Metabolomics Analysis Reveals Altered Metabolic Pathways and Response to Doxorubicin in Drug-Resistant Triple-Negative Breast Cancer Cells
Abstract
1. Introduction
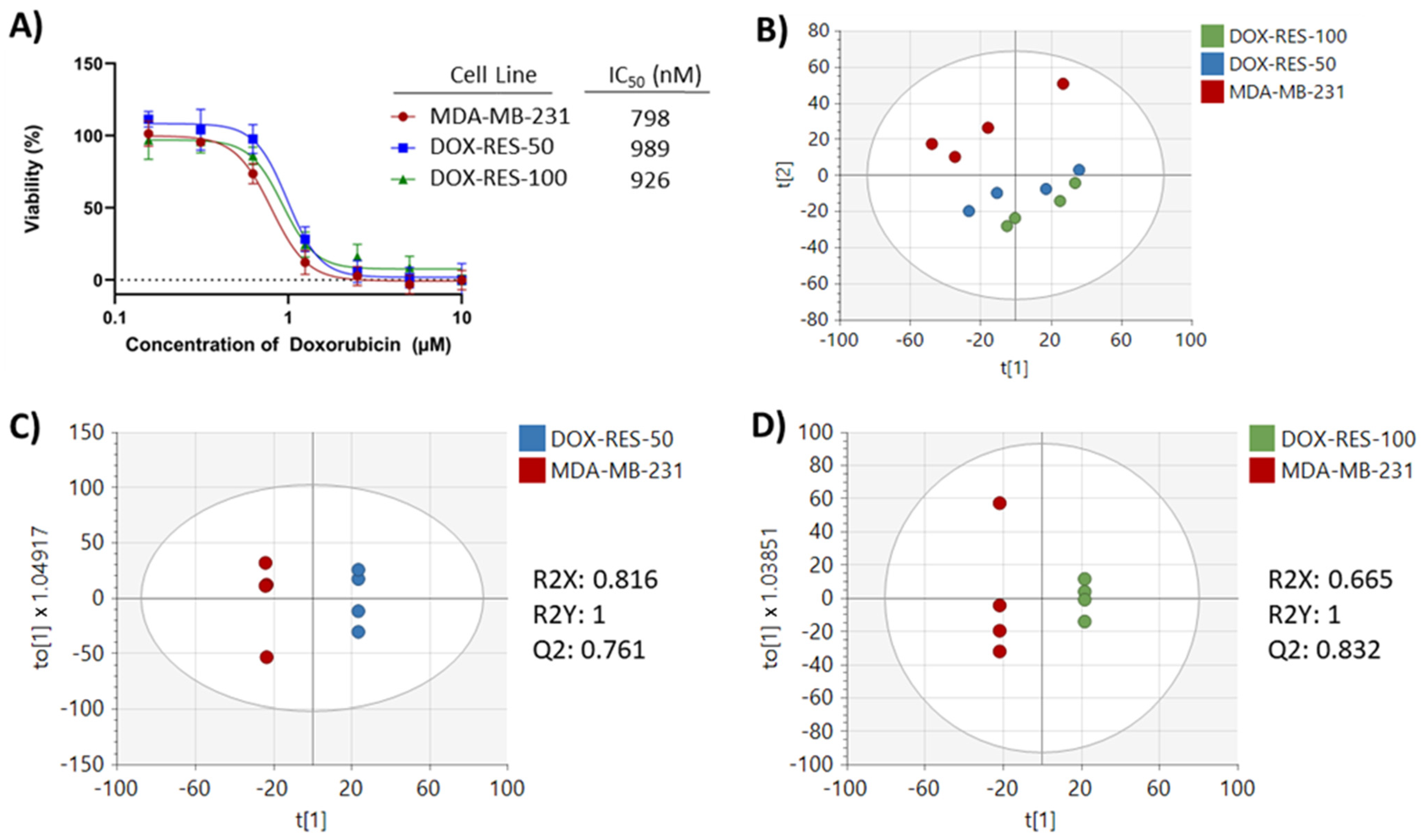
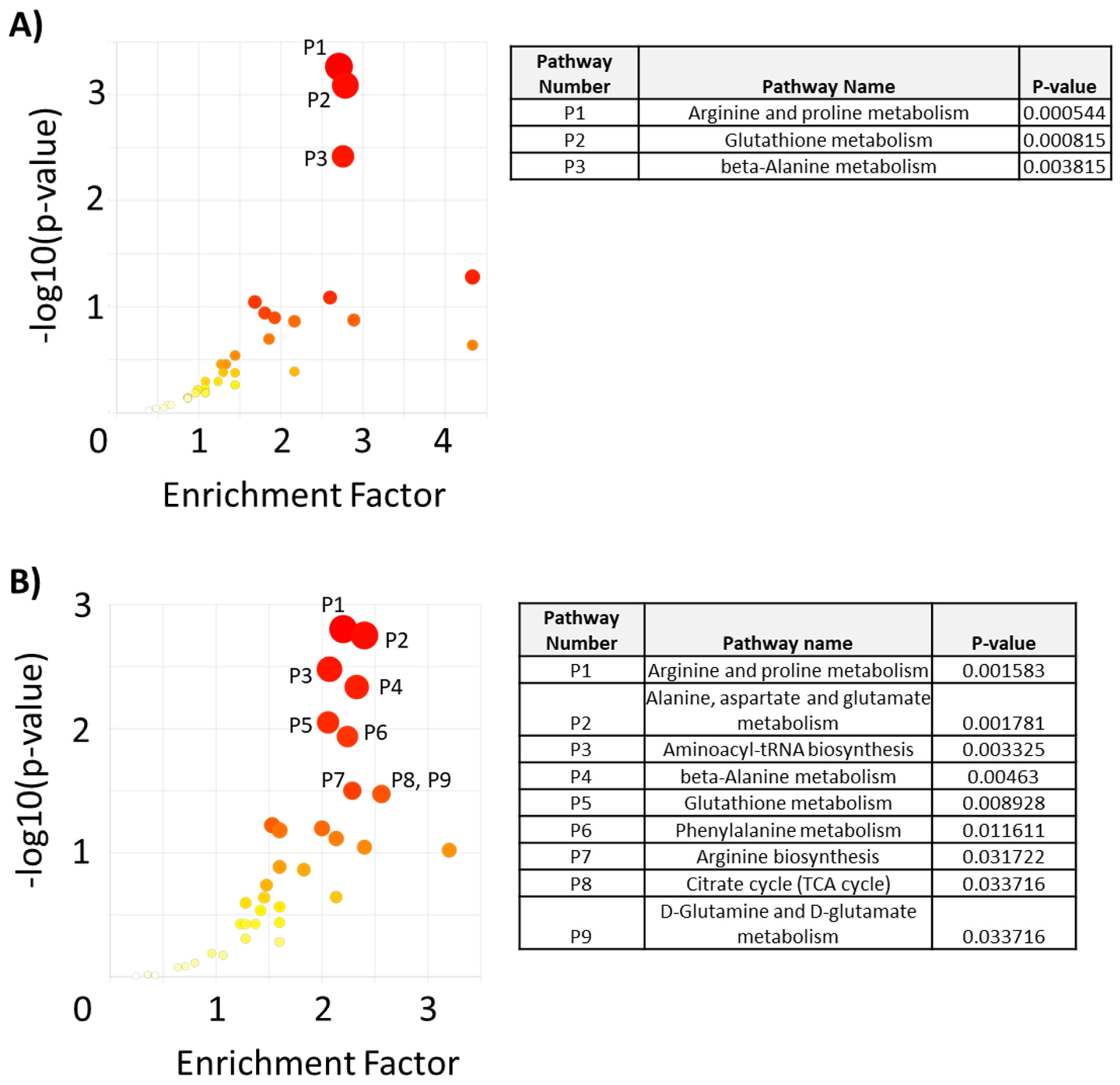
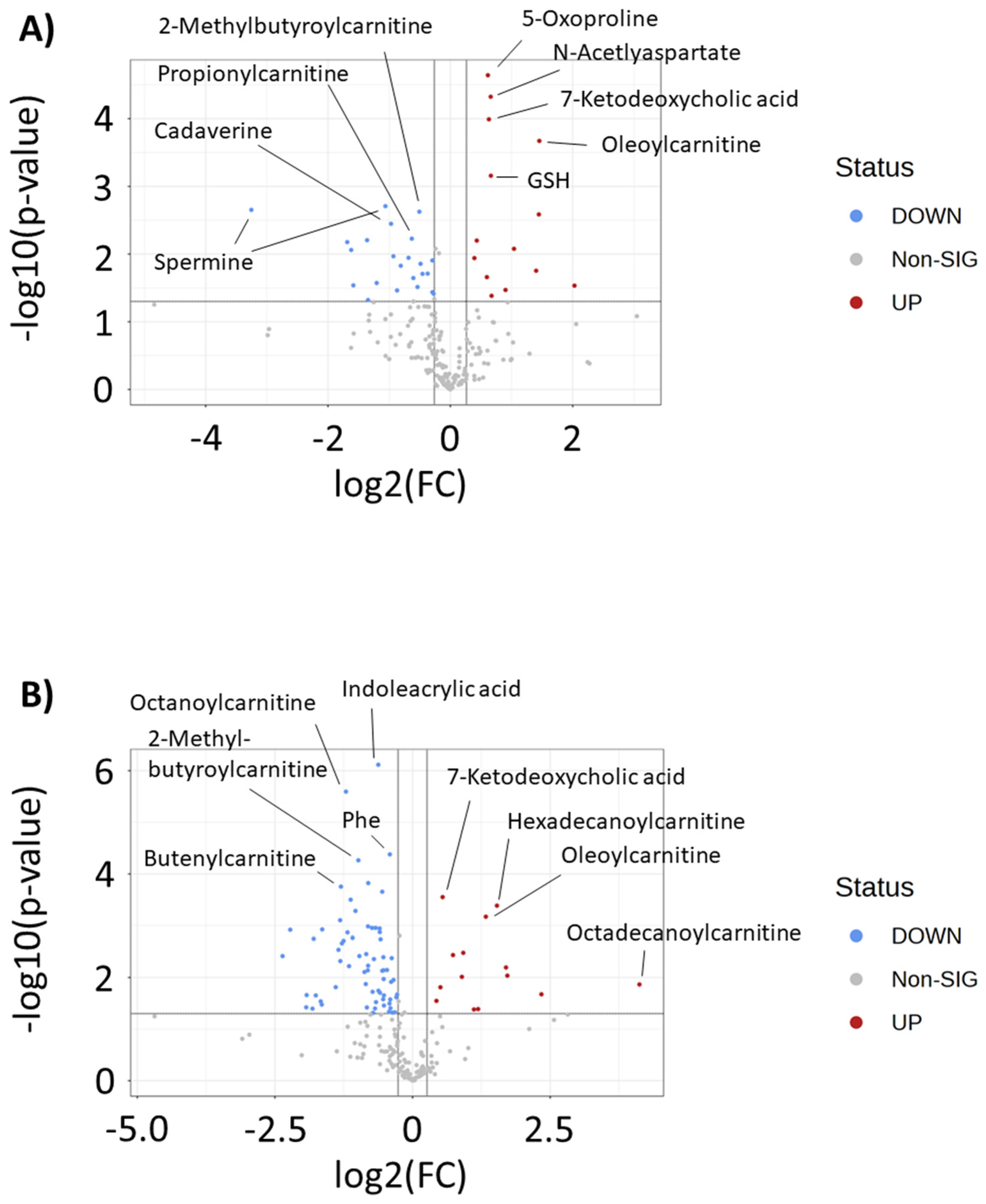
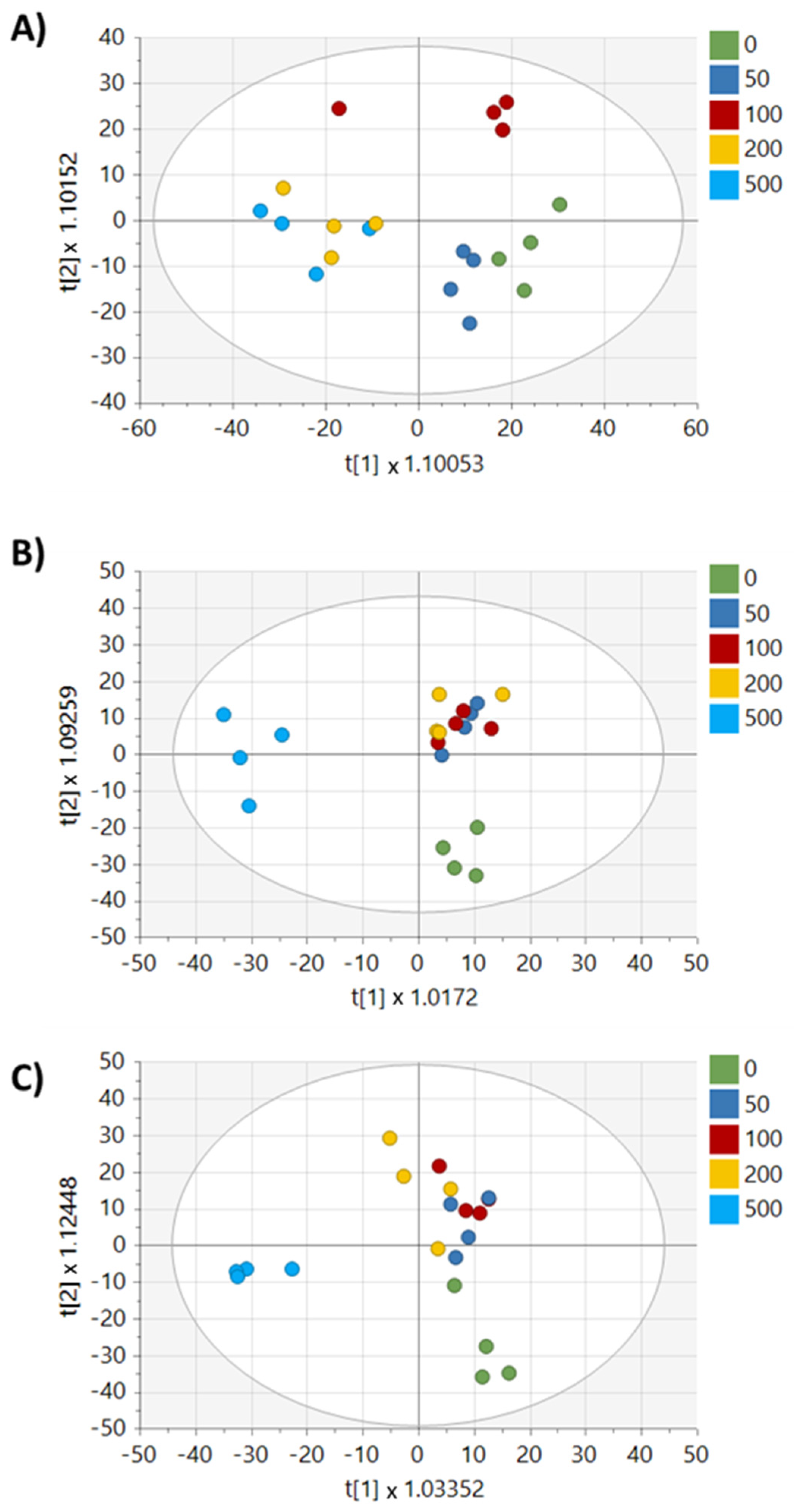
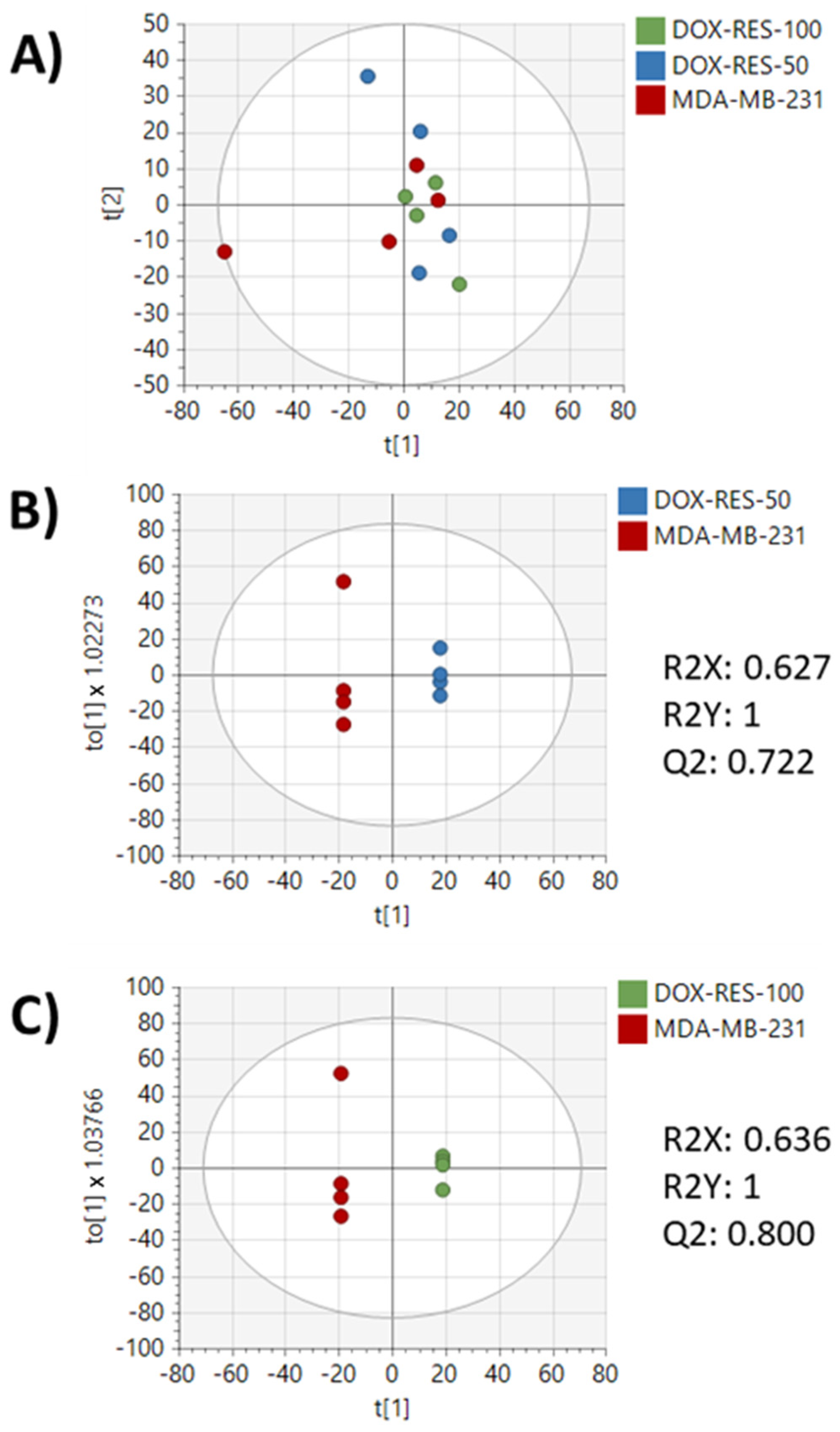
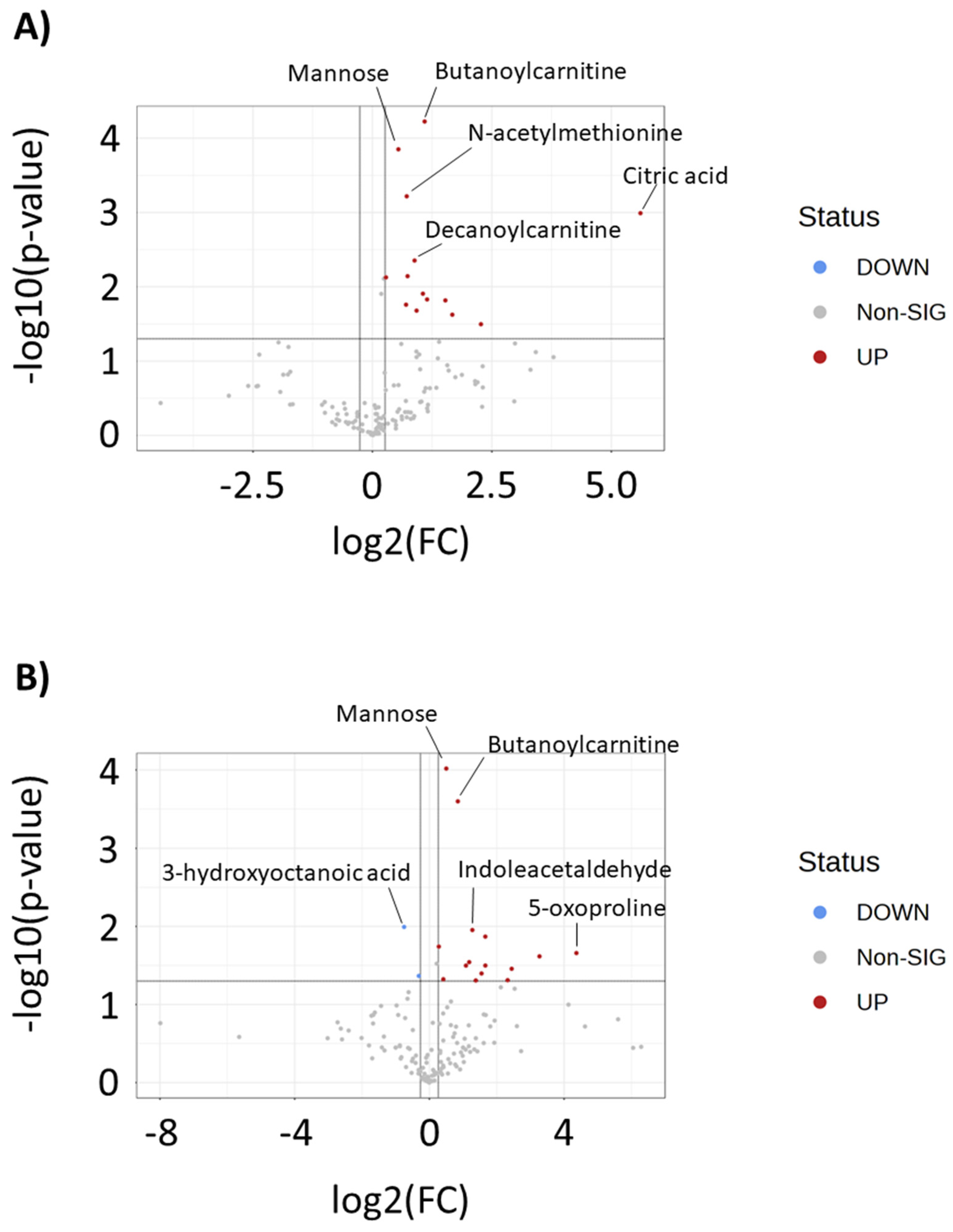
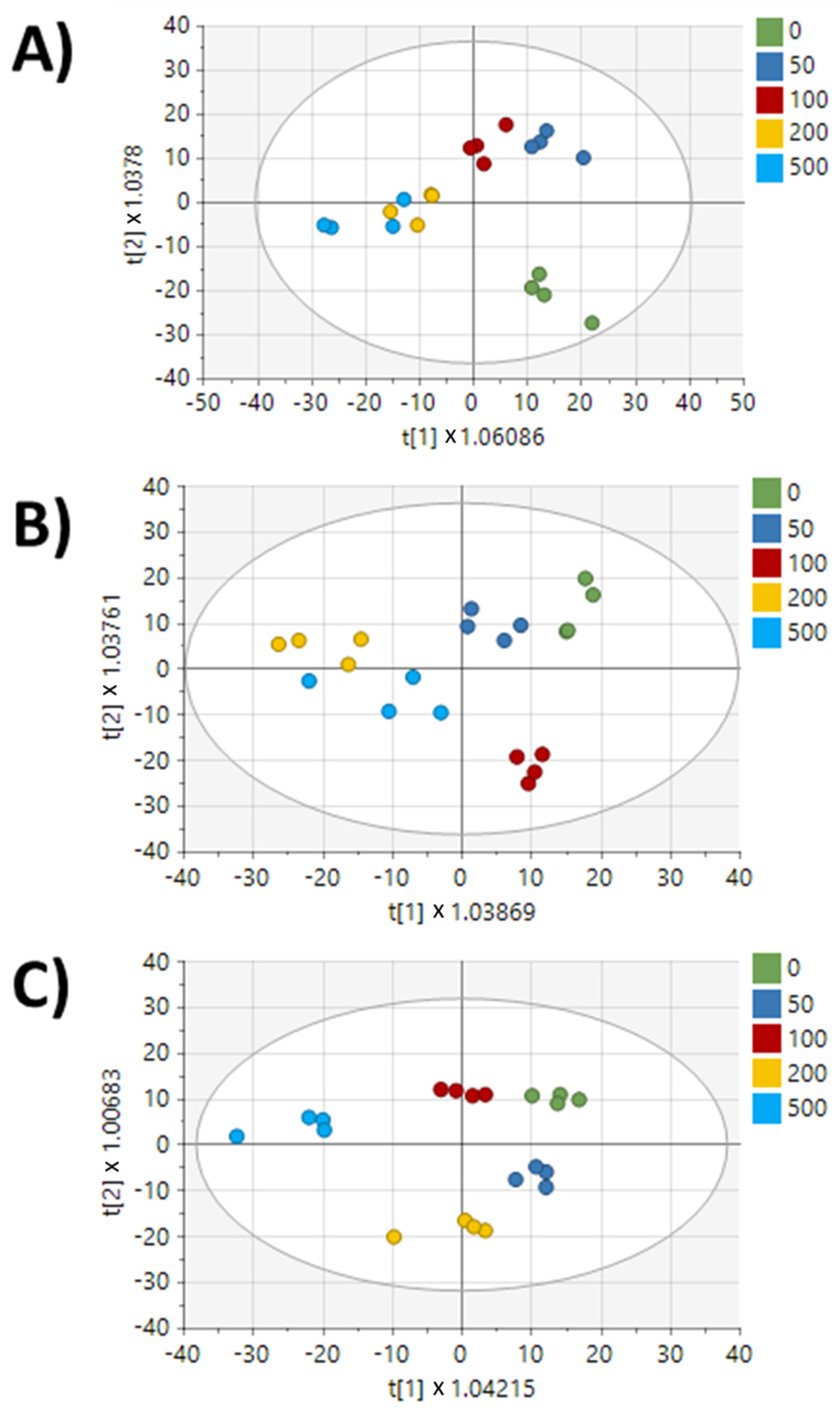
2. Results
3. Discussion
4. Methods
4.1. Chemical Reagents
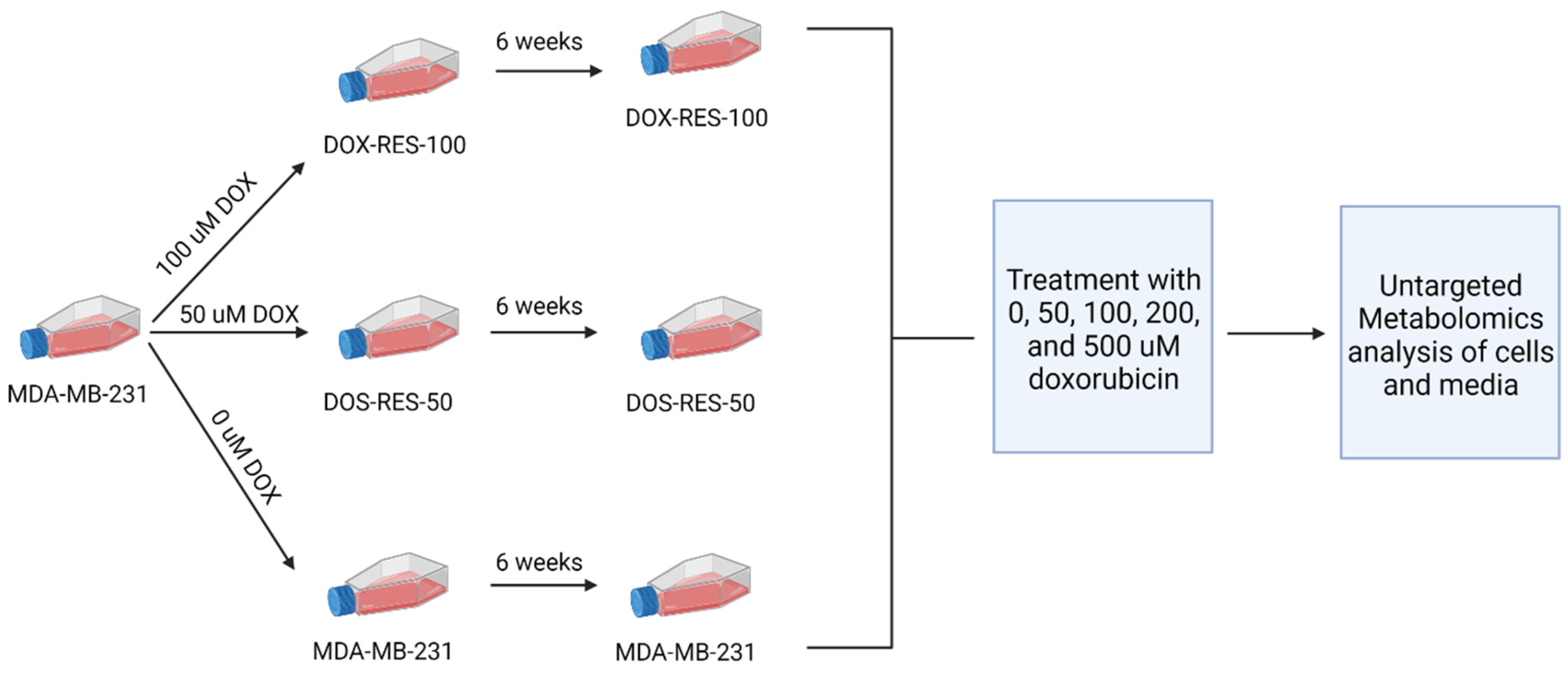
4.2. Cell Culture and Establishment of Doxorubicin Resistance
4.3. Doxorubicin Treatment and Metabolite Extraction
4.4. UHPLC-HRMS Metabolomics Data Acquisition and Preprocessing
4.5. Compound Identification/Annotation
4.6. Multivariate, Univariate, and Pathway Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Zagami, P.; Carey, L.A. Triple negative breast cancer: Pitfalls and progress. Breast Cancer 2022, 8, 95. [Google Scholar] [CrossRef]
- Echeverria, G.V.; Ge, Z.; Seth, S.; Zhang, X.; Jeter-jones, S.; Zhou, X.; Cai, S.; Tu, Y.; Mccoy, A.; Peoples, M.; et al. Resistance to neoadjuvant chemotherapy in triple negative breast cancer mediated by a reversible drug-tolerant state. Sci. Transl. Med. 2019, 11, eaav0936. [Google Scholar] [CrossRef]
- Lips, E.H.; Michaut, M.; Hoogstraat, M.; Mulder, L.; Besselink, N.J.M.; Koudijs, M.J.; Cuppen, E.; Voest, E.E.; Bernards, R.; Nederlof, P.M.; et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Sabrina Sapari, N.; Miao, H.; Hartman, M.; Loh, M.; Chng, W.J.; Iau, P.; Ahmad Buhari, S.; Soong, R.; Lee, S.C. High-throughput mutation profiling changes before and 3 weeks after chemotherapy in newly diagnosed breast cancer patients. PLoS ONE 2015, 10, e0142466. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Furlanetto, J.; Loibl, S. Optimal Systemic Treatment for Early Triple-Negative Breast Cancer. Breast Care 2020, 15, 217–226. [Google Scholar] [CrossRef]
- Mirzaei, S.; Gholami, M.H.; Hashemi, F.; Zabolian, A.; Farahani, M.V.; Hushmandi, K.; Zarrabi, A.; Goldman, A.; Ashrafizadeh, M.; Orive, G. Advances in understanding the role of P-gp in doxorubicin resistance: Molecular pathways, therapeutic strategies, and prospects. Drug Discov. Today 2022, 27, 436–455. [Google Scholar] [CrossRef]
- Huang, J.F.; Wen, C.J.; Zhao, G.Z.; Dai, Y.; Li, Y.; Wu, L.X.; Zhou, H.H. Overexpression of ABCB4 contributes to acquired doxorubicin resistance in breast cancer cells in vitro. Cancer Chemother. Pharmacol. 2018, 82, 199–210. [Google Scholar] [CrossRef]
