Characterization of Arabidopsis thaliana Coq9 in the CoQ Biosynthetic Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Reagents
2.2. Phylogenetic Analysis
2.3. Beta-Glucuronidase (GUS) Reporter Assay
2.4. Subcellular Localization
2.5. Quantitative Reverse-Transcription PCR
2.6. Yeast Strain and Complementation Assays
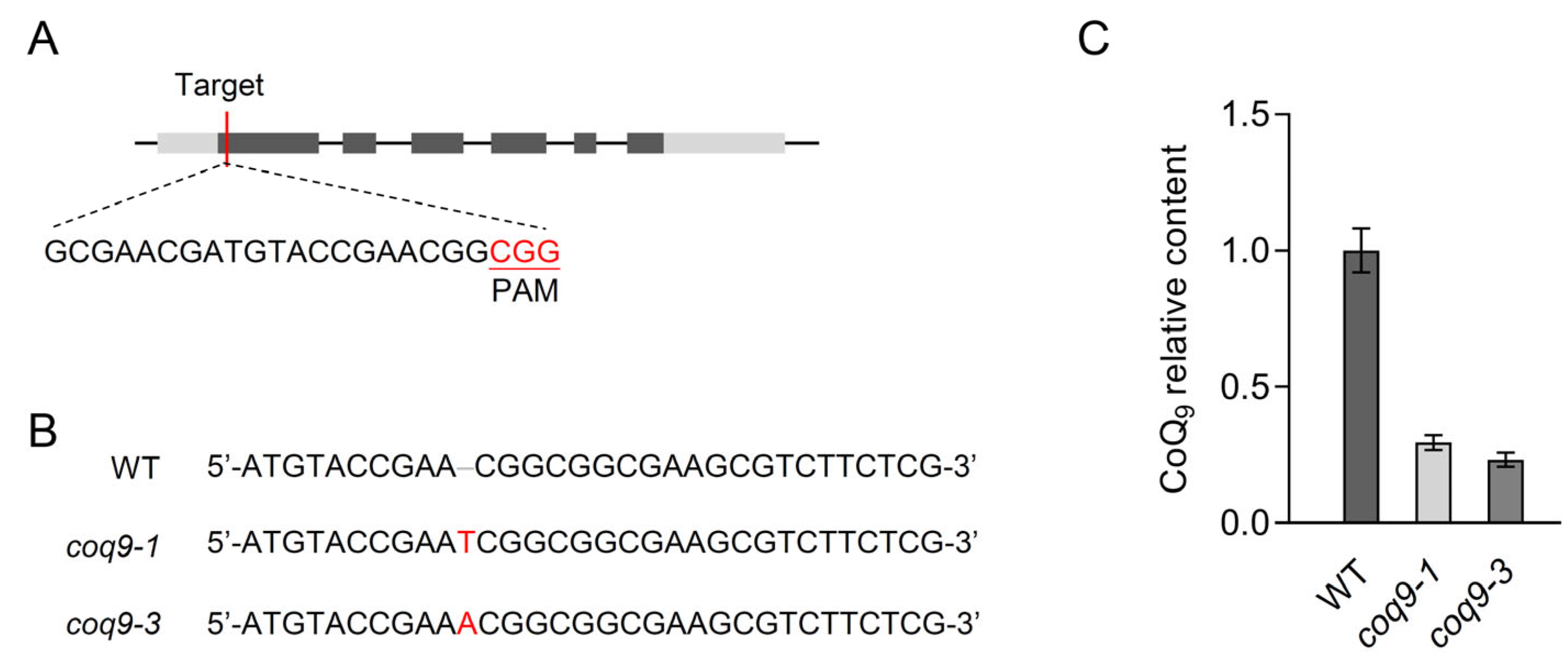
2.7. Generation of coq9 Mutants Using CRISPR-Cas9
2.8. Analysis of CoQ Contents
3. Results
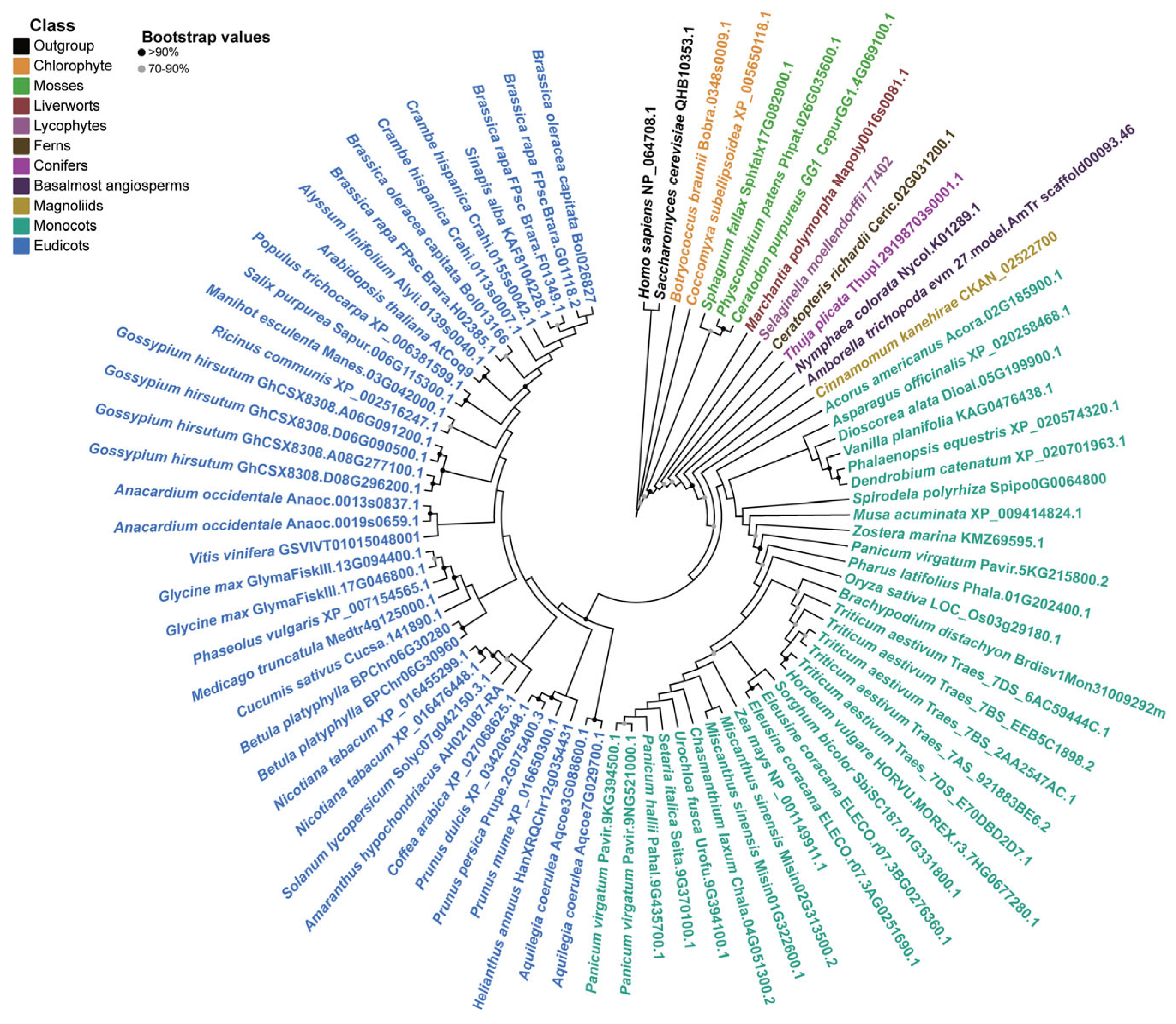
3.1. Phylogenetic Analysis of Plant Coq9
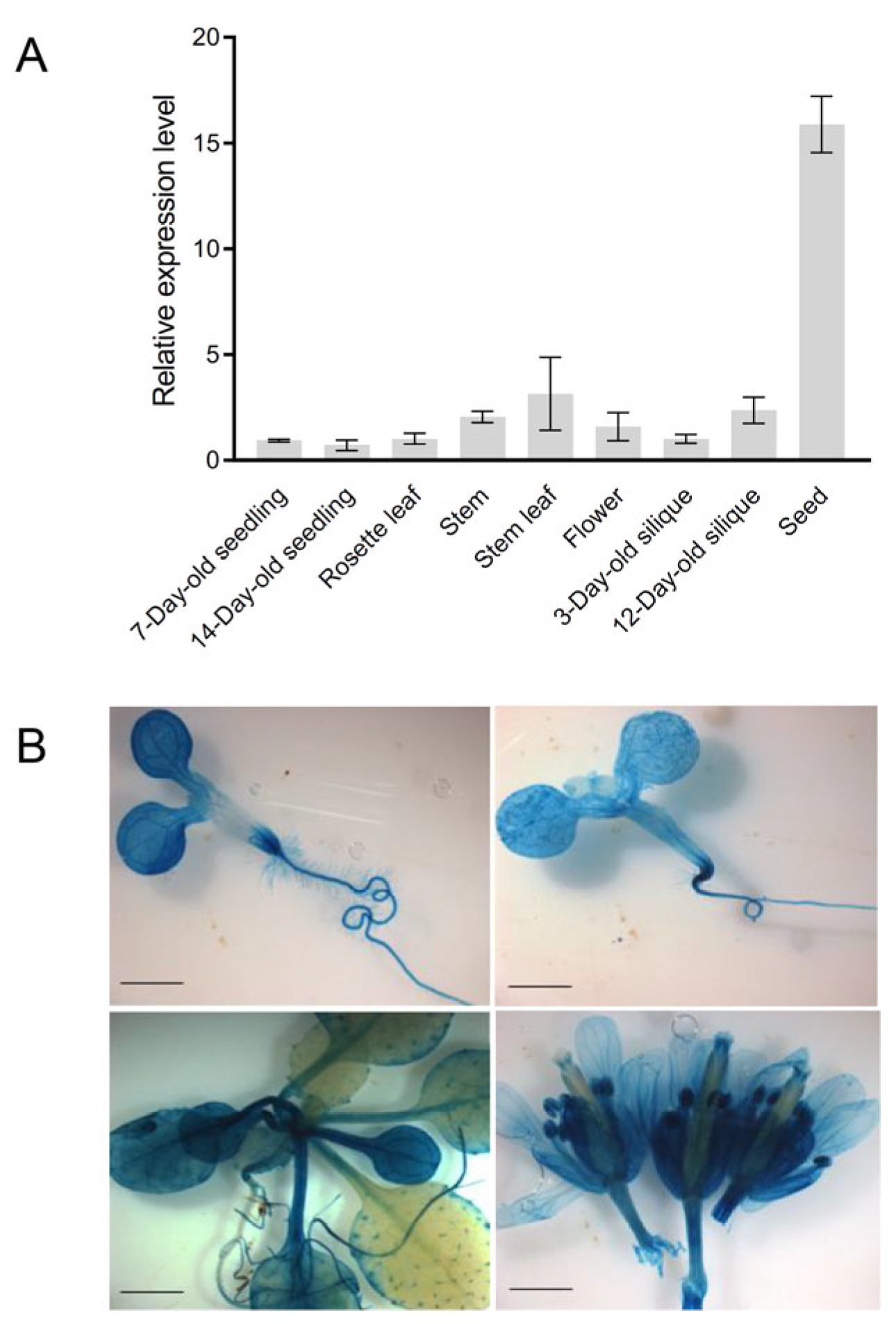
3.2. Expression Patterns of AtCoq9
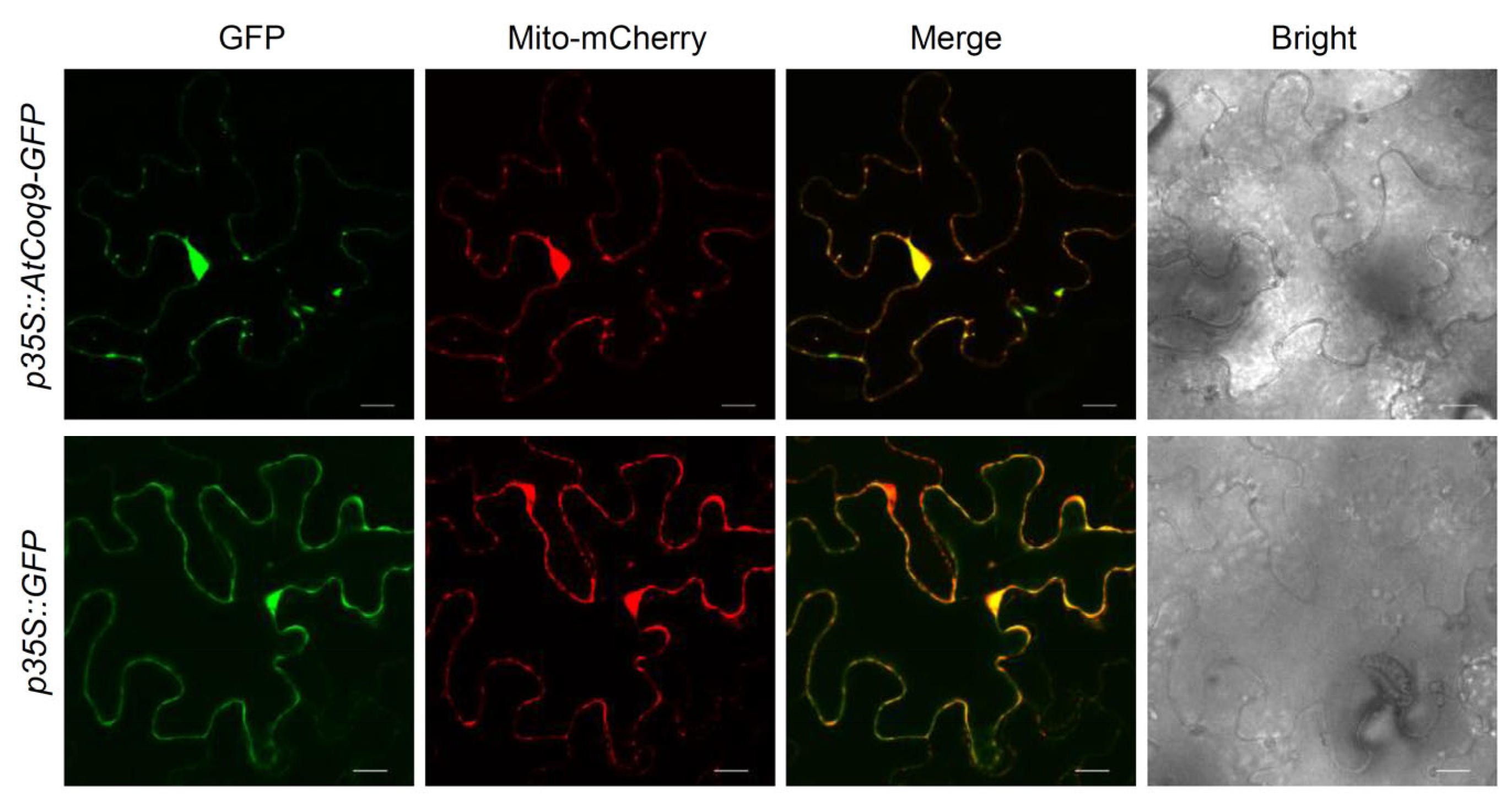
3.3. AtCoq9 Is Localized in Mitochondria
3.4. AtCoq9 Rescues the Yeast coq9 Point Mutant
3.5. AtCoq9 Defective Mutants Contained Less CoQ
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banerjee, R.; Purhonen, J.; Kallijarvi, J. The mitochondrial coenzyme Q junction and complex III: Biochemistry and pathophysiology. FEBS J. 2021, 289, 6936–6958. [Google Scholar] [CrossRef] [PubMed]
- Guerra, R.M.; Pagliarini, D.J. Coenzyme Q biochemistry and biosynthesis. Trends Biochem. Sci. 2023, 48, 463–476. [Google Scholar] [CrossRef]
- Awad, A.M.; Bradley, M.C.; Fernandez-Del-Rio, L.; Nag, A.; Tsui, H.S.; Clarke, C.F. Coenzyme Q10 deficiencies: Pathways in yeast and humans. Essays Biochem. 2018, 62, 361–376. [Google Scholar] [CrossRef]
