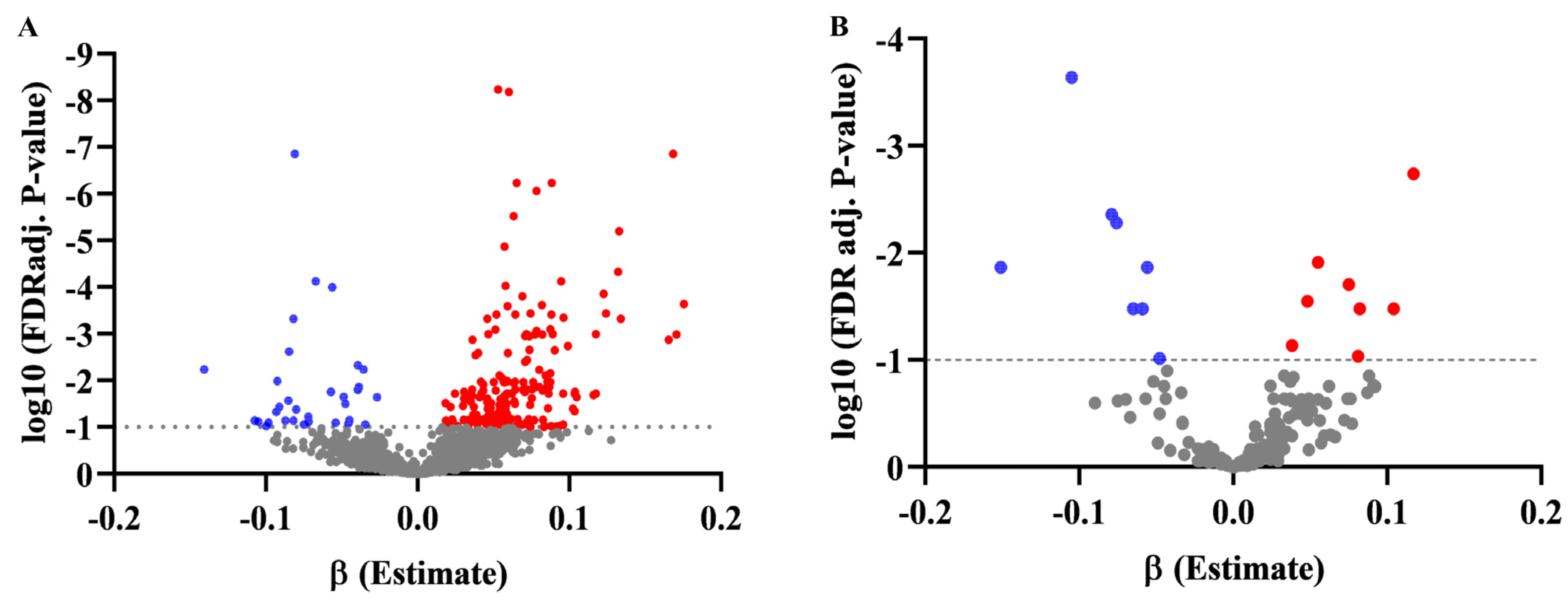
3.3. Association of Metabolites with BMI Z-Score in Discovery and Validation Set, and Changes in BMI at the 1-Year Follow-Up
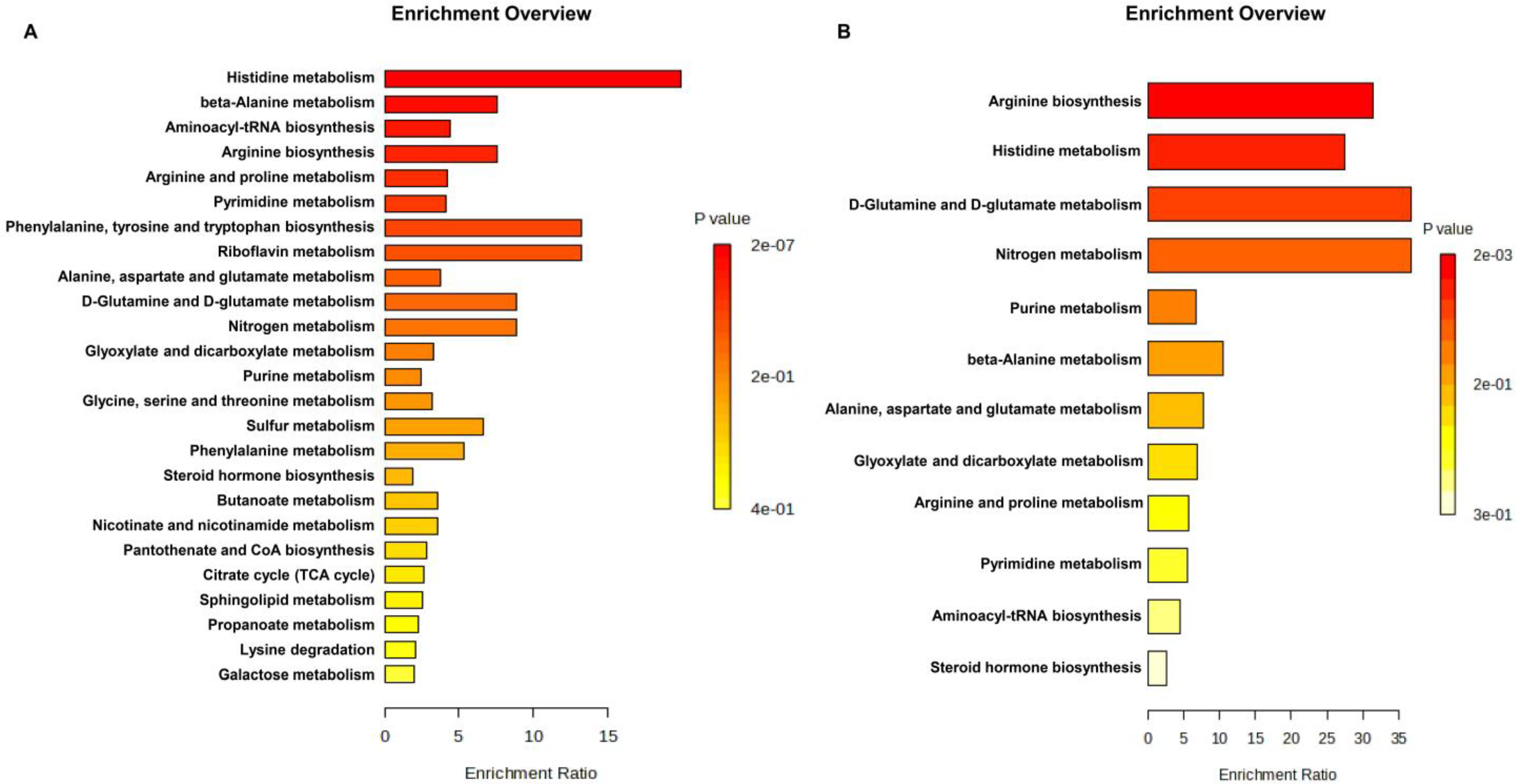
3.4. Differential Mapping of Metabolites in Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMI | Body mass index |
| UPLC–QTOF-MS | Ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry |
| FDR | False-discovery-rate |
| BTH | Buckeye Teen Health Study |
| OSU | Ohio State University |
| HPLC | High-performance liquid chromatography |
| ACN | Acetonitrile |
| 4-NBA | 4-nitrobenzoic acid |
| ESI | Electrospray ionization |
| QC | Quality control |
| CV | Coefficient of variation |
| MSTUS | Mass spectrometry total usable signal |
| HMDB | Human metabolome database |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CV | Coefficient of variation |
| MetPA | Metabolomic pathway enrichment analysis |
| AAs | Amino acids |
| BCAAs | Branched-chain amino acids |
| IGF-1 | Insulin-like growth factor-1 |
| cGP | cyclic Glycine-Proline |
| NEAAs | Non-essential amino acids |
| ROS | Reactive oxygen species |
| GSH | Glutathione |
| CVD | Cardiovascular diseases |
References
- Szczerbinski, L.; Wojciechowska, G.; Olichwier, A.; Taylor, M.A.; Puchta, U.; Konopka, P.; Paszko, A.; Citko, A.; Goscik, J.; Fiehn, O.; et al. Untargeted Metabolomics Analysis of the Serum Metabolic Signature of Childhood Obesity. Nutrients 2022, 14, 214. [Google Scholar] [CrossRef] [PubMed]
- Stierman, B.; Afful, J.; Carroll, M.D.; Chen, T.-C.; Davy, O.; Fink, S.; Fryar, C.D.; Gu, Q.; Hales, C.M.; Hughes, J.P. National Health and Nutrition Examination Survey 2017–March 2020 Prepandemic Data Files Development of Files and Prevalence Estimates for Selected Health Outcomes. Natl. Health Stat. Rep. 2021, 158. [Google Scholar]
- Simmonds, M.; Llewellyn, A.; Owen, C.G.; Woolacott, N. Predicting adult obesity from childhood obesity: A systematic review and meta-analysis. Obes. Rev. 2016, 17, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.; Yeo, G.S. The bigger picture of FTO—The first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2014, 10, 51–61. [Google Scholar] [CrossRef]
- Kahan, L.G.; Mehrzad, R. Chapter 10-Environmental factors related to the obesity epidemic. In Obesity; Mehrzad, R., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 117–139. [Google Scholar] [CrossRef]
- Pérez, L.M.; García, K.; Herrera, R. Psychological, behavioral and familial factors in obese Cuban children and adolescents. MEDICC Rev. 2013, 15, 24–28. [Google Scholar]
- Karatsoreos, I.N.; Thaler, J.P.; Borgland, S.L.; Champagne, F.A.; Hurd, Y.L.; Hill, M.N. Food for thought: Hormonal, experiential, and neural influences on feeding and obesity. J. Neurosci. 2013, 33, 17610–17616. [Google Scholar] [CrossRef][Green Version]
- Romieu, I.; Dossus, L.; Barquera, S.; Blottière, H.M.; Franks, P.W.; Gunter, M.; Hwalla, N.; Hursting, S.D.; Leitzmann, M.; Margetts, B. Energy balance and obesity: What are the main drivers? Cancer Causes Control 2017, 28, 247–258. [Google Scholar] [CrossRef]
- Bardugo, A.; Fishman, B.; Libruder, C.; Tanne, D.; Ram, A.; Hershkovitz, Y.; Zucker, I.; Furer, A.; Gilon, R.; Chodick, G.; et al. Body Mass Index in 1.9 Million Adolescents and Stroke in Young Adulthood. Stroke 2021, 52, 2043–2052. [Google Scholar] [CrossRef]
- Handakas, E.; Lau, C.H.; Alfano, R.; Chatzi, V.L.; Plusquin, M.; Vineis, P.; Robinson, O. A systematic review of metabolomic studies of childhood obesity: State of the evidence for metabolic determinants and consequences. Obes. Rev. 2022, 23, e13384. [Google Scholar] [CrossRef]
- Hruby, A.; Hu, F.B. The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics 2015, 33, 673–689. [Google Scholar] [CrossRef]
- Steinberger, J.; Daniels, S.R. Obesity, Insulin Resistance, Diabetes, and Cardiovascular Risk in Children. Circulation 2003, 107, 1448–1453. [Google Scholar] [CrossRef] [PubMed]
- Daneshzad, E.; Rostami, S.; Aghamahdi, F.; Mahdavi-Gorabi, A.; Qorbani, M. Association of cardiometabolic risk factors with insulin resistance in overweight and obese children. BMC Endocr. Disord. 2022, 22, 320. [Google Scholar] [CrossRef]
- Vanderwall, C.; Randall Clark, R.; Eickhoff, J.; Carrel, A.L. BMI is a poor predictor of adiposity in young overweight and obese children. BMC Pediatr. 2017, 17, 135. [Google Scholar] [CrossRef]
- Vehrs, P.R.; Fellingham, G.W.; McAferty, A.; Kelsey, L. Trends in BMI Percentile and Body Fat Percentage in Children 12 to 17 Years of Age. Children 2022, 9, 744. [Google Scholar] [CrossRef]