- Barata, I.S.; Gomes, B.C.; Rodrigues, A.S.; Rueff, J.; Kranendonk, M.; Esteves, F. The Complex Dynamic of Phase I Drug Metabolism in the Early Stages of Doxorubicin Resistance in Breast Cancer Cells. Genes 2022, 13, 1977. [Google Scholar] [CrossRef]
- Nedeljkovi, M.; Damjanovic, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer—How We Can Rise to the Challenge. Cell 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.R.; Lu, D.Y.; Lin, H.Y.; Yeh, W.L. Mesenchymal stem cell-induced doxorubicin resistance in triple negative breast cancer. Biomed Res. Int. 2014, 2014, 532161. [Google Scholar] [CrossRef]
- Zaal, E.A.; Berkers, C.R. The influence of metabolism on drug response in cancer. Front. Oncol. 2018, 8, 500. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Zhao, Y.; Butler, E.B.; Tan, M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013, 4, e532. [Google Scholar] [CrossRef]
- Zhang, Z.; Bao, C.; Jiang, L.; Wang, S.; Wang, K.; Lu, C.; Fang, H. When cancer drug resistance meets metabolomics (bulk, single-cell and/or spatial): Progress, potential, and perspective. Front. Oncol. 2023, 12, 1054233. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.; Eustace, A.J.; Busschots, S.; Breen, L.; Crown, J.; Clynes, M.; O’Donovan, N.; Stordal, B. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: A practical guide with case studies. Front. Oncol. 2014, 4, 40. [Google Scholar] [CrossRef]
- Kuo, M.T.; Chen, H.H.W.; Feun, L.G.; Savaraj, N. Targeting the proline–glutamine–asparagine–arginine metabolic axis in amino acid starvation cancer therapy. Pharmaceuticals 2021, 14, 72. [Google Scholar] [CrossRef]
- Hatem, E.; El Banna, N.; Huang, M.E. Multifaceted Roles of Glutathione and Glutathione-Based Systems in Carcinogenesis and Anticancer Drug Resistance. Antioxid. Redox Signal. 2017, 27, 1217–1234. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.C.; Han, J.M. Amino acid metabolism in cancer drug resistance. Cells 2022, 11, 140. [Google Scholar] [CrossRef]
- Pandurangan, M.; Enkhtaivan, G.; Mistry, B.; Patel, R.V.; Moon, S.; Kim, D.H. β-Alanine intercede metabolic recovery for amelioration of human cervical and renal tumors. Amino Acids 2017, 49, 1373–1380. [Google Scholar] [CrossRef]
- Dambrova, M.; Makrecka-Kuka, M.; Kuka, J.; Vilskersts, R.; Nordberg, D.; Attwood, M.M.; Smesny, S.; Sen, Z.D.; Guo, A.C.; Oler, E.; et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol. Rev. 2022, 74, 506–551. [Google Scholar] [CrossRef] [PubMed]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity review-article. Cell Death Dis. 2018, 9, 228. [Google Scholar] [CrossRef]
- Koves, T.R.; Ussher, J.R.; Noland, R.C.; Slentz, D.; Mosedale, M.; Ilkayeva, O.; Bain, J.; Stevens, R.; Dyck, J.R.B.; Newgard, C.B.; et al. Mitochondrial Overload and Incomplete Fatty Acid Oxidation Contribute to Skeletal Muscle Insulin Resistance. Cell Metab. 2008, 7, 45–56. [Google Scholar] [CrossRef]
