Abstract
Aflatoxin pollution poses great harm to human and animal health and causes huge economic losses. The biological detoxification method that utilizes microorganisms and their secreted enzymes to degrade aflatoxin has the advantages of strong specificity, high efficiency, and no pollution inflicted onto the environment. In this study, Bacillus subtilis WJ6 with a high efficiency in aflatoxin B1 degradation was screened and identified through molecular identification, physiological, and biochemical methods. The fermentation broth, cell-free supernatant, and cell suspension degraded 81.57%, 73.27%, and 8.39% of AFB1, respectively. The comparative transcriptomics analysis indicated that AFB1 led to 60 up-regulated genes and 31 down-regulated genes in B. subtilis WJ6. A gene ontology (GO) analysis showed that the function classifications of cell aggregation, the organizational aspect, and the structural molecule activity were all of large proportions among the up-regulated genes. The down-regulated gene expression was mainly related to the multi-organism process function under the fermentation condition. Therefore, B. subtilis WJ6 degraded AFB1 through secreted extracellular enzymes with the up-regulated genes of structural molecule activity and down-regulated genes of multi-organism process function.
1. Introduction
Aspergillus flavus and Aspergillus parasiticus naturally produce secondary mycotoxin metabolites, aflatoxins [1], of which aflatoxin B1 (AFB1) is the most toxic and carcinogenic due to its potential to induce liver toxicity and oxidative stress [2]. AFB1 pollution occurring during the processes of production, harvest, transportation, processing, and the storage of cereals, milk, and nuts usually causes serious public security problems and huge economic losses [3]. To reduce AFB1 contamination in food and feed, various strategies of physical, chemical, and biological approaches have been applied in the removal and degradation of AFB1 [4]. Although physical means (such as high temperature, ultraviolet rays, and microwave) and chemical means (such as ammoniation, neutral electric oxidation water, and sodium sulfite) have been applied in AFB1 degradation and removal, these two approaches have the disadvantages of an excessive energy consumption, serious nutrient loss, and even chemical residue pollution [5,6,7]. The microbial degradation and removal of AFB1 have the advantages of the conversion of toxins into low-toxic or nontoxic products, strong specificity, high efficiency, low energy consumption, and low toxicity without damage to nutrients [8].
The microbial degradation of AFB1 was carried out by microorganisms (such as Bacillus licheniformis [9], Fusarium oxysporum [10], Levilactobacillus brevis [11], and Streptomyces cacaoi [12]) and enzymes (such as catalase [13], laccase [14]). The possible mechanisms of AFB1 degradation and removal are: (1) adsorption by secondary metabolites N-acetylmuramic acid, N-acetylglucosamine, and peptidoglycan in the cell wall [15] and (2) degradation catalyzed by secreted extracellular or intracellular proteins [16]. The reported references with regard to the degradation of AFB1 mainly focused on the specific products of microorganisms, such as the protein extract of Lactobacillus helveticus [17], the extractable oxidase components of Rhodococcus pyridinvorans [18], Bacillus licheniformis extracellular fraction [19], and CotA laccase in B. licheniformis [20]. In practice, microorganisms usually simultaneously degrade and remove AFB1 through various enzymes for degradation and composite components for adsorption [21].
In this study, a strain of Bacillus subtilis WJ6 (B. subtilis WJ6) that degraded AFB1 was isolated and identified. The possible causes for AFB1 degradation were investigated by determining intracellular and extracellular enzymes and other components in the fermentation broth. Furthermore, the molecular mechanism of AFB1 degradation by B. subtilis WJ6 was explored based on a comparative transcriptomics approach at the mRNA expression level.
2. Material and Methods
2.1. Apparatus, Chemicals, and Kits
AFB1 with a purity of 99% was from Pribolab Company (China). Coumarin was from Sigma-Aldrich (Germany). The microscope with a magnification of 400 times (eyepiece of 10 times, objective lens of 40 times) was from OLYMPUS Corporation (CX33 model, Japan). A dual-beam ultraviolet–visible spectrophotometer was the TU-1901 model from Beijing Puxi General Instrument Co., Ltd. (China). High-performance liquid chromatography (HPLC) was manufactured by Waters Alliance (e2695 model, USA). Gram staining kits and other chemical reagents were from Solarbio Company (China) and Aladdin (China). The screening medium was prepared with 0.25 g KH2PO4, 0.25 g MgSO4 · 7H2O, 0.5 g KNO3, 0.5 g (NH4) 2SO4, 0.005 g CaCl2, 0.003 g FeCl3 · 6H2O, 1 g coumarin, and 15 g agar, diluted to 1 L using double-distilled water.
2.2. Isolation and Molecular Identification of AFB1-Degrading Strain
A strain degrading AFB1 was screened from rotten feed contaminated with aflatoxins as follows. An amount of 5 g of sample was added to 95 mL of sterile saline, and the mixture was cultured at 37 °C with a shaking speed of 180 rpm for 2 h. Single colonies were screened on the prepared screening medium. After 12 h, single colonies were picked out for purification and further culture. In the logarithmic phase, the bacterial suspension was used for DNA extraction using Bacterial Genomic DNA Extraction Kit with the following processes. The mixture of binding solution and proteinase K was used to rapidly lyse cells and inactivate nuclease in cells. Additionally, genomic DNA was then selectively adsorbed on the silicon matrix membrane in the centrifuge column in the state of highly disordered salt. Through a series of rapid rinsing centrifugation steps and inhibitor removal, rinsing solutions were used to remove impurities such as cell metabolites and proteins. The low salt elution buffer was used to elute pure genomic DNA from the silicon matrix membrane.