- Kawamukai, M. Biosynthesis and applications of prenylquinones. Biosci. Biotechnol. Biochem. 2018, 82, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Stefely, J.A.; Pagliarini, D.J. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem. Sci. 2017, 42, 824–843. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.J.; Hu, M.; Yang, L.; Chen, X.Y. How plants synthesize coenzyme Q. Plant Commun. 2022, 3, 100341. [Google Scholar] [CrossRef]
- Xu, J.J.; Zhang, X.F.; Jiang, Y.; Fan, H.; Li, J.X.; Li, C.Y.; Zhao, Q.; Yang, L.; Hu, Y.H.; Martin, C.; et al. A unique flavoenzyme operates in ubiquinone biosynthesis in photosynthesis-related eukaryotes. Sci. Adv. 2021, 7, eabl3594. [Google Scholar] [CrossRef]
- Latimer, S.; Keene, S.A.; Stutts, L.R.; Berger, A.; Bernert, A.C.; Soubeyrand, E.; Wright, J.; Clarke, C.F.; Block, A.K.; Colquhoun, T.A.; et al. A dedicated flavin-dependent monooxygenase catalyzes the hydroxylation of demethoxyubiquinone into ubiquinone (coenzyme Q) in Arabidopsis. J. Biol. Chem. 2021, 297, 101283. [Google Scholar] [CrossRef]
- Soubeyrand, E.; Johnson, T.S.; Latimer, S.; Block, A.; Kim, J.; Colquhoun, T.A.; Butelli, E.; Martin, C.; Wilson, M.A.; Basset, G.J. The Peroxidative Cleavage of Kaempferol Contributes to the Biosynthesis of the Benzenoid Moiety of Ubiquinone in Plants. Plant Cell 2018, 30, 2910–2921. [Google Scholar] [CrossRef]