- Muoio, D.M.; Neufer, P.D. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012, 15, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Rushing, B.R.; Wiggs, A.; Molina, S.; Schroder, M.; Sumner, S. Metabolomics Analysis Reveals Novel Targets of Chemosensitizing Polyphenols and Omega-3 Polyunsaturated Fatty Acids in Triple Negative Breast Cancer Cells. Int. J. Mol. Med. 2023, 24, 4406. [Google Scholar] [CrossRef]
- Wajner, M.; Amaral, A.U. Mitochondrial dysfunction in fatty acid oxidation disorders: Insights from human and animal studies. Biosci. Rep. 2016, 36, e00281. [Google Scholar] [CrossRef]
- Furuno, T.; Kanno, T.; Arita, K.; Asami, M.; Utsumi, T.; Doi, Y.; Inoue, M.; Utsumi, K. Roles of long chain fatty acids and carnitine in mitochondrial membrane permeability transition. Biochem. Pharmacol. 2001, 62, 1037–1046. [Google Scholar] [CrossRef]
- Nicoletto, R.E.; Ofner, C.M. Cytotoxic mechanisms of doxorubicin at clinically relevant concentrations in breast cancer cells. Cancer Chemother. Pharmacol. 2022, 89, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Wu, P. 5′-Methylthioadenosine and Cancer: Old Molecules, New Understanding. J. Cancer 2019, 10, 927–936. [Google Scholar] [CrossRef]
- Bigaud, E.; Corrales, F.J. Methylthioadenosine (MTA) regulates liver cells proteome and methylproteome: Implications in liver biology and disease. Mol. Cell. Proteom. 2016, 15, 1498–1510. [Google Scholar] [CrossRef] [PubMed]
- Rushing, B.R.; Schroder, M.; Sumner, S.C.J. Comparison of Lysis and Detachment Sample Preparation Methods for Cultured Triple-Negative Breast Cancer Cells Using UHPLC–HRMS-Based Metabolomics. Metabolites 2022, 12, 168. [Google Scholar] [CrossRef]
- Rushing, B.R.; Tilley, S.; Molina, S.; Schroder, M.; Sumner, S. Commonalities in Metabolic Reprogramming between Tobacco Use and Oral Cancer. Int. J. Environ. Res. Public Health 2022, 19, 10261. [Google Scholar] [CrossRef]
- Rushing, B.R.; Fogle, H.M.; Sharma, J.; You, M.; Mccormac, J.P.; Molina, S.; Sumner, S.; Krupenko, N.I.; Krupenko, S.A. Exploratory Metabolomics Underscores the Folate Enzyme ALDH1L1 as a Regulator of Glycine and Methylation Reactions. Molecules 2022, 27, 8394. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Rushing, B.; Schroder, M.; Sumner, S.; Kay, C.D. Exploring the Contribution of (Poly)phenols to the Dietary Exposome Using High Resolution Mass Spectrometry Untargeted Metabolomics. Mol. Nutr. Food Res. 2022, 66, 2100922. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Rushing, B.R.; Harris, S.E.; McRitchie, S.L.; Jones, J.C.; Dominguez, D.; Sumner, S.J.; Dohlman, H.G. Multi-omics analysis of glucose-mediated signaling by a moonlighting Gβ protein Asc1/RACK1. PLoS Genet. 2021, 17, e1009640. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Rushing, B.R.; Hall, M.S.; Helke, K.L.; McRitchie, S.L.; Krupenko, N.I.; Sumner, S.J.; Krupenko, S.A. Sex-Specific Metabolic Effects of Dietary Folate Withdrawal in Wild-Type and Aldh1l1 Knockout Mice. Metabolites 2022, 12, 454. [Google Scholar] [CrossRef]
- Välikangas, T.; Suomi, T.; Elo, L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, 388–396. [Google Scholar] [CrossRef]
- Bender, R.; Lange, S. Adjusting for multiple testing—When and how? J. Clin. Epidemiol. 2001, 54, 343–349. [Google Scholar] [CrossRef]
| Metabolite (Name_Ontology Level_Retention Time_Mass) | MDA-MB-231 | DOX-RES-50 | DOX-RES-100 | |||
|---|---|---|---|---|---|---|
| Correlation | p-Value | Correlation | p-Value | Correlation | p-Value | |
| Methylthioadenosine_OL1_4.92_297.0893 n | −0.81 | 0.00001554 | −0.72 | 0.0003888 | −0.68 | 0.0010676 |
| N-Acetylmethionine_OL1_4.71_191.0610 n | −0.77 | 0.00007602 | −0.50 | 0.023791 | −0.33 | 0.15296 |
| Anthranilate_OL1_5.84_137.0474 n | −0.76 | 0.00009184 | 0.15 | 0.53421 | −0.27 | 0.24814 |
| 12-Hydroxydodecanoic Acid_OL2b_5.12_261.1439 m/z | −0.76 | 0.00011148 | −0.30 | 0.19307 | 0.29 | 0.21246 |
| 2-Octenedioic Acid_OL2b_1.44_137.0594 m/z | −0.74 | 0.00019629 | −0.31 | 0.17985 | −0.37 | 0.11085 |
| Indoleacrylic Acid_OL2b_4.34_187.0632 n | −0.74 | 0.00020622 | −0.35 | 0.13308 | −0.50 | 0.023743 |
| 2-Aminocaprylic Acid_OL1_6.37_160.1329 m/z | −0.72 | 0.00037329 | −0.39 | 0.088065 | −0.49 | 0.029253 |
| Adenosine 2′,3′-Cyclic Phosphate_OL2b_2.91_330.0592 m/z | 0.71 | 0.00047735 | 0.21 | 0.37223 | 0.83 | 0.00000680 |
| Trans-3-Hydroxycinnamate_OL2b_1.56_164.0473 n | −0.70 | 0.00053789 | −0.42 | 0.062772 | −0.44 | 0.051265 |
| Pyridoxine_OL1_1.27_169.0735 n | −0.70 | 0.00054082 | 0.09 | 0.70269 | 0.06 | 0.79124 |
| Spermidine_OL2b_3.42_146.1649 m/z | −0.68 | 0.00099902 | −0.14 | 0.54993 | 0.16 | 0.5026 |
| Cytidine_OL1_0.69_243.0848 n | 0.67 | 0.0013015 | 0.18 | 0.44717 | 0.16 | 0.50397 |
| Threonine_OL2a_0.67_158.0211 m/z | −0.66 | 0.0015141 | −0.47 | 0.037284 | −0.37 | 0.10919 |
| 2-Octenedioic Acid_OL2b_3.72_172.0730 n | −0.66 | 0.0016946 | −0.34 | 0.13976 | 0.30 | 0.19228 |
| 5-Hydroxyindoleacetate_OL2b_2.91_191.0575 n | −0.66 | 0.001717 | −0.28 | 0.23609 | −0.20 | 0.38949 |
| Indoleacetaldehyde_OL1_4.34_160.0754 m/z | −0.65 | 0.0017981 | −0.15 | 0.5315 | −0.26 | 0.27521 |
| N-Acetylaspartate_OL2a_0.83_198.0368 m/z | 0.65 | 0.0018161 | 0.27 | 0.2445 | 0.43 | 0.059134 |