- Grossman, D.C.; Bibbins-Domingo, K.; Curry, S.J.; Barry, M.J.; Davidson, K.W.; Doubeni, C.A.; Epling, J.W.; Kemper, A.R.; Krist, A.H.; Kurth, A.E. Screening for obesity in children and adolescents: US Preventive Services Task Force recommendation statement. Jama 2017, 317, 2417–2426. [Google Scholar]
- Cirulli, E.T.; Guo, L.; Swisher, C.L.; Shah, N.; Huang, L.; Napier, L.A.; Kirkness, E.F.; Spector, T.D.; Caskey, C.T.; Thorens, B. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019, 29, 488–500.e482. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward personalized medicine. Mass. Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Yu, E.Y.-W.; Ren, Z.; Mehrkanoon, S.; Stehouwer, C.D.A.; van Greevenbroek, M.M.J.; Eussen, S.J.P.M.; Zeegers, M.P.; Wesselius, A. Plasma metabolomic profiling of dietary patterns associated with glucose metabolism status: The Maastricht Study. BMC Med. 2022, 20, 450. [Google Scholar] [CrossRef]
- Sohn, M.J.; Chae, W.; Ko, J.S.; Cho, J.Y.; Kim, J.E.; Choi, J.Y.; Jang, H.B.; Lee, H.J.; Park, S.I.; Park, K.H.; et al. Metabolomic Signatures for the Effects of Weight Loss Interventions on Severe Obesity in Children and Adolescents. Metabolites 2021, 12, 27. [Google Scholar] [CrossRef]
- Rigamonti, A.E.; Frigerio, G.; Caroli, D.; De Col, A.; Cella, S.G.; Sartorio, A.; Fustinoni, S. A Metabolomics-Based Investigation of the Effects of a Short-Term Body Weight Reduction Program in a Cohort of Adolescents with Obesity: A Prospective Interventional Clinical Study. Nutrients 2023, 15, 529. [Google Scholar] [CrossRef]
- Brachem, C.; Langenau, J.; Weinhold, L.; Schmid, M.; Nöthlings, U.; Oluwagbemigun, K. Associations of BMI and Body Fat with Urine Metabolome in Adolescents Are Sex-Specific: A Cross-Sectional Study. Metabolites 2020, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Concepcion, J.; Chen, K.; Saito, R.; Gangoiti, J.; Mendez, E.; Nikita, M.E.; Barshop, B.A.; Natarajan, L.; Sharma, K.; Kim, J.J. Identification of pathognomonic purine synthesis biomarkers by metabolomic profiling of adolescents with obesity and type 2 diabetes. PLoS ONE 2020, 15, e0234970. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.-H.E.; Siskos, A.P.; Maitre, L.; Robinson, O.; Athersuch, T.J.; Want, E.J.; Urquiza, J.; Casas, M.; Vafeiadi, M.; Roumeliotaki, T. Determinants of the urinary and serum metabolome in children from six European populations. BMC Med. 2018, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Pierri, L.; Landolfi, A.; Marciano, F.; Bisogno, A.; Belmonte, F.; Palladino, C.; Guercio Nuzio, S.; Campiglia, P.; Vajro, P. Urinary Metabolomics in Pediatric Obesity and NAFLD Identifies Metabolic Pathways/Metabolites Related to Dietary Habits and Gut-Liver Axis Perturbations. Nutrients 2017, 9, 485. [Google Scholar] [CrossRef]
- Cho, K.; Moon, J.S.; Kang, J.H.; Jang, H.B.; Lee, H.J.; Park, S.I.; Yu, K.S.; Cho, J.Y. Combined untargeted and targeted metabolomic profiling reveals urinary biomarkers for discriminating obese from normal-weight adolescents. Pediatr. Obes. 2017, 12, 93–101. [Google Scholar] [CrossRef]
- Pathmasiri, W.; Pratt, K.; Collier, D.; Lutes, L.; McRitchie, S.; Sumner, S. Integrating metabolomic signatures and psychosocial parameters in responsivity to an immersion treatment model for adolescent obesity. Metabolomics 2012, 8, 1037–1051. [Google Scholar] [CrossRef]
- Leal-Witt, M.J.; Ramon-Krauel, M.; Samino, S.; Llobet, M.; Cuadras, D.; Jimenez-Chillaron, J.C.; Yanes, O.; Lerin, C. Untargeted metabolomics identifies a plasma sphingolipid-related signature associated with lifestyle intervention in prepubertal children with obesity. Int. J. Obes. 2018, 42, 72–78. [Google Scholar] [CrossRef]
- Villarreal-Pérez, J.Z.; Villarreal-Martínez, J.Z.; Lavalle-González, F.J.; Torres-Sepúlveda, M.d.R.; Ruiz-Herrera, C.; Cerda-Flores, R.M.; Castillo-García, E.R.; Rodríguez-Sánchez, I.P.; Martínez de Villarreal, L.E. Plasma and urine metabolic profiles are reflective of altered beta-oxidation in non-diabetic obese subjects and patients with type 2 diabetes mellitus. Diabetol. Metab. Syndr. 2014, 6, 129. [Google Scholar] [CrossRef]
- Robinson, O.; Chadeau Hyam, M.; Karaman, I.; Climaco Pinto, R.; Ala-Korpela, M.; Handakas, E.; Fiorito, G.; Gao, H.; Heard, A.; Jarvelin, M.R.; et al. Determinants of accelerated metabolomic and epigenetic aging in a UK cohort. Aging Cell 2020, 19, e13149. [Google Scholar] [CrossRef]