The universal primers for the V3 region in the 16S-rRNA gene were F341: 5-CCTACGGGGAGGCAGCAG-3 and R518: 5-ATTACCCGGGCTGG-3. PCR amplification was carried out using the following parameters: 3 min at 95 °C initial denaturation; 15 s at 95 °C, 15 s at 56 °C, 90 s at 72 °C; 25 cycle numbers; extension time of 5 min at 72 °C. The amplified DNA was sequenced by Sangon Biotech (Shanghai, China). BLAST and MEGA11 were used for gene alignment and neighbor-joining phylogenetic tree drawing, respectively.
2.3. Identification Based on Physical and Chemical Properties
In this study, the physical and chemical properties, including catalase test, oxidase test, nitrate reduction test, starch hydrolysis test, arginine dihydrolase test, and V-P determination test of the bacterium were performed based on Bergey’s manual [22]. Cell staining was performed at the bacterial logarithmic phase according to the instruction of the Gram staining kit. The cell morphology was observed and recorded under the microscope at 400-fold magnification using M Shot Image Analysis System Software. The bacterial species were finally identified based on their molecular, physical, and chemical properties.
2.4. Bacterium Growth and Proliferation
To explore cell growth and proliferation, the concentrations of B. subtilis WJ6 were determined in the presence or absence of AFB1. The suspension samples were taken out for measurement during the 72 h fermentation. The OD600 nm values of B. subtilis WJ6 were measured using a UV spectrophotometer [23]. In addition, the growth and proliferation of the cell were investigated under pH values of 5 to 9.
2.5. AFB1 Degradation by B. Subtilis WJ6
The efficiency of AFB1 degradation by B. subtilis WJ6 was investigated using different components of fermentation suspension. In this study, the whole suspension was divided into the following three states based on the presence of bacteria: “fermentation broth” without any treatment after fermentation, “cell-free supernatant” without strain cells after centrifugation, and “cell suspension”, only strain cells after centrifugation. After incubating B. subtilis WJ6 at 37 °C for 24 h under a shaking speed of 200 rpm, the cell-free supernatant and cell suspension in the fermentation broth were separated by centrifugation at 8000 rpm for 10 min at 4 °C. AFB1 was added to the systems of cell-free supernatant, cell suspension AFB1, and fermentation broth to a final concentration of 5 μg/mL. After incubating at 37 °C for 48 h under a shaking speed of 200 rpm, a 1:1 ratio of dichloromethane and suspension (v/v) was added three times to extract the AFB1. After drying with nitrogen, the residue was redissolved in 1 mL of acetonitrile and then filtered through a 0.22 μm filter membrane.
2.6. HPLC Approach Determining AFB1 Content
HPLC approach was used to determine AFB1 concentration under modified conditions based on previous reports [24]. The detailed HPLC parameters were: 43:57 of acetonitrile and water mobile phase ratio (v/v), Waters 2489 of UV detector, C18 chromatographic column of Waters XBridge (4.6 × 250 mm, 5 μm), 365 nm of detection wavelength, 1 mL/min of flow rate, 20 μL of injection volume, and 30 °C of column temperature.
2.7. Investigation of AFB1 Degradation under Different Catalytic Conditions
B. subtilis WJ6 cells were cultured for 48 h at 37 °C with a shaking speed of 200 rpm. The supernatant of B. subtilis WJ6 fermentation broth was obtained through centrifugation at 8000 rpm for 10 min at 4 °C. The effect of culture time on the degradation of AFB1 was investigated using the supernatant added to a final AFB1 concentration of 5 μg/mL during fermentation of 48 h. In addition, the effects of temperature and pH values on the AFB1 degradation in the supernatant were also investigated to analyze the degradation efficiency of B. subtilis WJ6.
2.8. Effects of Lignin and Coumarin on AFB1 Degradation of B. Subtilis WJ6
AFB1-degrading enzyme generally catalyzed lignin and coumarin due to their similar structures of phenolic model compounds [25,26,27]. In this study, as the sole carbon sources, lignin and coumarin were used to induce the expression of AFB1-degrading enzyme during the fermentation of B. subtilis WJ6. The effect of lignin and coumarin concentrations on the proliferation of B. subtilis WJ6 was investigated by the determination of OD600 nm values in the broth during fermentation. After determining the effect of lignin and coumarin concentrations on cell proliferation, the lignin and coumarin concentrations with slight inhibition on B. subtilis WJ6 proliferation were added to the broth of B. subtilis WJ6 for AFB1 degradation. The fermentation of B. subtilis WJ6 was carried out at 37 °C with a shaking speed of 200 rpm in the presence of 5 g/L of lignin/coumarin, 5 g/L lignin, and 5 g/L coumarin as the sole carbon source. The broth supernatant of B. subtilis WJ6 was obtained after centrifugation at 8000 rpm for 10 min at 4 °C. The broth supernatant containing an initial AFB1 concentration of 5 μg/mL was prepared and incubated to degrade AFB1 at 37 °C for 48 h. The percentage of AFB1 degradation was calculated in the broth supernatant by the measurement of residual AFB1 content using the HPLC approach [24].