| Leucine_OL1_1.70_131.0944 n | −0.65 | 0.0019876 | −0.38 | 0.097942 | −0.41 | 0.071259 |
| Hydroxyphenyllactate_OL1_4.59_205.0468 m/z | −0.65 | 0.0020371 | −0.23 | 0.33412 | −0.44 | 0.05153 |
| Indoleacetaldehyde_OL2b_2.46_159.0682 n | −0.63 | 0.0030782 | 0.06 | 0.78868 | 0.06 | 0.78725 |
| Octanoyl-L-Carnitine_OL1_9.20_288.2162 m/z | −0.63 | 0.0031425 | −0.42 | 0.067438 | −0.25 | 0.29713 |
| Isoleucyl-Leucine_OL2b_5.43_245.1852 m/z | −0.62 | 0.0035538 | −0.66 | 0.0016847 | −0.08 | 0.73277 |
| L-Tryptophan_OL1_4.34_204.0897 n | −0.62 | 0.0036778 | −0.24 | 0.30013 | −0.38 | 0.1017 |
| L-Phenylalanine_OL1_3.22_165.0787 n | −0.61 | 0.0042322 | −0.48 | 0.033566 | −0.54 | 0.014204 |
| Deoxyadenosine_OL2b_3.57_252.1084 m/z | −0.60 | 0.0048732 | −0.20 | 0.39106 | −0.29 | 0.22223 |
| L-Methionine_OL2a_1.07_172.0400 m/z | −0.59 | 0.0057758 | −0.37 | 0.11265 | −0.25 | 0.28305 |
| Pantothenate_OL1_4.04_219.1102 n | −0.58 | 0.0068454 | 0.12 | 0.60768 | 0.24 | 0.31459 |
| Trans-Cinnamic Acid_OL2b_3.24_148.0522 n | −0.58 | 0.0069352 | −0.35 | 0.12974 | −0.42 | 0.063941 |
| Pyridoxal_OL2b_2.81_150.0548 m/z | −0.58 | 0.0079745 | 0.22 | 0.36202 | 0.07 | 0.77067 |
| Leucyl-Leucine_OL1_5.89_245.1852 m/z | −0.57 | 0.0083982 | −0.34 | 0.14013 | −0.28 | 0.23375 |
| Indole-3-Aldehyde_OL2b_4.34_146.0599 m/z | −0.56 | 0.0094943 | −0.24 | 0.31034 | −0.33 | 0.15284 |
| 2-Octenedioic Acid_OL2a_6.83_195.0622 m/z | 0.56 | 0.010025 | −0.17 | 0.47878 | −0.38 | 0.093781 |
| N-Acetylaspartate_OL1_1.11_175.0478 n | 0.54 | 0.013375 | 0.61 | 0.0046458 | 0.50 | 0.025329 |
| Spermidine_OL2a_0.65_145.1575 n | −0.54 | 0.013553 | −0.14 | 0.56286 | 0.05 | 0.82905 |
| Phenylacetylglycine_OL1_5.92_193.0737 n | −0.54 | 0.014787 | 0.16 | 0.48874 | 0.15 | 0.5273 |
| Dodecenoylcarnitine_OL1_11.80_342.2630 m/z | −0.53 | 0.015577 | −0.39 | 0.087782 | −0.09 | 0.70138 |
| Indole-3-Ethanol_OL2b_4.34_144.0806 m/z | −0.52 | 0.017664 | −0.22 | 0.34956 | −0.26 | 0.26126 |
| Guanine_OL2b_3.62_152.0563 m/z | −0.51 | 0.020217 | −0.21 | 0.36735 | −0.18 | 0.45806 |
| L-Tyrosine_OL2b_3.37_182.0807 m/z | −0.51 | 0.020866 | −0.09 | 0.70388 | −0.20 | 0.40599 |
| Threonine_OL1_1.07_102.0548 m/z | −0.51 | 0.021699 | 0.14 | 0.56117 | −0.14 | 0.56762 |
| Butenylcarnitine_OL2a_2.71_262.1642 m/z | −0.51 | 0.022923 | 0.17 | 0.47043 | 0.35 | 0.126 |
| Creatine_OL2a_0.51_263.1455 m/z | −0.50 | 0.024141 | 0.39 | 0.093473 | 0.85 | 0.00000202 |
| 5-Oxoproline_OL2b_0.77_129.0424 n | −0.49 | 0.026792 | −0.10 | 0.68462 | −0.04 | 0.85349 |
| S-Adenosylhomocysteine_OL1_2.09_384.1212 n | −0.48 | 0.030318 | 0.21 | 0.38359 | −0.07 | 0.75567 |