- Friedman, K.L.; Roberts, M.E.; Keller-Hamilton, B.; Yates, K.A.; Paskett, E.D.; Berman, M.L.; Slater, M.D.; Lu, B.; Ferketich, A.K. Attitudes toward Tobacco, Alcohol, and Non-Alcoholic Beverage Advertisement Themes among Adolescent Boys. Subst. Use Misuse 2018, 53, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Keller-Hamilton, B.; Muff, J.; Blue, T.; Lu, B.; Slater, M.D.; Roberts, M.E.; Ferketich, A.K. Tobacco and Alcohol on Television: A Content Analysis of Male Adolescents' Favorite Shows. Prev. Chronic Dis. 2018, 15, E134. [Google Scholar] [CrossRef] [PubMed]
- Keller-Hamilton, B.; Roberts, M.E.; Slater, M.D.; Ferketich, A.K. Memorability of Cigarette Advertisements Making "Natural" Claims Among Adolescents. Tob. Regul. Sci. 2019, 5, 326–331. [Google Scholar] [CrossRef]
- Keller-Hamilton, B.; Lu, B.; Roberts, M.E.; Berman, M.L.; Root, E.D.; Ferketich, A.K. Electronic cigarette use and risk of cigarette and smokeless tobacco initiation among adolescent boys: A propensity score matched analysis. Addict. Behav. 2021, 114, 106770. [Google Scholar] [CrossRef] [PubMed]
- Burgoon, M.L.; Albani, T.; Keller-Hamilton, B.; Lu, B.; Roberts, M.E.; Craigmile, P.F.; Browning, C.; Xi, W.; Ferketich, A.K. Exposures to the tobacco retail environment among adolescent boys in urban and rural environments. Am. J. Drug Alcohol. Abus. 2019, 45, 217–226. [Google Scholar] [CrossRef]
- Nixon, D.E.; Ferketich, A.K.; Slater, M.D.; Mays, D.; Keller-Hamilton, B. Prospective associations between attitudes toward alcohol advertisements and alcohol use behaviors among adolescent boys. Addict. Behav. Rep. 2022, 15, 100428. [Google Scholar] [CrossRef]
- Roberts, M.E.; Keller-Hamilton, B.; Ferketich, A.K. Testing if attitudes mediate the association between advertising exposure and adolescent tobacco use. Addict. Behav. 2022, 134, 107415. [Google Scholar] [CrossRef]
- Mahieu, N.G.; Genenbacher, J.L.; Patti, G.J. A roadmap for the XCMS family of software solutions in metabolomics. Curr. Opin. Chem. Biol. 2016, 30, 87–93. [Google Scholar] [CrossRef]
- Faquih, T.; van Smeden, M.; Luo, J.; le Cessie, S.; Kastenmüller, G.; Krumsiek, J.; Noordam, R.; van Heemst, D.; Rosendaal, F.R.; van Hylckama Vlieg, A.; et al. A Workflow for Missing Values Imputation of Untargeted Metabolomics Data. Metabolites 2020, 10, 486. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. WIREs Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Metabolomic Data Processing, Analysis, and Interpretation Using MetaboAnalyst. Curr. Protoc. Bioinform. 2011, Chapter 14. 14.10.11–14.10.48. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making Metabolomics More Meaningful. Nucleic Acids Research 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Sorrow, P.; Maguire, R.; Murphy, S.K.; Belcher, S.M.; Hoyo, C. Elevated metabolites of acetaminophen in cord blood of children with obesity. Pediatr. Obes. 2019, 14, e12465. [Google Scholar] [CrossRef]
- Butte, N.F.; Liu, Y.; Zakeri, I.F.; Mohney, R.P.; Mehta, N.; Voruganti, V.S.; Göring, H.; Cole, S.A.; Comuzzie, A.G. Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am. J. Clin. Nutr. 2015, 102, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.; Yu, Z.; Kleber, M.; Singmann, P.; Holzapfel, C.; He, Y.; Mittelstrass, K.; Polonikov, A.; Prehn, C.; Römisch-Margl, W.; et al. Childhood Obesity Is Associated with Changes in the Serum Metabolite Profile. Obes. Facts 2012, 5, 660–670. [Google Scholar] [CrossRef]
- Gawlik, A.; Shmoish, M.; Hartmann, M.F.; Malecka-Tendera, E.; Wudy, S.A.; Hochberg, Z. Steroid Metabolomic Disease Signature of Nonsyndromic Childhood Obesity. J. Clin. Endocrinol. Metab. 2016, 101, 4329–4337. [Google Scholar] [CrossRef]
- Takashina, C.; Tsujino, I.; Watanabe, T.; Sakaue, S.; Ikeda, D.; Yamada, A.; Sato, T.; Ohira, H.; Otsuka, Y.; Oyama-Manabe, N.; et al. Associations among the plasma amino acid profile, obesity, and glucose metabolism in Japanese adults with normal glucose tolerance. Nutr Metab (Lond) 2016, 13, 5. [Google Scholar] [CrossRef]
- Gogna, N.; Krishna, M.; Oommen, A.M.; Dorai, K. Investigating correlations in the altered metabolic profiles of obese and diabetic subjects in a South Indian Asian population using an NMR-based metabolomic approach. Mol. BioSystems 2015, 11, 595–606. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.Y.; Kim, O.Y.; Ham, B.M.; Kim, H.-J.; Kwon, D.Y.; Jang, Y.; Lee, J.H. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC− Q-TOF MS). J. Proteome Res. 2010, 9, 4368–4375. [Google Scholar] [CrossRef]