2.9. Comparative Transcriptome Analysis of B. Subtilis WJ6
B. subtilis WJ6 cells were cultured in LB liquid medium containing 1 μg/mL of final AFB1 concentration at 37 °C under a shaking speed of 200 rpm. The strain was cultured in LB liquid medium without the addition of AFB1 as the control. When the cell concentrations reached 1 OD600 value, the cells were collected by centrifugation at 8000 rpm for 10 min at 4 °C for comparative transcriptome analysis. The total RNA of B. subtilis WJ6 was extracted using Invitrogen TRIzol. The RNA concentration and integrity were measured using the Qubit RNA detection kit from Life Technologies Corporation (Gaithersburg, MD, USA). The RNA Seq library was constructed by Sangon Biotech (Shanghai, China). Illumina Hiseq 2500 platform was used for sequencing. FastQC was used to evaluate the quality of the original sequencing data. DESeq2 was used to analyze gene expression differences and visualize the results of gene expression differences. Differential expression gene (DEG) for analysis of gene differences, gene ontology (GO) annotate, and Kyoto Encyclopedia of Genes and Genomes (KEGG) for gene function definition were performed according to previous references [28,29].
2.10. Statistical Analysis
All the data above were from the experiments conducted three times. The results were expressed as mean ± standard deviation (SD). Software Origin 2023 was used to analyze the data and create curves and bar charts.
3. Results
3.1. Colony Isolation and Identification Based on 16S-rRNA Analysis
A strain of AFB1-degrading bacteria, screened from rotten feed contaminated with aflatoxins, was inoculated onto LB solid medium containing 10 μg/mL of AFB1. The colony morphologies were observed and recorded over a 72 h culture period. Figure 1 indicated that the diameter of the colony increased to 7 mm after 24 h of cultivation from 3 mm after 12 h of cultivation. The morphology of the colony changed to a milky-white color after a culture of 36 h from a transparent color, which was observed after a culture of 12 h. Through gene amplification and sequencing, the 16S-rRNA of the strain screened in this study had a size of 1456 bp. After sequencing, the strain was further identified using the molecular comparison on the NCBI website.
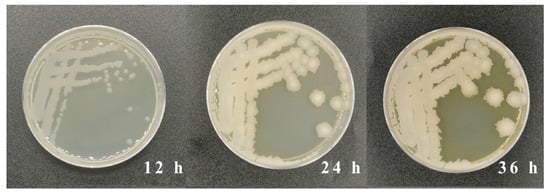
Figure 1.
Colony morphology on LB solid media containing 10 μg/L of AFB1 after culture of 12, 24, and 36 h.
3.2. Colony Identification Based on Physiological and Biochemical Characteristics
A phylogenetic tree was constructed using 16S-rRNA nucleotide sequences from the colony in this study and 13 other Bacillus strains (Figure 2a). The result showed that the genetic distance homology of 16S-rRNA of the colony was close to B. subtilis, while the homology of this colony had a far genetic distance from other strains, such as B. nematocidal, B. amyloliquefaciens, and B. halotolerans. The physiological and biochemical characteristics of the colony were performed to further identify the strain. The result of microscopy showed that the color of the colony was purple after Gram staining, which represented that the strain was Gram-positive bacteria (Figure 2b). Further, the colony was identified through physiological and biochemical experiments. The colony was aerobic, and a catalase test, oxidase, nitrate reduction to nitrite, starch hydrolysis, and V-P test results were all positive. In addition, an arginine dihydrolase test exhibited negative results. Therefore, the strain screened in this study was identified as B. subtilis (B. subtilis WJ6) based on the comprehensive analysis of the molecular identification and biological and biochemical characteristics.
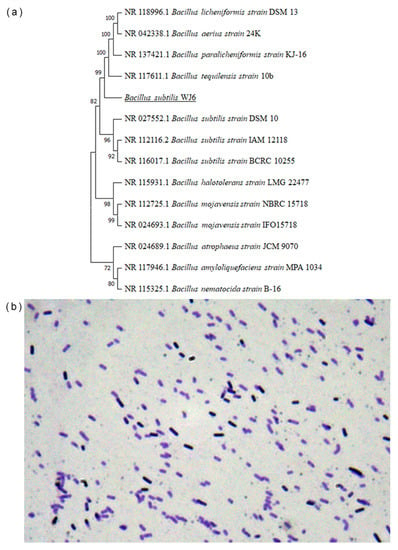
Figure 2.
Colony genetic distance based on the 16S-rRNA sequence (a) and Gram staining identification of colony cells (b).
3.3. Growth Characteristics of B. Subtilis WJ6
The cell concentrations of B. subtilis WJ6 were determined under different pH values ranging from 5 to 9 (Figure 3). The results showed that B. subtilis WJ6 was suitable for growth and proliferation in neutral environments. Both acidic and alkaline environments inhibited the reproduction of B. subtilis WJ6. The OD600 nm values were 3.46 and 8.04 under pH5 and 9 after a fermentation of 72 h, which were 0.38- and 0.89-fold compared to that at pH7 (9.08), respectively. Additionally, the cell concentration of B. subtilis WJ6 was also measured after the addition of 5 μg/mL AFB1. The result indicated that the addition of 5 μg/mL AFB1 did not affect the cell proliferation under the condition of pH7.
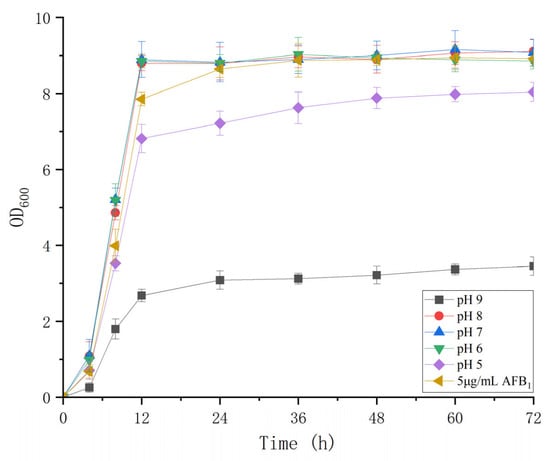
Figure 3.