| Propionyl-L-Carnitine_OL1_2.09_218.1387 m/z | −0.48 | 0.033559 | 0.34 | 0.1442 | 0.15 | 0.52079 |
| Trigonelline_OL2b_5.41_138.0546 m/z | 0.47 | 0.035034 | −0.35 | 0.12744 | −0.02 | 0.93339 |
| Creatine_OL2a_0.49_132.0766 m/z | −0.47 | 0.036563 | 0.43 | 0.055497 | 0.82 | 0.00000987 |
| L-Phenylalanine_OL2a_3.22_331.1643 m/z | −0.47 | 0.0368 | −0.38 | 0.10306 | −0.51 | 0.022902 |
| N-Acetylhistidine_OL2b_2.24_198.0872 m/z | −0.46 | 0.039432 | 0.28 | 0.23266 | −0.02 | 0.92921 |
| Spermidine_OL2a_0.49_73.5862 m/z | −0.46 | 0.040849 | 0.10 | 0.66657 | 0.22 | 0.35548 |
| Pantetheine_OL2b_9.30_261.1260 m/z | 0.46 | 0.041443 | −0.08 | 0.74182 | 0.18 | 0.43803 |
| Acetyl-Dl-Carnitine_OL1_1.01_204.1229 m/z | −0.46 | 0.041794 | −0.16 | 0.49485 | 0.13 | 0.57551 |
| 1-Aminocyclopropanecarboxylic Acid_OL2a_0.77_84.0443 m/z | −0.46 | 0.043552 | −0.02 | 0.93018 | 0.00 | 0.99278 |
| Hexanoylcarnitine_OL1_6.23_260.1850 m/z | −0.45 | 0.047391 | −0.24 | 0.2993 | 0.19 | 0.4278 |
| 4-Acetamidobutanoic Acid_OL2b_3.37_146.0809 m/z | −0.41 | 0.075098 | 0.51 | 0.020527 | 0.38 | 0.099942 |
| Hexanoylcarnitine_OL1_6.53_260.1850 m/z | −0.40 | 0.079541 | −0.33 | 0.15004 | −0.55 | 0.011742 |
| Maleic Acid_OL2a_1.11_134.0445 m/z | 0.39 | 0.087469 | 0.52 | 0.017879 | 0.41 | 0.074901 |
| Glutamyl-Valine_OL2b_3.72_247.1284 m/z | −0.39 | 0.090678 | 0.39 | 0.090925 | 0.53 | 0.016339 |
| 2-Hydroxytetradecanoic Acid_OL2a_14.99_227.1998 m/z | 0.38 | 0.099925 | −0.06 | 0.81747 | 0.51 | 0.021926 |
| Acetyl-DL-Carnitine_OL1_0.71_203.1156 n | −0.36 | 0.11621 | 0.46 | 0.039795 | 0.00 | 0.99876 |
| Butanoylcarnitine_OL1_3.64_231.1467 n | −0.34 | 0.14514 | 0.41 | 0.072478 | 0.48 | 0.032961 |
| Glycoursodeoxycholic Acid_OL2b_14.05_449.3136 n | −0.32 | 0.16414 | 0.47 | 0.037037 | −0.09 | 0.71422 |
| Sorbitol_OL1_0.65_182.0786 n | 0.30 | 0.19459 | −0.07 | 0.78117 | 0.47 | 0.037825 |
| Betaine_OL2a_0.67_140.0680 m/z | −0.30 | 0.20182 | −0.56 | 0.01107 | −0.03 | 0.90492 |
| Cadaverine_OL2a_0.83_102.1150 m/z | −0.30 | 0.20579 | 0.16 | 0.49012 | 0.55 | 0.011389 |
| 2-Methylbutyroylcarnitine_OL1_4.84_246.1696 m/z | −0.28 | 0.23419 | 0.55 | 0.012568 | 0.38 | 0.095206 |
| Tetradecenoyl-L-Carnitine_OL1_12.82_370.2941 m/z | −0.27 | 0.24147 | −0.61 | 0.0043994 | 0.00 | 0.99629 |
| Creatinine_OL1_0.67_136.0479 m/z | 0.27 | 0.24167 | 0.20 | 0.38662 | 0.45 | 0.046976 |
| Citric Acid_OL2a_0.98_176.0083 m/z | −0.27 | 0.25513 | 0.27 | 0.24076 | −0.45 | 0.048451 |
| Spermine_OL2b_1.97_202.2156 n | −0.25 | 0.27928 | 0.17 | 0.48187 | 0.49 | 0.028223 |
| 7-Ketodeoxycholic Acid_OL2b_15.51_406.2712n | 0.25 | 0.28049 | −0.22 | 0.34463 | −0.47 | 0.038013 |
| Indolelactic Acid_OL1_7.14_205.0735 n | −0.25 | 0.28598 | −0.26 | 0.26977 | −0.68 | 0.00088093 |