- Mihalik, S.J.; Michaliszyn, S.F.; de las Heras, J.; Bacha, F.; Lee, S.; Chace, D.H.; DeJesus, V.R.; Vockley, J.; Arslanian, S.A. Metabolomic Profiling of Fatty Acid and Amino Acid Metabolism in Youth With Obesity and Type 2 Diabetes: Evidence for enhanced mitochondrial oxidation. Diabetes Care 2012, 35, 605–611. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- Perng, W.; Rifas-Shiman, S.L.; Hivert, M.F.; Chavarro, J.E.; Oken, E. Branched Chain Amino Acids, Androgen Hormones, and Metabolic Risk Across Early Adolescence: A Prospective Study in Project Viva. Obes. (Silver Spring) 2018, 26, 916–926. [Google Scholar] [CrossRef]
- Kang, M.; Yoo, H.J.; Kim, M.; Kim, M.; Lee, J.H. Metabolomics identifies increases in the acylcarnitine profiles in the plasma of overweight subjects in response to mild weight loss: A randomized, controlled design study. Lipids Health Dis. 2018, 17, 237. [Google Scholar] [CrossRef]
- Dambrova, M.; Makrecka-Kuka, M.; Kuka, J.; Vilskersts, R.; Nordberg, D.; Attwood, M.M.; Smesny, S.; Sen, Z.D.; Guo, A.C.; Oler, E.; et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol. Rev. 2022, 74, 506–551. [Google Scholar] [CrossRef]
- Papandreou, C.; García-Gavilán, J.; Camacho-Barcia, L.; Hansen, T.T.; Sjödin, A.; Harrold, J.A.; Halford, J.C.G.; Bulló, M. Circulating Metabolites Associated with Body Fat and Lean Mass in Adults with Overweight/Obesity. Metabolites 2021, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Ganti, S.; Taylor, S.L.; Kim, K.; Hoppel, C.L.; Guo, L.; Yang, J.; Evans, C.; Weiss, R.H. Urinary acylcarnitines are altered in human kidney cancer. Int. J. Cancer 2012, 130, 2791–2800. [Google Scholar] [CrossRef]
- Li, S.; Gao, D.; Jiang, Y. Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma. Metabolites 2019, 9, 36. [Google Scholar] [CrossRef]
- Tchernof, A.; Labrie, F.; Bélanger, A.; Prud’homme, D.; Bouchard, C.; Tremblay, A.; Nadeau, A.; Després, J.-P. Androstane-3α,17β-Diol Glucuronide as a Steroid Correlate of Visceral Obesity in Men*. J. Clin. Endocrinol. Metab. 1997, 82, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Dalesio, N.M.; Lee, C.K.K.; Hendrix, C.W.; Kerns, N.; Hsu, A.; Clarke, W.; Collaco, J.M.; McGrath-Morrow, S.; Yaster, M.; Brown, R.H.; et al. Effects of Obstructive Sleep Apnea and Obesity on Morphine Pharmacokinetics in Children. Anesth. Analg. 2020, 131, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, L.; Barnard, L.; Baranowski, E.S.; Gilligan, L.C.; Taylor, A.E.; Arlt, W.; Shackleton, C.H.L.; Storbeck, K.-H. Human steroid biosynthesis, metabolism and excretion are differentially reflected by serum and urine steroid metabolomes: A comprehensive review. J. Steroid Biochem. Mol. Biol. 2019, 194, 105439. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.C.; Matthews, C.E.; Sampson, J.N.; Stolzenberg-Solomon, R.Z.; Zheng, W.; Cai, Q.; Tan, Y.T.; Chow, W.-H.; Ji, B.-T.; Liu, D.K. Human metabolic correlates of body mass index. Metabolomics 2014, 10, 259–269. [Google Scholar] [CrossRef]
- Vandenput, L.; Mellström, D.; Lorentzon, M.; Swanson, C.; Karlsson, M.K.; Brandberg, J.; Lönn, L.; Orwoll, E.; Smith, U.; Labrie, F.; et al. Androgens and Glucuronidated Androgen Metabolites Are Associated with Metabolic Risk Factors in Men. J. Clin. Endocrinol. Metab. 2007, 92, 4130–4137. [Google Scholar] [CrossRef][Green Version]
- Kelly, B.; Pearce, E.L. Amino assets: How amino acids support immunity. Cell Metab. 2020, 32, 154–175. [Google Scholar] [CrossRef]
- Yamakado, M.; Tanaka, T.; Nagao, K.; Ishizaka, Y.; Mitushima, T.; Tani, M.; Toda, A.; Toda, E.; Okada, M.; Miyano, H.; et al. Plasma amino acid profile is associated with visceral fat accumulation in obese Japanese subjects. Clin. Obes. 2012, 2, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Chadwick, J.Q.; Teague, A.M.; Tullier, M.A.; Wolbert, L.; Coleman, C.; Copeland, K.C. Effect of obesity and exercise training on plasma amino acids and amino metabolites in American Indian adolescents. J. Clin. Endocrinol. Metab. 2019, 104, 3249–3261. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hou, Y.; Dai, Z.; Hu, C.-A.A.; Wu, G. Metabolism, nutrition, and redox signaling of hydroxyproline. Antioxid. Redox Signal. 2019, 30, 674–682. [Google Scholar] [CrossRef]
- Wu, G. Important roles of dietary taurine, creatine, carnosine, anserine and 4-hydroxyproline in human nutrition and health. Amino Acids 2020, 52, 329–360. [Google Scholar] [CrossRef]