- Block, A.; Widhalm, J.R.; Fatihi, A.; Cahoon, R.E.; Wamboldt, Y.; Elowsky, C.; Mackenzie, S.A.; Cahoon, E.B.; Chapple, C.; Dudareva, N.; et al. The Origin and Biosynthesis of the Benzenoid Moiety of Ubiquinone (Coenzyme Q) in Arabidopsis. Plant Cell 2014, 26, 1938–1948. [Google Scholar] [CrossRef]
- Marbois, B.N.; Clarke, C.F. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 1996, 271, 2995–3004. [Google Scholar] [CrossRef] [PubMed]
- Manicki, M.; Aydin, H.; Abriata, L.A.; Overmyer, K.A.; Guerra, R.M.; Coon, J.J.; Dal Peraro, M.; Frost, A.; Pagliarini, D.J. Structure and functionality of a multimeric human COQ7:COQ9 complex. Mol. Cell 2022, 82, 4307–4323.e10. [Google Scholar] [CrossRef]
- Lohman, D.C.; Aydin, D.; Von Bank, H.C.; Smith, R.W.; Linke, V.; Weisenhorn, E.; McDevitt, M.T.; Hutchins, P.; Wilkerson, E.M.; Wancewicz, B.; et al. An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis. Mol. Cell 2019, 73, 763–774.e10. [Google Scholar] [CrossRef] [PubMed]
- Lohman, D.C.; Forouhar, F.; Beebe, E.T.; Stefely, M.S.; Minogue, C.E.; Ulbrich, A.; Stefely, J.A.; Sukumar, S.; Luna-Sanchez, M.; Jochem, A.; et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, E4697–E4705. [Google Scholar] [CrossRef] [PubMed]