Cell proliferation of B. subtilis WJ6 under different pH values and 5 μg/L of AFB1.
3.4. AFB1 Degradation Investigation of B. Subtilis WJ6 Fermentation Broth
The effect of cell fermentation liquid, cell-free supernatant, and cell suspension from B. subtilis WJ6 fermentation broth on the AFB1 degradation was investigated by measuring AFB1 content after catalysis (Figure 4A). The results showed that the cell fermentation liquid and cell-free supernatant had the highest AFB1 degradation efficiencies after a catalysis of 48 h with AFB1 degradation rates of 81.57% and 73.27%, respectively. Among these three groups, the cell suspension had the lowest degradation rate of 8.39%. HPLC measurement showed that the AFB1 content decreased with cell-free supernatant after treatment for 24 h based on the residual amount of AFB1 (Figure 4B). Therefore, the source of the degradation of aflatoxin could be located in the fermentation broth according to the test results.
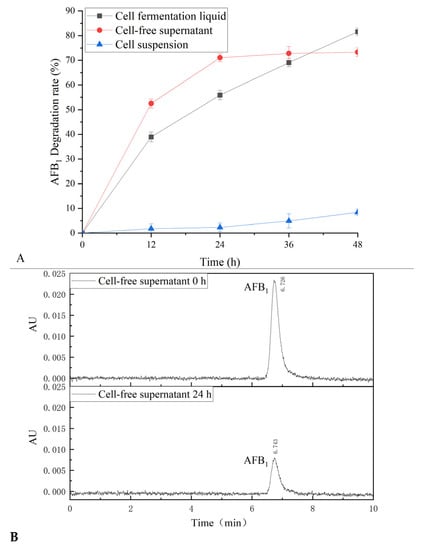
Figure 4.
AFB1 degradation of B. subtilis WJ6 (A) and HPLC determination of AFB1 degradation (B).
3.5. Cell-Free Supernatant Affecting AFB1 Degradation under Different Conditions
The effect of culture time, pH, and temperature on AFB1 degradation was investigated in B. subtilis WJ6 cell-free supernatant by measuring AFB1 concentration (Figure 5). The results indicated that the AFB1 degradation rate of cell-free supernatant was close to the highest when the fermentation time was 24 h during the period of 48 h (Figure 5A). The effect of temperatures on AFB1 degradation was investigated using B. subtilis WJ6 cell-free supernatant as a catalyst (Figure 5B). The result showed that under the condition of 40 °C the AFB1 degradation rate was 58.5%, which was the highest among the tested temperatures of 25, 30, 35, 40, and 45 °C. Additionally, the effect of pH values on the AFB1 degradation was also investigated in the cell-free supernatant from the fermentation broth of B. subtilis WJ6 (Figure 5C). The AFB1 degradation rate reached 63.27% under the pH value of 7, which was the highest among the values of 5–9. The result indicated that cell-free supernatant under pH-neutral conditions had the highest catalytic degradation efficiency of AFB1. Therefore, under the conditions of 24 h fermentation, 40 °C, and pH 7, B. subtilis WJ6 cell-free supernatant possessed the highest AFB1 degradation efficiency.
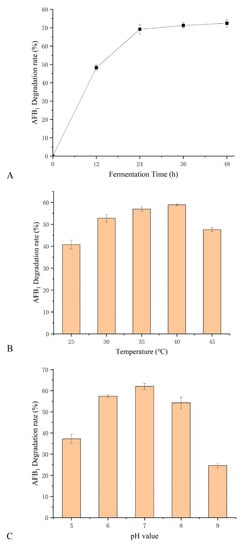
Figure 5.
Effect of fermentation time (A), temperature (B), and pH values (C) on AFB1 degradation in cell-free supernatant from B. subtilis WJ6.
3.6. Effect of Lignin and Coumarin Concentrations on the Cell Proliferation of B. Subtilis WJ6
The effect of lignin and coumarin concentrations on the cell proliferation of B. subtilis WJ6 was investigated by measuring the cell density after a fermentation of 36 h (Figure 6). The result indicated that the concentrations of lignin and coumarin from 0–5 g/L had a slight inhibitory effect on the cell proliferation of B. subtilis WJ6. OD600 nm values decreased to 8.4 and 7.8 from the initial OD600 nm value of 9 using 5 g/L of lignin and coumarin as the sole carbon source, respectively. While the lignin and coumarin concentrations of 5–10 g/L could remarkably enhance the inhibitory effect on the cell production of B. subtilis WJ6. OD600 nm values decreased to 7 and 6.5 using 10 g/L of lignin and coumarin, respectively. Thus, after a comprehensive consideration of the dose and its impact on cell growth, 5 g/L of lignin and coumarin could be suitable conditions to induce the expression of AFB1-degrading enzymes.