- Wang, C.; Feng, R.; Sun, D.; Li, Y.; Bi, X.; Sun, C. Metabolic profiling of urine in young obese men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC/Q-TOF MS). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011, 879, 2871–2876. [Google Scholar] [CrossRef]
- Singh-Mallah, G.; Singh, K.; McMahon, C.D.; Harris, P.; Brimble, M.A.; Thorstensen, E.; Guan, J. Maternally Administered Cyclic Glycine-Proline Increases Insulin-Like Growth Factor-1 Bioavailability and Novelty Recognition in Developing Offspring. Endocrinology 2016, 157, 3130–3139. [Google Scholar] [CrossRef]
- Qu, C.; Peng, Y.; Liu, S. Ferroptosis Biology and Implication in Cancers. Front. Mol. Biosci. 2022, 9, 892957. [Google Scholar] [CrossRef]
- Haam, J.H.; Kim, Y.S.; Cho, D.Y.; Chun, H.; Choi, S.W.; Lee, Y.K.; Lim, S.W.; Koo, H.S.; Kim, M.J. Elevated levels of urine isocitrate, hydroxymethylglutarate, and formiminoglutamate are associated with arterial stiffness in Korean adults. Sci. Rep. 2021, 11, 10180. [Google Scholar] [CrossRef]
- Rose, D.P. Value of Detection of Formiminoglutamic Acid in Urine for the Diagnosis of Malabsorption States. Br. Med. J. 1965, 1, 1031–1034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Verhaar, M.C.; Stroes, E.; Rabelink, T.J. Folates and Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 6–13. [Google Scholar] [CrossRef]
- Luhby, A.L.; Cooperman, J.M.; Teller, D.N. Urinary excretion of formiminoglutamic acid: Application in diagnosis of clinical folic acid deficiency. Am. J. Clin. Nutr. 1959, 7, 397–406. [Google Scholar] [CrossRef] [PubMed]
- McKnight, J.R.; Satterfield, M.C.; Jobgen, W.S.; Smith, S.B.; Spencer, T.E.; Meininger, C.J.; McNeal, C.J.; Wu, G. Beneficial effects of L-arginine on reducing obesity: Potential mechanisms and important implications for human health. Amino Acids 2010, 39, 349–357. [Google Scholar] [CrossRef]
- Fu, W.J.; Haynes, T.E.; Kohli, R.; Hu, J.; Shi, W.; Spencer, T.E.; Carroll, R.J.; Meininger, C.J.; Wu, G. Dietary L-arginine supplementation reduces fat mass in Zucker diabetic fatty rats. J. Nutr. 2005, 135, 714–721. [Google Scholar] [CrossRef]
- Lucotti, P.; Setola, E.; Monti, L.D.; Galluccio, E.; Costa, S.; Sandoli, E.P.; Fermo, I.; Rabaiotti, G.; Gatti, R.; Piatti, P. Beneficial effects of a long-term oral L-arginine treatment added to a hypocaloric diet and exercise training program in obese, insulin-resistant type 2 diabetic patients. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E906–E912. [Google Scholar] [CrossRef] [PubMed]
- Pahlavani, N.; Entezari, M.; Nasiri, M.; Miri, A.; Rezaie, M.; Bagheri-Bidakhavidi, M.; Sadeghi, O. The effect of l-arginine supplementation on body composition and performance in male athletes: A double-blinded randomized clinical trial. Eur. J. Clin. Nutr. 2017, 71, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Marc Rhoads, J.; Carey Satterfield, M.; Smith, S.B.; Spencer, T.E.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef]

| Parameter | Discovery Set (N = 235) | Validation Set (N = 125) | Replication Set (N = 81) |
|---|---|---|---|
| Age (Year) (Mean ± SD) Range | 14.80 ± 1.42 (11.14–16.99) | 14.84 ± 1.31 (11.06–16.99) | 16.08 ± 1.20 (12.9–18) |
| Height (Inch) (Mean ± SD) Range | 66.82 ± 4.10 (56.00–75.67) | 66.97 ± 3.87 (57.25–75.25) | 68.88 ± 3.18 (60–75) |
| Weight (lb) (Mean ± SD) Range | 158.50 ± 54.36 (72.93–334.13) | 149.85 ± 45.14 (78.2–304.6) | 170.64 ± 50.59 (79–320) |
| BMI (Kg/m2) (Mean ± SD) Range | 24.64 ± 7.13 (15.3–49) | 23.28 ± 6.01 (15.5–44.10) | 25.16 ± 6.91 (14.4–45.9) |
| BMI percentile (Mean ± SD) Range | 68.2 ± 30.43 (2–99) | 66.54 ± 30.39 (2–99) | 68.82 ± 29.11 (1–99) |
| BMI z-score (Mean ± SD) Range | 0.82 ± 1.23 (−2.07–3.01) | 0.60 ± 1.18 (−2.04–2.84) | 0.78 ± 1.23 (−3.06–2.96) |
| Positive change in BMI (Kg/m2) Mean/Median (Range) | --- | --- | 0.54/0.6 (−11.6–5.8) |
| Obesity (n (%)) Underweight (<5 percentile) Healthy weight (5 to <85 percentile) Overweight (>85 to <95 percentile) Obese (>95 percentile) | 3 (1.28%) 129 (54.89%) 29 (12.34%) 74 (31.49%) | 4 (3.20%) 70 (56.00%) 23 (18.40%) 28 (22.40%) | 3 (3.70%) 47 (58.02%) 12 (14.81%) 19 (23.46%) |
| Race (n (%)) White Black Hispanic Multiracial Others | 176 (74.47%) 36 (15.35%) 8 (3.39%) 12 (5.08%) 4 (1.69%) | 99 (79.20%) 13 (10.40%) 4 (3.20%) 7 (5.60%) 2 (1.60%) | 58 (71.60%) 13 (16.05%) 4 (4.94%) 5 (6.17%) 1 (1.23%) |
| County (n (%)) Franklin Non-Franklin | 121 (51.49%) 114 (48.51%) | 64 (51.20%) 61 (48.80%) | 44(54.32%) 37(45.68%) |