- He, C.H.; Black, D.S.; Nguyen, T.P.; Wang, C.; Srinivasan, C.; Clarke, C.F. Yeast Coq9 controls deamination of coenzyme Q intermediates that derive from para-aminobenzoic acid. Biochim. Biophys. Acta 2015, 1851, 1227–1239. [Google Scholar] [CrossRef]
- Hsieh, E.J.; Gin, P.; Gulmezian, M.; Tran, U.C.; Saiki, R.; Marbois, B.N.; Clarke, C.F. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 2007, 463, 19–26. [Google Scholar] [CrossRef]
- Johnson, A.; Gin, P.; Marbois, B.N.; Hsieh, E.J.; Wu, M.; Barros, M.H.; Clarke, C.F.; Tzagoloff, A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 31397–31404. [Google Scholar] [CrossRef]
- He, C.H.; Black, D.S.; Allan, C.M.; Meunier, B.; Rahman, S.; Clarke, C.F. Human COQ9 Rescues a coq9 Yeast Mutant by Enhancing Coenzyme Q Biosynthesis from 4-Hydroxybenzoic Acid and Stabilizing the CoQ-Synthome. Front. Physiol. 2017, 8, 463. [Google Scholar] [CrossRef]
- Duncan, A.J.; Bitner-Glindzicz, M.; Meunier, B.; Costello, H.; Hargreaves, I.P.; Lopez, L.C.; Hirano, M.; Quinzii, C.M.; Sadowski, M.I.; Hardy, J.; et al. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: A potentially treatable form of mitochondrial disease. Am. J. Hum. Genet. 2009, 84, 558–566. [Google Scholar] [CrossRef]
- Garcia-Corzo, L.; Luna-Sanchez, M.; Doerrier, C.; Garcia, J.A.; Guaras, A.; Acin-Perez, R.; Bullejos-Peregrin, J.; Lopez, A.; Escames, G.; Enriquez, J.A.; et al. Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum. Mol. Genet. 2013, 22, 1233–1248. [Google Scholar] [CrossRef]
