A Mass Spectrometry Database for Sea Cucumber Triterpene Glycosides
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Preparation
2.3. Data Acquisition
2.4. Data Processing and Spectral Library Constitution
2.5. LC-MS Analysis of the Eupentacta Fraudatrix Extract
3. Results and Discussion
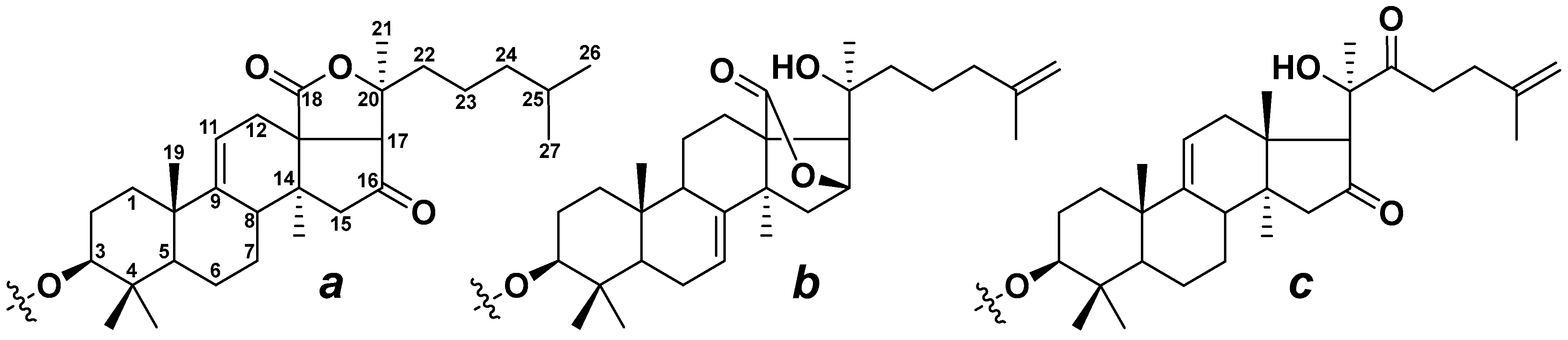
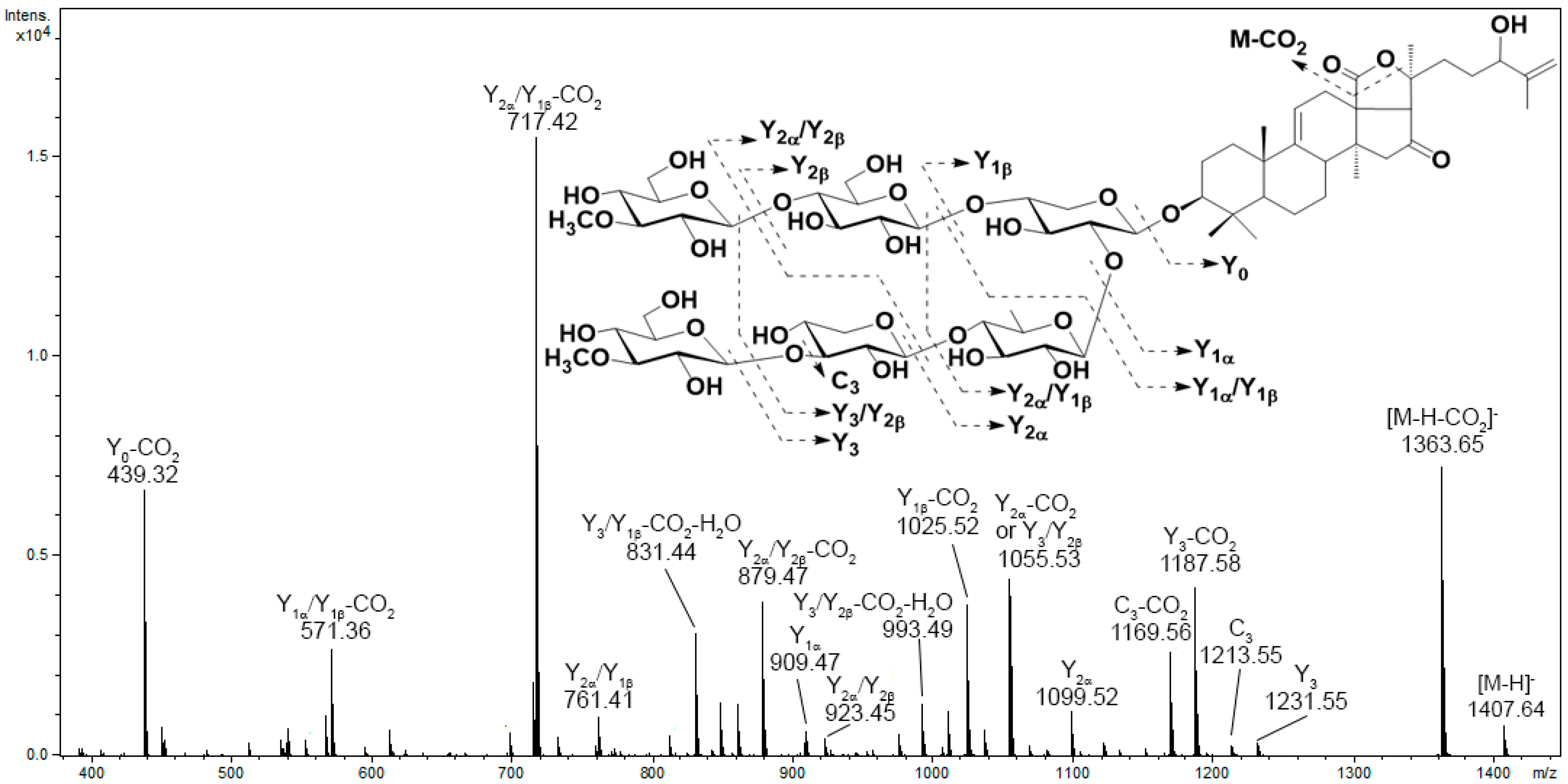
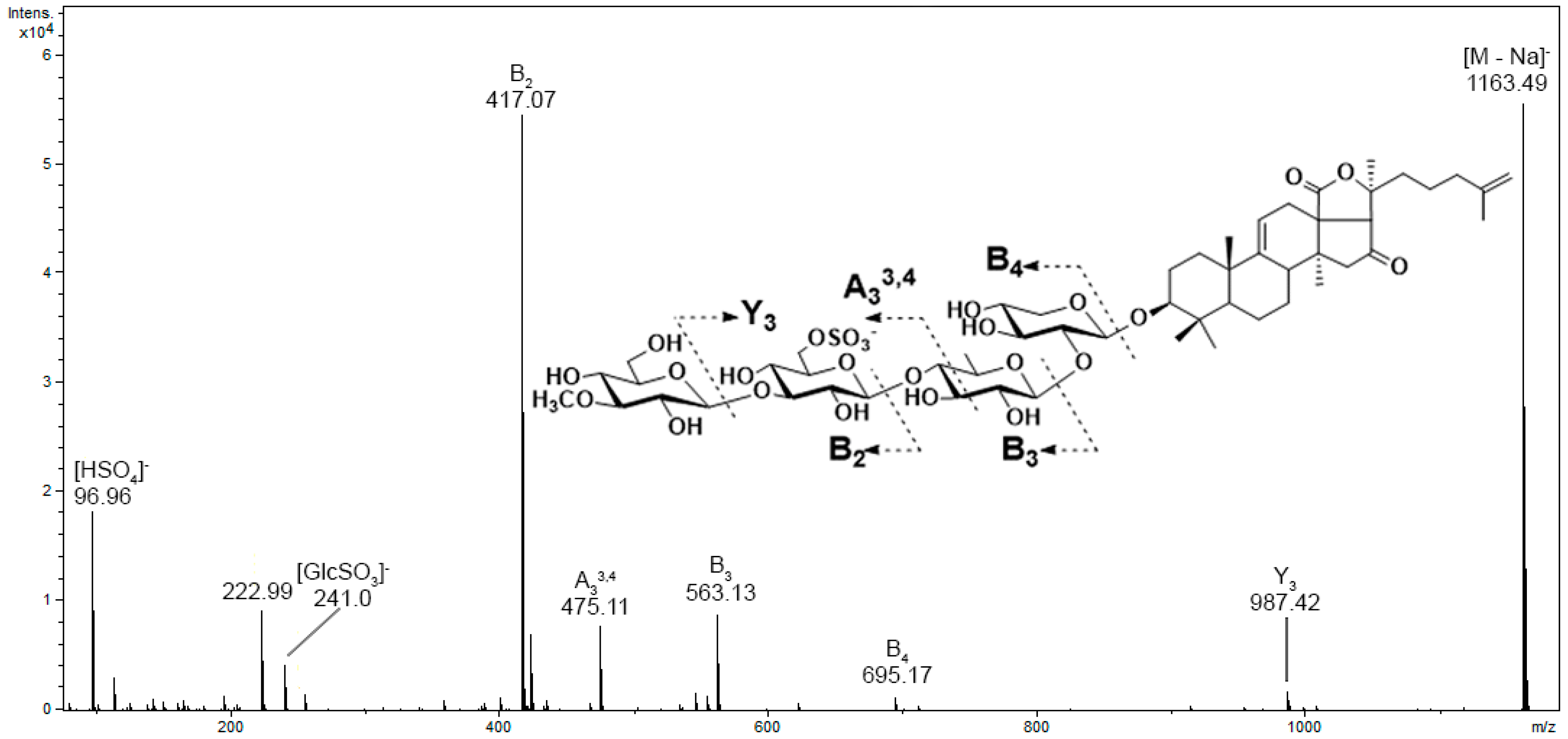
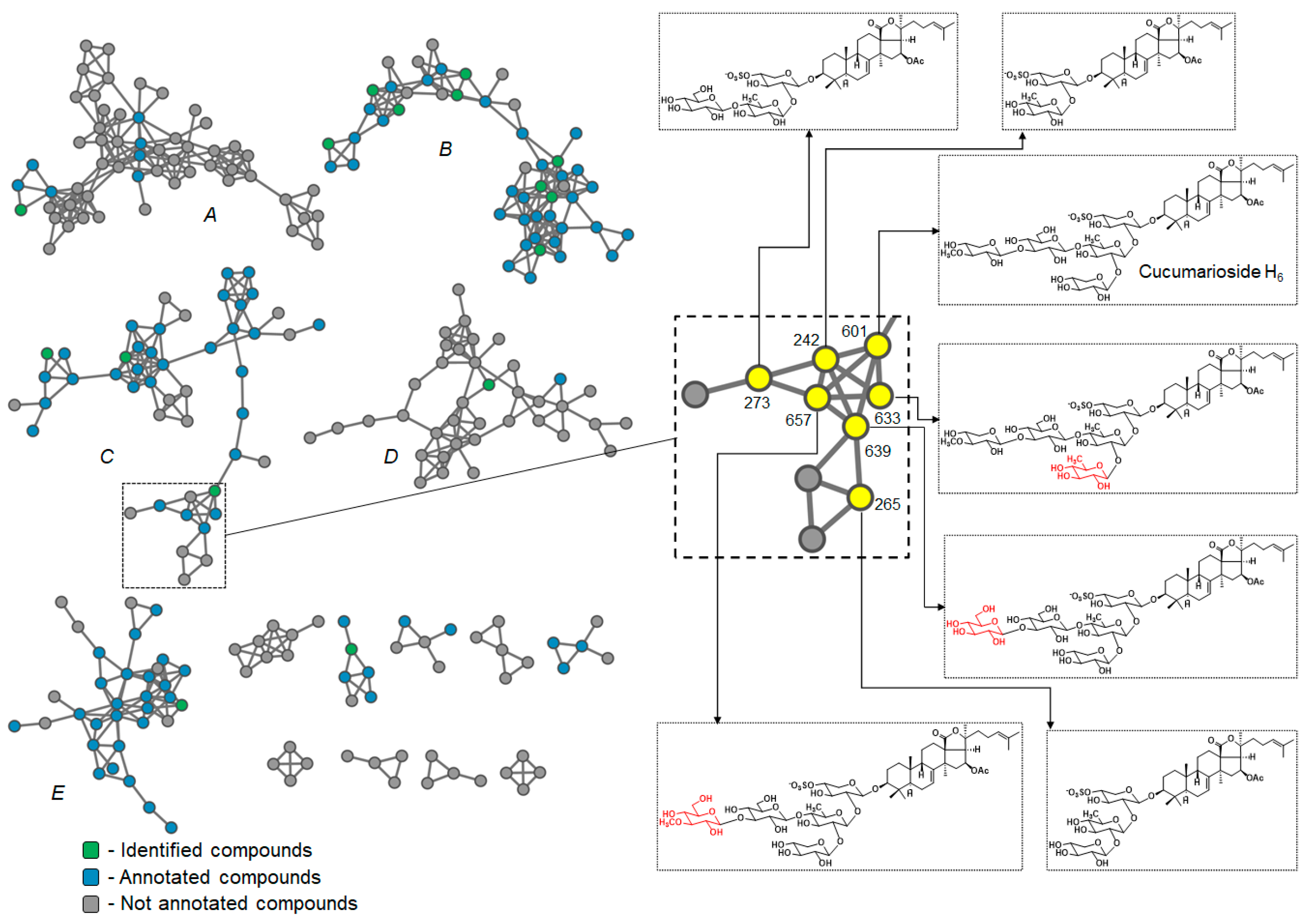
3.1. Analysis of the Mass Spectrometry Data
3.2. Analysis of the Chromatographic Behavior of Triterpene Glycosides
3.3. LC-MS Analysis of the Eupentacta fraudatrix Extract
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, S.; Himaya, S.W.A. Triterpene glycosides from sea cucumbers and their biological activities. Adv. Food Nutr. Res. 2012, 65, 297–319. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.R.; Freitas, A.C.; Rocha-Santos, T.A.P.; Duarte, A.C. Bioactive compounds derived from echinoderms. RSC Adv. 2014, 4, 29365–29382. [Google Scholar] [CrossRef]