| Total energy intake (kcals) (Mean/Median) Range | 1883.84/1745.54 (204.05–5209.14) | 1937.77/1873.41 (462.5–5549.48) | 1778.03/1507.12 (236.91–4589.2) |
| ID | Metabolite | Mode | MZ | RT | Adduct | HMDB ID | Super-Class | Class | Sub-Class | Discovery Set | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate (95%CI) | p-Value | FDR adj | ||||||||||
| neg_FT17207 | 3′-Sialyllactose | Neg | 632.2 | 0.67 | M − H | HMDB0000825 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.05 (0.03–0.06) | <0.0001 | 5.82 × 10−9 |
| pos_FT29354 | 3′-Sialyllactose | Pos | 656.2 | 0.67 | M + Na | HMDB0000825 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.06 (0.04–0.07) | <0.0001 | 6.56 × 10−9 |
| pos_FT04847 | Estrone sulfate | Pos | 176.06 | 0.58 | M + 2H | HMDB0001425 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Sulfated steroids | −0.08 (−0.1–−0.05) | <0.0001 | 1.38 × 10−7 |
| pos_FT13024 | N-Ribosylhistidine | Pos | 288.11 | 0.53 | M + H | HMDB0002089 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.16 (0.11–0.21) | <0.0001 | 1.38 × 10−7 |
| pos_FT04858 | Citrulline | Pos | 176.1 | 0.78 | M + H | HMDB0000904 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.06 (0.04–0.08) | <0.0001 | 5.77 × 10−7 |
| neg_FT15762 | PA(22:5(4Z,7Z,10Z,13Z,19Z)-O(16,17)/2:0) | Neg | 539.24 | 6.53 | M − H | HMDB0266570 | 0.08 (0.06–0.11) | <0.0001 | 5.77 × 10−7 | |||
| pos_FT27056 | Tetrahydroaldosterone-3-glucuronide | Pos | 563.24 | 6.57 | M + Na | HMDB0010357 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Steroidal glycosides | 0.07 (0.05–0.1) | <0.0001 | 8.64 × 10−7 |
| neg_FT01967 | Citrulline | Neg | 174.08 | 0.81 | M − H | HMDB0000904 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.13 (0.08–0.17) | <0.0001 | 6.28 × 10−6 |
| neg_FT15806 | Cortolone-3-glucuronide | Neg | 541.26 | 6.18 | M − H | HMDB0010320 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Steroidal glycosides | 0.05 (0.03–0.07) | <0.0001 | 1.34 × 10−5 |
| neg_FT01921 | Formiminoglutamic acid | Neg | 173.05 | 1.31 | M − H | HMDB0000854 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.13 (0.08–0.18) | <0.0001 | 4.66 × 10−5 |
| pos_FT04673 | Glycylproline | Pos | 173.09 | 0.75 | M + H | HMDB0000721 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.06 (−0.09–−0.04) | <0.0001 | 7.46 × 10−5 |
| neg_FT02197 | Galactitol | Neg | 181.07 | 1.81 | M − H | HMDB0000107 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.09 (0.05–0.13) | <0.0001 | 7.46 × 10−5 |
| pos_FT27104 | Cortolone-3-glucuronide | Pos | 565.26 | 6.1 | M + Na | HMDB0010320 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Steroidal glycosides | 0.05 (0.03–0.08) | <0.0001 | 9.30 × 10−5 |
| pos_FT09630 | 4-Vinylsyringol | Pos | 243.09 | 0.73 | M + H | HMDB0301746 | Phenylpropanoids and polyketides | Stilbenes | −0.05 (−0.07–−0.03) | <0.0001 | 9.96 × 10−5 | |
| neg_FT07183 | Fludiazepam | Neg | 301.05 | 2.36 | M − H | HMDB0015513 | Organoheterocyclic compounds | Benzodiazepines | 1,4-benzodiazepines | 0.12 (0.07–0.17) | <0.0001 | 1.38 × 10−4 |
| neg_FT02892 | Adipoylglycine | Neg | 202.07 | 3.23 | M − H | HMDB0240731 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.06 (0.04–0.09) | <0.0001 | 1.57 × 10−4 |
| neg_FT11170 | 6-Hydroxymelatonin glucuronide | Neg | 389.18 | 6.34 | M + Cl | HMDB0060786 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.17 (0.1–0.24) | <0.0001 | 2.29 × 10−4 |
| neg_FT15862 | N-Acetylgalactosaminyl lactose | Neg | 544.18 | 0.71 | M − H | HMDB0041622 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.08 (0.04–0.11) | <0.0001 | 2.39 × 10−4 |
| neg_FT02148 | 3-Chlorotyrosine | Neg | 180.06 | 1.81 | M + Cl | HMDB0001885 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.05 (0.03–0.08) | <0.0001 | 2.55 × 10−4 |
| pos_FT18573 | Cephalexin | Pos | 370.08 | 5.82 | M + Na | HMDB0014707 | Organoheterocyclic compounds | Lactams | Beta lactams | 0.12 (0.07–0.17) | <0.0001 | 3.66 × 10−4 |
| neg_FT08222 | Dihyroxy-1H-indole glucuronide I | Neg | 324.07 | 3.91 | M − H | HMDB0059997 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.07 (0.04–0.1) | <0.0001 | 3.66 × 10−4 |
| neg_FT14008 | 3-alpha-hydroxy-5-alpha-androstane-17-one 3-D-glucuronide | Neg | 465.24 | 7.84 | M − H | HMDB0010365 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Steroidal glycosides | 0.06 (0.03–0.09) | <0.0001 | 3.84 × 10−4 |
| neg_FT14066 | Clozapine glucuronide | Neg | 467.19 | 6.92 | M + Cl | HMDB0060901 | Organic oxygen compounds | Organooxygen compounds | Carbonyl compounds | 0.08 (0.05–0.12) | <0.0001 | 3.84 × 10−4 |