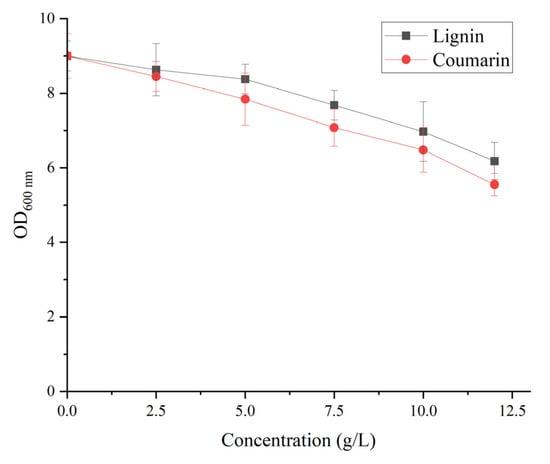
Figure 6.
Effect of different lignin and coumarin concentrations on the cell proliferation of B. subtilis WJ6.
3.7. AFB1 Degradation of B. Subtilis WJ6 in the Presence of Lignin and Coumarin
In this study, the effect of 5 g/L lignin and coumarin on AFB1 degradation in the broth from B. subtilis WJ6 was investigated by measuring the residual content of AFB1 after catalysis (Figure 7). The results indicated that 5 g/L coumarin treatment had the highest degradation percentage of AFB1 (85.3%) among the four groups, which was 1.1-fold of the control (77.8%). In addition, 5 g/L lignin treatment resulted in the lowest degradation percentage of AFB1 (73.3%), which was 0.9-fold of the control (without the addition of coumarin and lignin). Therefore, coumarin had more AFB1 degradation capability than lignin in the broth supernatant of B. subtilis WJ6.
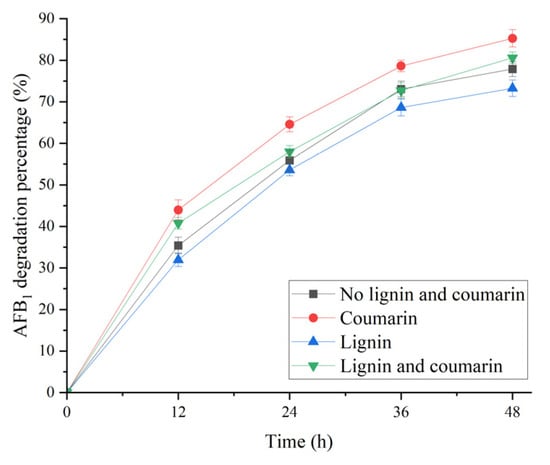
Figure 7.
Effect of treatment time on AFB1 degradation percentages of B. subtilis WJ6 in the presence of 5 g/L lignin and coumarin.
3.8. Differentially Expressed Genes (DEGs)
DEGs data had been submitted in Supplementary File S1. The scatter diagram of differential gene expression was drawn based on the transcriptomics sequencing (Figure 8). The results showed that B. subtilis WJ6 in the presence of AFB1 caused 91 differentially expressed genes, including 60 up-regulated genes and 31 down-regulated genes. Thus, AFB1 in the fermentation broth resulted in more up-regulated genes than down-regulated genes in B. subtilis WJ6 at the transcriptome level.
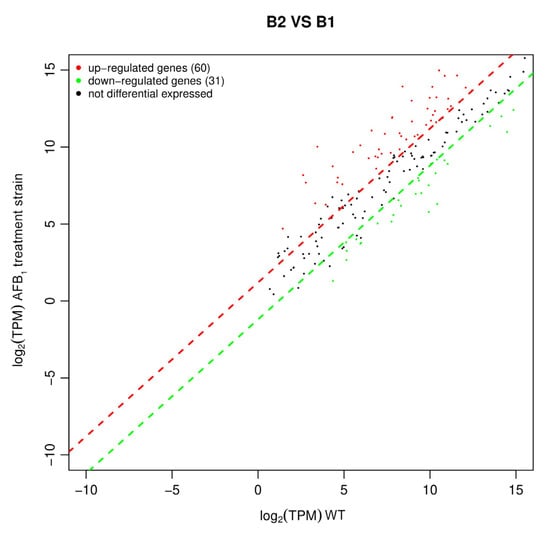
Figure 8.
Up-regulated and down-regulated genes of B. subtilis WJ6 based on DEGs.
3.9. GO Annotation of Differentially Expressed Genes
The GO annotation analysis of differentially expressed genes was performed using data from transcriptomics sequencing (Figure 9). A total of 29 gene function classifications were divided into three gene function classification groups: biological processes (15 gene classifications), cell components (9 gene classifications), and molecular functions (5 gene classifications). Among the up-regulated genes, in each classification, 50%, 49%, and 56.8% of DEGs participated in cell aggregation, the organizational part, and structural molecule activity, respectively. In addition, among the down-regulated genes, 15.4% of DEGs participated in the multi-organism process. Therefore, AFB1 could result in up-regulated gene expression related to functions of cell aggregation, the organizational part, and structural molecule activity, and down-regulated gene expression related to the multi-organism process function under the fermentation condition.
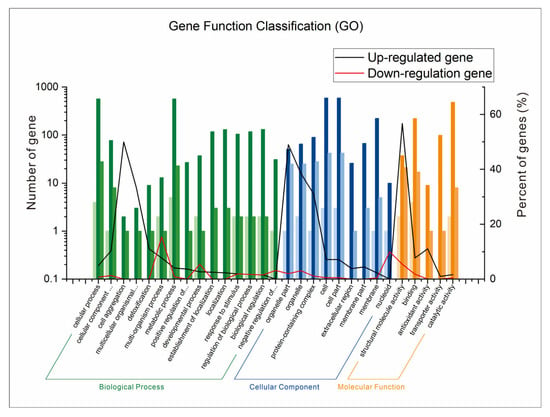
Figure 9.