| 2-Hydroxytetradecanoic Acid_OL2a_15.04_283.1663 m/z | 0.25 | 0.29295 | −0.12 | 0.60035 | 0.55 | 0.012847 |
| Glutarate_OL1_2.66_115.0388 m/z | −0.24 | 0.30605 | 0.52 | 0.018071 | 0.55 | 0.011919 |
| Cytosine_OL1_1.04_112.0503 m/z | 0.22 | 0.34136 | 0.83 | 0.00000607 | 0.61 | 0.0043897 |
| B-Nicotinamide Adenine Dinucleotide_OL2a_3.06_333.5691 m/z | −0.18 | 0.44516 | 0.06 | 0.81435 | 0.51 | 0.02153 |
| Cadaverine_OL2a_0.40_102.1150 m/z | −0.18 | 0.4511 | 0.21 | 0.36841 | 0.59 | 0.0061138 |
| Xanthine_OL1_1.54_153.0403 m/z | −0.15 | 0.52016 | 0.69 | 0.0007609 | 0.28 | 0.23164 |
| Creatine_OL1_0.69_131.0693 n | 0.04 | 0.8519 | 0.25 | 0.27987 | 0.46 | 0.039218 |
| Docosatrienoic Acid_OL2a_16.73_299.2728 m/z | 0.00 | 0.99386 | −0.58 | 0.0073867 | 0.26 | 0.27377 |
| Metabolite (Name_Ontology Level_Retention Time_Mass) | MDA-MB-231 | DOX-RES-50 | DOX-RES-100 | |||
|---|---|---|---|---|---|---|
| Correlation | p-Value | Correlation | p-Value | Correlation | p-Value | |
| Cytosine_OL1_1.04_112.0503 m/z | 0.84937 | 0.00000217 | 0.59474 | 0.0056768 | 0.67691 | 0.001046 |
| Methylthioadenosine_OL1_4.92_297.0893 n | −0.79527 | 0.0000278 | −0.87672 | 0.000000396 | −0.79282 | 3.06 × 10−5 |
| Malic Acid_OL2a_0.86_157.0103 m/z | 0.58391 | 0.0068684 | 0.0039314 | 0.98688 | −0.020673 | 0.93106 |
| Threonine_OL1_1.07_102.0548 m/z | 0.56734 | 0.009083 | 0.29776 | 0.2023 | −0.040839 | 0.86426 |
| Butanoylcarnitine_OL1_3.64_231.1467 n | 0.56564 | 0.0093398 | 0.015986 | 0.94667 | 0.16808 | 0.47874 |
| Xanthine_OL1_1.54_153.0403 m/z | 0.55436 | 0.011196 | 0.43665 | 0.054237 | 0.41854 | 0.066265 |
| N-Methyl-D-Aspartic Acid_OL2a_0.65_147.0526 n | −0.54143 | 0.013681 | −0.25443 | 0.27903 | 0.39605 | 0.083868 |
| Glutarate_OL2b_4.07_115.0388 m/z | 0.53065 | 0.016078 | 0.32205 | 0.16614 | −0.20547 | 0.38482 |
| Indoleacetaldehyde_OL1_4.34_160.0754 m/z | 0.52999 | 0.016234 | 0.094866 | 0.69075 | 0.10551 | 0.65797 |
| 3-(Carbamoylamino)Propanoic Acid_OL2a_1.07_177.0240 m/z | −0.52048 | 0.018636 | 0.4759 | 0.033922 | −0.045774 | 0.84803 |
| N-Acetylserine_OL2a_1.16_148.0600 m/z | −0.51527 | 0.020066 | −0.1719 | 0.46865 | 0.0052969 | 0.98232 |
| 12-Hydroxydodecanoic Acid_OL2b_5.12_261.1439 m/z | 0.51409 | 0.020401 | −0.23781 | 0.31268 | −0.064887 | 0.78579 |
| Indoleacetaldehyde_OL2b_2.46_159.0682 n | 0.50304 | 0.02377 | −0.21828 | 0.35521 | −0.22613 | 0.33773 |
| Nicotinamide_OL1_1.23_123.0551 m/z | 0.48868 | 0.028787 | 0.10396 | 0.66271 | −0.20153 | 0.39419 |
| Fructose_OL2a_0.88_113.0206 m/z | −0.48047 | 0.03201 | −0.22229 | 0.34621 | −0.037041 | 0.87679 |
| L-Phenylalanine_OL2a_3.22_331.1643 m/z | 0.47757 | 0.033214 | 0.32444 | 0.16283 | 0.28265 | 0.22724 |