| neg_FT01695 | Quinolinic acid | Neg | 166.01 | 1.17 | M − H | HMDB0000232 | Organoheterocyclic compounds | Pyridines and derivatives | Pyridinecarboxylic acids and derivatives | 0.05 (0.02–0.07) | <0.0001 | 3.84 × 10−4 |
| neg_FT08456 | Hydroxytyrosol 3′-glucuronide | Neg | 329.08 | 3.78 | M − H | HMDB0240531 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.09 (0.05–0.13) | <0.0001 | 4.50 × 10−4 |
| neg_FT01863 | Glycylproline | Neg | 171.07 | 0.91 | M − H | HMDB0000721 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.08 (−0.11–−0.04) | <0.0001 | 4.68 × 10−4 |
| neg_FT09551 | 5-Caffeoylquinic acid | Neg | 353.08 | 6.84 | M − H | HMDB0240477 | Organic oxygen compounds | Organooxygen compounds | Alcohols and polyols | 0.13 (0.07–0.19) | <0.0001 | 4.68 × 10−4 |
| neg_FT00341 | (R)-3-Hydroxyisobutyric acid | Neg | 103.04 | 2.02 | M − H | HMDB0000336 | Organic acids and derivatives | Hydroxy acids and derivatives | Beta hydroxy acids and derivatives | 0.04 (0.02–0.06) | <0.0001 | 4.75 × 10−4 |
| neg_FT02346 | 1-(Malonylamino)cyclopropanecarboxylic acid | Neg | 186.04 | 3.48 | M − H | HMDB0031700 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.08 (0.04–0.12) | <0.0001 | 7.96 × 10−4 |
| neg_FT14465 | 11-beta-Hydroxyandrosterone-3-glucuronide | Neg | 481.24 | 6.63 | M − H | HMDB0010351 | Organoheterocyclic compounds | Indoles and derivatives | Hydroxyindoles | 0.05 (0.02–0.07) | <0.0001 | 8.07 × 10−4 |
| ID | Metabolite | Mode | MZ | RT | Adduct | HMDB ID | Super-Class | Class | Sub-Class | Validation Set | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate (95%CI) | p-Value | FDR adj | ||||||||||
| pos_FT04673 | Glycylproline | Pos | 173.09 | 0.75 | M + H | HMDB0000721 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.105 (−0.145–0.064) | <0.0001 | 2.29 × 10−4 |
| neg_FT01967 | Citrulline | Neg | 174.08 | 0.81 | M − H | HMDB0000904 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.117 (0.065–0.168) | <0.0001 | 0.002 |
| pos_FT09630 | 4-Vinylsyringol | Pos | 243.09 | 0.73 | M + H | HMDB0301746 | Phenylpropanoids and polyketides | Stilbenes | −0.079 (−0.117–0.041) | <0.0001 | 0.004 | |
| neg_FT01863 | Glycylproline | Neg | 171.07 | 0.91 | M − H | HMDB0000721 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.076 (−0.114–0.039) | <0.0001 | 0.005 |
| neg_FT17207 | 3′-Sialyllactose | Neg | 632.2 | 0.67 | M − H | HMDB0000825 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.055 (0.026–0.084) | 0.0003 | 0.012 |
| pos_FT04847 | Estrone sulfate | Pos | 176.06 | 0.58 | M + 2H | HMDB0001425 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Sulfated steroids | −0.056 (−0.087–0.025) | 0.0004 | 0.014 |
| pos_FT10083 | Carnosine | Pos | 249.09 | 0.49 | M + Na | HMDB0000033 | Organic acids and derivatives | Peptidomimetics | Hybrid peptides | −0.151 (−0.234–0.069) | 0.0004 | 0.014 |
| neg_FT01921 | Formiminoglutamic acid | Neg | 173.05 | 1.31 | M − H | HMDB0000854 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.075 (0.032–0.118) | 0.0007 | 0.020 |
| pos_FT04858 | Citrulline | Pos | 176.1 | 0.78 | M + H | HMDB0000904 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.048 (0.019–0.077) | 0.001 | 0.028 |
| neg_FT00788 | 4-Hydroxyproline | Neg | 130.05 | 1.4 | M − H | HMDB0000725 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.082 (0.031–0.132) | 0.002 | 0.033 |
| pos_FT09878 | Hydroxyprolyl-Asparagine | Pos | 246.1 | 0.69 | M + H | HMDB0028858 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.059 (−0.097–0.022) | 0.002 | 0.033 |
| pos_FT13592 | 2-Hexenoylcarnitine | Pos | 296.12 | 6.85 | M + K | HMDB0013161 | Lipids and lipid-like molecules | Fatty Acyls | Fatty acid esters | 0.104 (0.04–0.167) | 0.002 | 0.033 |
| pos_FT03157 | L-Glutamine | Pos | 147.07 | 0.52 | M + H | HMDB0000641 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.065 (−0.106–0.024) | 0.002 | 0.033 |
| neg_FT05602 | Inosine | Neg | 267.07 | 0.67 | M − H | HMDB0000195 | Nucleosides, nucleotides, and analogues | Purine nucleosides | 0.038 (0.011–0.064) | 0.005 | 0.073 | |
| neg_FT08407 | N-(2-Hydroxyphenyl)acetamide glucuronide | Neg | 328.06 | 3.81 | M − H | HMDB0240542 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.081 (0.023–0.139) | 0.006 | 0.092 |
| pos_FT15625 | Galactosylhydroxylysine | Pos | 325.16 | 0.5 | M + H | HMDB0000600 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.048 (−0.084–0.013) | 0.007 | 0.096 |