GO annotation analysis of differentially expressed genes of B. subtilis WJ6.
3.10. KEGG Enrichment Analysis Based on Differential Gene Function
KEGG enrichment was analyzed based on the differential gene function using the cluster profiler plot analysis approach (Figure 10). KEGG enrichment of the up-regulated genes showed that a total of 30 gene function classifications were given. The up-regulated genes related to ribosome had the highest proportion of differential genes (60.4%) with 29 gene numbers (Figure 10A). All the proportions of differential genes in 29 other differential function classifications were less than 20%. In addition, the KEGG enrichment of the down-regulated genes was also analyzed (Figure 10B). A total of nine gene function classifications were used to construct the KEGG enrichment map. The down-regulated genes related to ribosome had the highest rich factor of 0.4 among these function classifications. Thus, AFB1 caused more up-regulated genes of B. subtilis WJ6 than down-regulated genes based on KEGG enrichment.
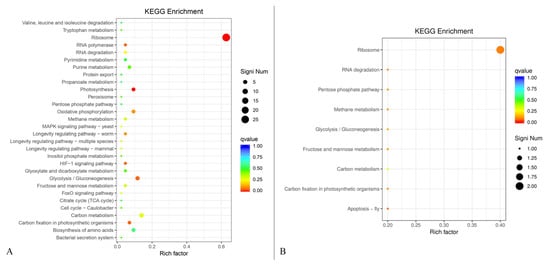
Figure 10.
Up-regulated genes (A) and up-regulated genes (B) of B. subtilis WJ6 based on KEGG enrichment analysis.
4. Discussions
AFB1, a natural toxin produced by food-contaminant fungi, is a threat to human and animal health. The microbial degradation of AFB1 was achieved via biological adsorption, living organisms, supernatants, and purified enzyme approaches [30]. Previous references showed that AFB1 could be degraded by microorganisms via the enzymes secreted in the fermentation supernatant (Table 1). Multiple strains such as Trichoderma reesei [31], Stenotrophomonas acidoaminiphila [32], Lactobacillus plantarum [33], and Cerrena unicolor [34] possessed the capability of AFB1 degradation by extracellular enzyme expression in a liquid medium. The percentages of AFB1 degradation by different strains varied from 46.9% to 98.56% under the conditions of different treatment time and initial AFB1 concentrations. Bacillus strains possessed a strong capability of AFB1 degradation in the fermentation medium [35,36,37]. In this study, B. subtilis WJ6 could degrade 81.57% of AFB1 from the initial concentration of 5 μg/mL after treatment for 48 h through an extracellular enzyme in a liquid medium. The high AFB1 degradation rate generally needed a long treatment time and/or initial AFB1 concentration. For example, Stenotrophomonas sp. degraded 91.2% of AFB1 with a low initial AFB1 concentration of 0.047 μg/mL by a combination of enzymes and oxides [32]. In addition, C. unicolor was able to degrade 98.56% of AFB1 after a period of 14 d [34]. B. subtilis WJ6 possessed a higher efficiency of AFB1 degradation with a higher initial concentration of AFB1 and shorter catalytic time compared with the reported strains [35,38]. Additionally, this study also revealed the mechanism based on the up-regulated genes of structural molecule activity and the down-regulated genes of multi-organism process function. Therefore, B. subtilis WJ6 still had an advantage in AFB1 degradation based on the comprehensive comparison of substrate concentration, catalytic time, and degradation efficiency.

Table 1.
Microbial AFB1 degradation efficiency and possible degradation mechanisms.
Glycolysis and gluconeogenesis path analyses were performed using the DEGs data based on the transcriptomics (Supplementary File S2). Enzymes participating in the glycolysis/glucose neogenesis pathway exhibited significant overexpression. In the pentose phosphate pathway, the conversion of glyceraldehyde-3-P from D-fructose-1,6-P2 via glycerone-P and glyceraldehyde-3-P, represented significant overexpression. In addition, the enzyme for the conversion of glycerate-2-P and phosphoenolpyruvate also displayed significant overexpression. Therefore, gene expression related to AFB1 degradation could involve in the pentose phosphate pathway. The possible limitations of this study lacked the determination of specific AFB1 decomposition enzymes and the feasibility analysis of the multi-enzyme degradation of AFB1 using the synergistic approach. In addition, artificial intelligence and machine learning would be used to modify the AFB1-degrading enzyme with high activity and specificity, which was a key direction of AFB1 degradation in the future.
5. Conclusions
AFB1 microbial degradation is a promising field due to its guarantee of nutritional values, reduction in AFB1 without residual toxicity, and no modification of food or feed properties. In this study, B. subtilis WJ6, with the capability of AFB1 degradation, was identified using molecular, physiological, and biochemical properties. The cell-free supernatant after fermentation could effectively degrade AFB1 under the conditions of 40 °C and pH 7. A percentage of 81.57% was degraded in the cell-free supernatant of B. subtilis WJ6 with an initial AFB1 concentration of 5 μg/mL after treatment for 48 h. As the sole carbon source, coumarin had a higher AFB1-degrading capability than lignin in the broth from B. subtilis WJ6. Further, the result of comparative transcriptomics analysis indicated that AFB1 caused 60 up-regulated genes and 31 down-regulated genes in B. subtilis WJ6. GO annotation analysis showed that 50%, 49%, and 56.8% of DEGs participated in cell aggregation, the organizational part, and structural molecule activity among the up-regulated genes, respectively. KEGG analysis showed that the up-regulated genes related to ribosome had the highest proportion of differential genes (60.4%) with 29 gene numbers. AFB1-degrading B. subtilis WJ6 degraded AFB1 through the extracellular enzymes secreted in cell-free supernatant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo13070785/s1, The data of DEGs and glycolysis and gluconeogenesis path analysis using the DEGs data based on the transcriptomics have been listed in Supplementary Files S1 and S2, respectively.