| Pantothenate_OL1_4.04_219.1102 n | 0.46838 | 0.037262 | 0.082628 | 0.7291 | 0.0097354 | 0.96751 |
| 7-Ketodeoxycholic Acid_OL2b_15.51_406.2712 n | −0.45131 | 0.045786 | −0.59553 | 0.0055969 | −0.61972 | 0.003564 |
| Glutamyl-Valine_OL2b_3.72_247.1284 m/z | 0.44907 | 0.047004 | 0.061125 | 0.79795 | −0.076558 | 0.74836 |
| 1-Methyl-L-Histidine_OL2a_0.58_192.0737 m/z | 0.44537 | 0.049076 | −0.25272 | 0.28237 | 0.10562 | 0.65765 |
| Creatinine_OL1_0.65_114.0661 m/z | 0.39497 | 0.084797 | 0.051977 | 0.82772 | −0.47478 | 0.034403 |
| Sphinganine_OL2b_17.24_284.2941 m/z | 0.38174 | 0.096742 | 0.44605 | 0.048695 | 0.14411 | 0.54441 |
| Guanine_OL2b_3.62_152.0563 m/z | −0.29382 | 0.20862 | −0.49466 | 0.026606 | −0.37563 | 0.10266 |
| Prostaglandin B2_OL2b_15.92_357.2030 m/z | 0.2713 | 0.24726 | −0.44964 | 0.046693 | 0.18064 | 0.44597 |
| Histidine_OL1_0.58_155.0693 n | −0.18863 | 0.42576 | 0.61221 | 0.0041156 | 0.20709 | 0.381 |
| Arginine_OL1_0.58_175.1188 m/z | −0.14538 | 0.54083 | 0.58944 | 0.0062371 | 0.40546 | 0.076132 |
| Deoxyadenosine_OL2b_3.57_252.1084 m/z | −0.087792 | 0.71284 | −0.44602 | 0.048709 | −0.32382 | 0.16369 |
| L-Carnitine_OL2a_0.69_144.1017 m/z | 0.080419 | 0.73609 | 0.016996 | 0.9433 | 0.47174 | 0.035739 |
| Lysine_OL1_0.51_146.1053 n | −0.079963 | 0.73754 | 0.40311 | 0.078011 | 0.56567 | 0.009334 |
| 2-Hydroxypyridine_OL2b_1.23_96.0443 m/z | −0.034015 | 0.88679 | 0.56715 | 0.0091119 | 0.28764 | 0.21879 |
| Pipecolate_OL1_0.51_130.0861 m/z | −0.0041201 | 0.98625 | 0.42884 | 0.059206 | 0.51643 | 0.019741 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rushing, B.R.; Molina, S.; Sumner, S. Metabolomics Analysis Reveals Altered Metabolic Pathways and Response to Doxorubicin in Drug-Resistant Triple-Negative Breast Cancer Cells. Metabolites 2023, 13, 865. https://doi.org/10.3390/metabo13070865
Rushing BR, Molina S, Sumner S. Metabolomics Analysis Reveals Altered Metabolic Pathways and Response to Doxorubicin in Drug-Resistant Triple-Negative Breast Cancer Cells. Metabolites. 2023; 13(7):865. https://doi.org/10.3390/metabo13070865
Chicago/Turabian StyleRushing, Blake R., Sabrina Molina, and Susan Sumner. 2023. "Metabolomics Analysis Reveals Altered Metabolic Pathways and Response to Doxorubicin in Drug-Resistant Triple-Negative Breast Cancer Cells" Metabolites 13, no. 7: 865. https://doi.org/10.3390/metabo13070865
APA StyleRushing, B. R., Molina, S., & Sumner, S. (2023). Metabolomics Analysis Reveals Altered Metabolic Pathways and Response to Doxorubicin in Drug-Resistant Triple-Negative Breast Cancer Cells. Metabolites, 13(7), 865. https://doi.org/10.3390/metabo13070865