| ID | Metabolites | Mode | MZ | RT | Adduct | HMDB ID | Super-Class | Class | Sub-Class | Replication Set | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate (95%CI) | p-Value | FDR adj | ||||||||||
| neg_FT01863 | Glycylproline | Neg | 173.09 | 0.75 | M + H | HMDB0000721 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.018 (−0.029–0.007) | 0.002 | 0.031 |
| neg_FT17207 | 3′-Sialyllactose | Neg | 632.2 | 0.67 | M − H | HMDB0000825 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.009 (0.002–0.016) | 0.006 | 0.038 |
| neg_FT01921 | Formiminoglutamic acid | Neg | 173.05 | 1.31 | M − H | HMDB0000854 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.016 (0.004–0.028) | 0.008 | 0.038 |
| pos_FT04673 | Glycylproline | Pos | 171.07 | 0.91 | M − H | HMDB0000721 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.014 (−0.025–0.003) | 0.01 | 0.038 |
| neg_FT00788 | 4-Hydroxyproline | Neg | 130.05 | 1.4 | M − H | HMDB0000725 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.016 (0.003–0.03) | 0.016 | 0.043 |
| pos_FT04858 | Citrulline | Pos | 174.08 | 0.81 | M − H | HMDB0000904 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.01 (0.002–0.018) | 0.013 | 0.043 |
| pos_FT09630 | 4-Vinylsyringol | Pos | 243.09 | 0.73 | M + H | HMDB0301746 | Phenylpropanoids and polyketides | Stilbenes | −0.01 (−0.02–0.001) | 0.022 | 0.049 | |
| neg_FT01967 | Citrulline | Neg | 176.1 | 0.78 | M + H | HMDB0000904 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | 0.012 (0.001–0.023) | 0.025 | 0.049 |
| neg_FT05602 | Inosine | Neg | 267.07 | 0.67 | M − H | HMDB0000195 | Nucleosides, nucleotides, and analogues | Purine nucleosides | 0.005 (0.0004–0.01) | 0.033 | 0.058 | |
| pos_FT10083 | Carnosine | Pos | 249.09 | 0.49 | M + Na | HMDB0000033 | Organic acids and derivatives | Peptidomimetics | Hybrid peptides | −0.015 (−0.031–0.0004) | 0.056 | 0.082 |
| neg_FT08407 | N-(2-Hydroxyphenyl)acetamide glucuronide | Neg | 328.06 | 3.81 | M − H | HMDB0240542 | Organic oxygen compounds | Organooxygen compounds | Carbohydrates and carbohydrate conjugates | 0.015 (−0.0004–0.032) | 0.056 | 0.082 |
| pos_FT03157 | L-Glutamine | Pos | 147.07 | 0.52 | M + H | HMDB0000641 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.006 (−0.014–0.0001) | 0.086 | 0.114 |
| pos_FT04847 | Estrone sulfate | Pos | 176.06 | 0.58 | M + 2H | HMDB0001425 | Lipids and lipid-like molecules | Steroids and steroid derivatives | Sulfated steroids | −0.004 (−0.011–0.001) | 0.139 | 0.171 |
| pos_FT09878 | Hydroxyprolyl-Asparagine | Pos | 246.1 | 0.69 | M + H | HMDB0028858 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.001 (−0.009–0.006) | 0.751 | 0.858 |
| pos_FT15625 | Galactosylhydroxylysine | Pos | 325.16 | 0.5 | M + H | HMDB0000600 | Organic acids and derivatives | Carboxylic acids and derivatives | Amino acids, peptides, and analogues | −0.0008 (−0.008–0.007) | 0.83 | 0.885 |
| pos_FT13592 | 2-Hexenoylcarnitine | Pos | 296.12 | 6.85 | M + K | HMDB0013161 | Lipids and lipid-like molecules | Fatty Acyls | Fatty acid esters | 0.00 (−0.017–0.017) | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Kinnebrew, G.; Hsu, P.-C.; Weng, D.Y.; Song, M.-A.; Reisinger, S.A.; McElroy, J.P.; Keller-Hamilton, B.; Ferketich, A.K.; Freudenheim, J.L.; et al. Untargeted Metabolomics and Body Mass in Adolescents: A Cross-Sectional and Longitudinal Analysis. Metabolites 2023, 13, 899. https://doi.org/10.3390/metabo13080899
Singh A, Kinnebrew G, Hsu P-C, Weng DY, Song M-A, Reisinger SA, McElroy JP, Keller-Hamilton B, Ferketich AK, Freudenheim JL, et al. Untargeted Metabolomics and Body Mass in Adolescents: A Cross-Sectional and Longitudinal Analysis. Metabolites. 2023; 13(8):899. https://doi.org/10.3390/metabo13080899
Chicago/Turabian StyleSingh, Amarnath, Garrett Kinnebrew, Ping-Ching Hsu, Daniel Y. Weng, Min-Ae Song, Sarah A. Reisinger, Joseph P. McElroy, Brittney Keller-Hamilton, Amy K. Ferketich, Jo L. Freudenheim, and et al. 2023. "Untargeted Metabolomics and Body Mass in Adolescents: A Cross-Sectional and Longitudinal Analysis" Metabolites 13, no. 8: 899. https://doi.org/10.3390/metabo13080899
APA StyleSingh, A., Kinnebrew, G., Hsu, P.-C., Weng, D. Y., Song, M.-A., Reisinger, S. A., McElroy, J. P., Keller-Hamilton, B., Ferketich, A. K., Freudenheim, J. L., & Shields, P. G. (2023). Untargeted Metabolomics and Body Mass in Adolescents: A Cross-Sectional and Longitudinal Analysis. Metabolites, 13(8), 899. https://doi.org/10.3390/metabo13080899