Author Contributions
P.Y. provided conceptualization and wrote the manuscript; W.W. performed the experiment; D.Z. provided the software; L.C. and J.C. provided resources. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Hefei Municipal Natural Science Foundation (2022047) and Major Science and Technology Projects of Anhui Province (202003c08020001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data generated in the study were included in the present manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Demirhan, B.E.; Demirhan, B. Investigation of twelve significant mycotoxin contamination in nut-based products by the LC-MS/MS method. Metabolites 2022, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Da Silveira, A.R.; Rosa, V.F.; Sari, M.H.M.; Sampaio, T.B.; Dos Santos, J.T.; Jardim, N.S.; Müller, S.G.; Oliveira, M.S.; Nogueira, C.W.; Furian, A.F. Therapeutic potential of beta-caryophyllene against aflatoxin B1-Induced liver toxicity: Biochemical and molecular insights in rats. Chem.-Biol. Interactions 2021, 348, 109635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, X.; Cai, Y.; Zhou, W.-W. Mycotoxin contamination status of cereals in China and potential microbial decontamination methods. Metabolites 2023, 13, 551. [Google Scholar] [CrossRef]
- Song, C.; Yang, J.; Wang, Y.; Ding, G.; Guo, L.; Qin, J. Mechanisms and transformed products of aflatoxin B1 degradation under multiple treatments: A review. Crit. Rev. Food Sci. Nutr. 2022, 1–13. [Google Scholar] [CrossRef]
- Muaz, K.; Riaz, M.; de Oliveira, C.A.F.; Akhtar, S.; Ali, S.W.; Nadeem, H.; Park, S.; Balasubramanian, B. Aflatoxin M1 in milk and dairy products: Global occurrence and potential decontamination strategies. Toxin Rev. 2022, 41, 588–605. [Google Scholar] [CrossRef]
- Marshall, H.; Meneely, J.P.; Quinn, B.; Zhao, Y.; Bourke, P.; Gilmore, B.F.; Zhang, G.; Elliott, C.T. Novel decontamination approaches and their potential application for post-harvest aflatoxin control. Trends Food Sci. Technol. 2020, 106, 489–496. [Google Scholar] [CrossRef]
- Brekke, O.L.; Stringfellow, A.C.; Peplinski, A.J. Aflatoxin inactivation in corn by ammonia gas: Laboratory trials. J. Agric. Food Chem. 1978, 26, 1383–1389. [Google Scholar] [CrossRef]
- Xu, H.; Wang, L.; Sun, J.; Wang, L.; Guo, H.; Ye, Y.; Sun, X. Microbial detoxification of mycotoxins in food and feed. Crit. Rev. Food Sci. Nutr. 2022, 62, 4951–4969. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Wu, F.; Liu, F.; Wang, Q.; Zhang, X.; Selvaraj, J.N.; Zhao, Y.; Xing, F.; Yin, W.-B.; et al. A consensus ochratoxin a biosynthetic pathway: Insights from the genome sequence of Aspergillus ochraceus and a comparative genomic analysis. Appl. Environ. Microbiol. 2018, 84, e01009–e010018. [Google Scholar] [CrossRef]
- Shcherbakova, L.; Statsyuk, N.; Mikityuk, O.; Nazarova, T.; Dzhavakhiya, V. Aflatoxin B1 degradation by metabolites of Phoma glomerata PG41 isolated from natural substrate colonized by aflatoxigenic Aspergillus flavus. Jundishapur J. Microbiol. 2015, 8, e24324. [Google Scholar] [CrossRef]
- Gomaa, E.Z.; Abdelall, M.F.; El-Mahdy, O.M. Detoxification of aflatoxin B1 by antifungal compounds from Lactobacillus brevis and Lactobacillus paracasei, isolated from dairy products. Probiotics Antimicrob. Proteins 2018, 10, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Harkai, P.; Szabó, I.; Cserháti, M.; Krifaton, C.; Risa, A.; Radó, J.; Balázs, A.; Berta, K.; Kriszt, B. Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection. Int. Biodeterior. Biodegrad. 2016, 108, 48–56. [Google Scholar] [CrossRef]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Alberts, J.; Gelderblom, W.; Botha, A.; van Zyl, W. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Campagnollo, F.B.; Khaneghah, A.M.; Borges, L.L.; Bonato, M.A.; Fakhri, Y.; Barbalho, C.B.; Barbalho, R.L.; Corassin, C.H.; Oliveira, C.A. In vitro and in vivo capacity of yeast-based products to bind to aflatoxins B1 and M1 in media and foodstuffs: A systematic review and meta-analysis. Food Res. Int. 2020, 137, 109505. [Google Scholar] [CrossRef]
- Shetty, P.H.; Jespersen, L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 2006, 17, 48–55. [Google Scholar] [CrossRef]
- Ismail, A.; Levin, R.E.; Riaz, M.; Akhtar, S.; Gong, Y.Y.; de Oliveira, C.A. Effect of different microbial concentrations on binding of aflatoxin M1 and stability testing. Food Control 2017, 73, 492–496. [Google Scholar] [CrossRef]
- Deng, D.; Tang, J.; Liu, Z.; Tian, Z.; Song, M.; Cui, Y.; Rong, T.; Lu, H.; Yu, M.; Li, J.; et al. Functional characterization and whole-genome analysis of an aflatoxin-degrading Rhodococcus pyridinivorans strain. Biology 2022, 11, 774. [Google Scholar] [CrossRef]