- Toda, T.; Hayashi, K.; Ogiyama, Y.; Yokomi, K.; Nakagawa, T.; Kaino, T.; Kawamukai, M. Functional Conservation of Coenzyme Q Biosynthetic Genes among Yeasts, Plants, and Humans. PLoS ONE 2014, 9, e99038. [Google Scholar] [CrossRef]
- Rivero, L.; Scholl, R.; Holomuzki, N.; Crist, D.; Grotewold, E.; Brkljacic, J. Handling Arabidopsis Plants: Growth, Preservation of Seeds, Transformation, and Genetic Crosses. In Arabidopsis Protocols; Sanchez-Serrano, J.J., Salinas, J., Eds.; Humana Press: Totowa, NJ, USA, 2014; pp. 3–25. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. Gus Fusions—Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher-Plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of invivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Werner, S.; Engler, C.; Weber, E.; Gruetzner, R.; Marillonnet, S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 2012, 3, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Niehaus, M.; Straube, H.; Kunzler, P.; Rugen, N.; Hegermann, J.; Giavalisco, P.; Eubel, H.; Witte, C.P.; Herde, M. Rapid Affinity Purification of Tagged Plant Mitochondria (Mito-AP) for Metabolome and Proteome Analyses. Plant Physiol. 2020, 182, 1194–1210. [Google Scholar] [CrossRef] [PubMed]