- Khotimchenko, Y. Pharmacological potential of sea cucumbers. Int. J. Mol. Sci. 2018, 19, 1342. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Himaya, S.W.A.; Kim, S.K. Triterpenoids of marine origin as anti-cancer agents. Molecules 2013, 18, 7886–7909. [Google Scholar] [CrossRef] [PubMed]
- Aminin, D.L.; Pislyagin, E.A.; Menchinskaya, E.S.; Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I. Immunomodulatory and anticancer activity of sea cucumber triterpene glycosides. In Studies in Natural Products Chemistry; Rahman, A.-U., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 75–94. ISBN 9780444632944. [Google Scholar]
- Chludil, H.D.; Murray, A.P.; Seldes, A.M.; Maier, M.S. Biologically active triterpene glycosides from sea cucumbers (Holothuroidea, Echinodermata). In Studies in Natural Products Chemistry; Rahman, A.-U., Ed.; Elsevier Science Publisher: Amsterdam, The Netherlands, 2003; Volume 28, pp. 587–615. [Google Scholar]
- Kalinin, V.I.; Silchenko, A.S.; Avilov, S.A.; Stonik, V.A. Progress in the studies of triterpene glycosides from sea cucumbers (Holothuroidea, Echinodermata) between 2017 and 2021. Nat. Prod. Commun. 2021, 16, 1934578X211053934. [Google Scholar] [CrossRef]