- Rao, K.R.; Vipin, A.; Hariprasad, P.; Appaiah, K.A.; Venkateswaran, G. Biological detoxification of aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 2017, 71, 234–241. [Google Scholar] [CrossRef]
- Guo, Y.; Qin, X.; Tang, Y.; Ma, Q.; Zhang, J.; Zhao, L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020, 325, 126877. [Google Scholar] [CrossRef]
- Siahmoshteh, F.; Siciliano, I.; Banani, H.; Hamidi-Esfahani, Z.; Razzaghi-Abyaneh, M.; Gullino, M.L.; Spadaro, D. Efficacy of Bacillus subtilis and Bacillus amyloliquefaciens in the control of Aspergillus parasiticus growth and aflatoxins production on pistachio. Int. J. Food Microbiol. 2017, 254, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, H.; Li, Z.; Tan, Y.; Han, Y.; Wang, X.; Du, Z.; Liu, Y.; Yang, R.; Bai, Y.; et al. Safety evaluation of a novel strain of Bacteroides fragilis. Front. Microbiol. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, C.; Zhang, D.; Zhao, M.; Zheng, D.; Lyu, Y.; Cheng, W.; Guo, P.; Cui, Z. Effective degradation of aflatoxin B1 using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017, 224, 166–173. [Google Scholar] [CrossRef]
- Yang, P.; Lu, S.; Xiao, W.; Zheng, Z.; Jiang, S.; Jiang, S.; Jiang, S.; Cheng, J.; Zhang, D. Activity enhancement of Trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) by molecular docking and site-directed mutagenesis. Food Bioprod. Process. 2021, 129, 168–175. [Google Scholar] [CrossRef]
- Wang, X.; Qin, X.; Hao, Z.; Luo, H.; Yao, B.; Su, X. Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins 2019, 11, 566. [Google Scholar] [CrossRef]
- Qin, X.; Su, X.; Tu, T.; Zhang, J.; Wang, X.; Wang, Y.; Wang, Y.; Bai, Y.; Yao, B.; Luo, H.; et al. Enzymatic degradation of multiple major mycotoxins by dye-decolorizing peroxidase from Bacillus subtilis. Toxins 2021, 13, 429. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, H.; Yohannes, K.W.; Wan, Z.; Cao, Y.; Tron, T.; Lin, J.; Jiang, Y.; Li, H.; Wang, J. Degradation of aflatoxin B1 by a recombinant laccase from Trametes sp. C30 expressed in Saccharomyces cerevisiae: A mechanism assessment study in vitro and in vivo. Food Res. Int. 2021, 145, 110418. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Du, J.; Li, M.; Yuan, Z.; Guo, M.; Song, J.; Xie, X.; Chen, Y. A decision analysis model for KEGG pathway analysis. BMC Bioinform. 2016, 17, 407. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef]
- Yue, X.; Ren, X.; Fu, J.; Wei, N.; Altomare, C.; Haidukowski, M.; Logrieco, A.F.; Zhang, Q.; Li, P. Characterization and mechanism of aflatoxin degradation by a novel strain of Trichoderma reesei CGMCC3.5218. Front. Microbiol. 2022, 13, 1003039. [Google Scholar] [CrossRef]
- Cai, M.; Qian, Y.; Chen, N.; Ling, T.; Wang, J.; Jiang, H.; Wang, X.; Qi, K.; Zhou, Y. Detoxification of aflatoxin B1 by Stenotrophomonas sp. CW117 and characterization the thermophilic degradation process. Environ. Pollut. 2020, 261, 114178. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, Y.; Yang, Q. Antifungal properties and AFB1 detoxification activity of a new strain of Lactobacillus plantarum. J. Hazard. Mater. 2021, 414, 125569. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, R.; Ng, T.B.; Lai, Y.; Yang, J.; Ye, X. A new laccase of lac 2 from the white rot fungus Cerrena unicolor 6884 and lac 2-mediated degradation of aflatoxin B1. Toxins 2020, 12, 476. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Mavumengwana, V. Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 2016, 68, 92–96. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Sidu, S.; Tlou, M.G.; Mavumengwana, V. Aflatoxin B1 degradation by liquid cultures and lysates of three bacterial strains. Int. J. Food Microbiol. 2016, 233, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Fang, Q.; Liao, Z.; Xu, C.; Liang, Z.; Liu, T.; Zhong, Q.; Wang, L.; Fang, X.; Wang, J. Detoxification of aflatoxin B1 by a potential probiotic Bacillus amyloliquefaciens WF2020. Front. Microbiol. 2022, 13, 891091. [Google Scholar] [CrossRef]
- Petchkongkaew, A.; Taillandier, P.; Gasaluck, P.; Lebrihi, A. Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): Screening for aflatoxin B1 and ochratoxin A detoxification. J. Appl. Microbiol. 2008, 104, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Du, M.; Chen, J.; Liu, T.; Zheng, Y.; Liao, Z.; Zhong, Q.; Wang, L.; Fang, X.; Wang, J. Degradation and detoxification of aflatoxin B1 by tea-derived Aspergillus niger RAF106. Toxins 2020, 12, 777. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).