- Ducluzeau, A.L.; Wamboldt, Y.; Elowsky, C.G.; Mackenzie, S.A.; Schuurink, R.C.; Basset, G.J. Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 2012, 69, 366–375. [Google Scholar] [CrossRef]
- Okada, K.; Ohara, K.; Yazaki, K.; Nozaki, K.; Uchida, N.; Kawamukai, M.; Nojiri, H.; Yamane, H. The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol. Biol. 2004, 55, 567–577. [Google Scholar] [CrossRef]
- Meinke, D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2020, 226, 306–325. [Google Scholar] [CrossRef]
- Degli Esposti, M. A Journey across Genomes Uncovers the Origin of Ubiquinone in Cyanobacteria. Genome Biol. Evol. 2017, 9, 3039–3053. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Jiang, Y.; Xu, J.-J. Characterization of Arabidopsis thaliana Coq9 in the CoQ Biosynthetic Pathway. Metabolites 2023, 13, 813. https://doi.org/10.3390/metabo13070813
Hu M, Jiang Y, Xu J-J. Characterization of Arabidopsis thaliana Coq9 in the CoQ Biosynthetic Pathway. Metabolites. 2023; 13(7):813. https://doi.org/10.3390/metabo13070813
Chicago/Turabian StyleHu, Mei, Yan Jiang, and Jing-Jing Xu. 2023. "Characterization of Arabidopsis thaliana Coq9 in the CoQ Biosynthetic Pathway" Metabolites 13, no. 7: 813. https://doi.org/10.3390/metabo13070813
APA StyleHu, M., Jiang, Y., & Xu, J.-J. (2023). Characterization of Arabidopsis thaliana Coq9 in the CoQ Biosynthetic Pathway. Metabolites, 13(7), 813. https://doi.org/10.3390/metabo13070813