- Kalinin, V.I.; Avilov, S.A.; Silchenko, A.S.; Stonik, V.A.; Elyakov, G.B. Triterpene glycosides of sea cucumbers (Holothuroidea, Echinodermata) as taxonomic markers. Nat. Prod. Commun. 2015, 10, 21–26. [Google Scholar] [CrossRef]
- Kalinin, V.I.; Aminin, D.L.; Avilov, S.A.; Silchenko, A.S.; Stonik, V.A. Triterpene glycosides from sea cucumbers (Holothurioidea, Echinodermata). Biological activities and functions. In Studies in Natural Products Chemistry (Bioactive Natural Products); Rahman, A.-U., Ed.; Elsevier Science Publisher: Amsterdam, The Netherlands, 2008; Volume 35, pp. 135–196. ISBN 1572-5995. [Google Scholar]
- Careaga, V.P.; Maier, M.S. Cytotoxic triterpene glycosides from sea cucumbers. In Handbook of Anticancer Drugs from Marine Origin; Kim, S.-K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 515–528. ISBN 978-3-319-07145-9. [Google Scholar]
- Popov, R.S.; Ivanchina, N.V.; Dmitrenok, P.S. Application of MS-based metabolomic approaches in analysis of starfish and sea cucumber bioactive compounds. Mar. Drugs 2022, 20, 320. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Nuzillard, J.M.; Van Der Hooft, J.J.J.; Renault, J.H.; Bertrand, S. Accelerating metabolite identification in natural product research: Toward an ideal combination of liquid chromatography-high-resolution tandem mass spectrometry and NMR profiling, in silico databases, and chemometrics. Anal. Chem. 2019, 91, 704–742. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Smith, C.A.; Maille, G.O.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Lee, S.; Hwang, S.; Seo, M.; Shin, K.B.; Kim, K.H.; Park, G.W.; Kim, J.Y.; Yoo, J.S.; No, K.T. BMDMS-NP: A comprehensive ESI-MS/MS spectral library of natural compounds. Phytochemistry 2020, 177, 112427. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Olivier-Jimenez, D.; Chollet-Krugler, M.; Rondeau, D.; Beniddir, M.A.; Ferron, S.; Delhaye, T.; Allard, P.M.; Wolfender, J.L.; Sipman, H.J.M.; Lücking, R.; et al. A database of high-resolution MS/MS spectra for lichen metabolites. Sci. Data 2019, 6, 294. [Google Scholar] [CrossRef] [PubMed]
- Fox Ramos, A.E.; Le Pogam, P.; Fox Alcover, C.; Otogo N’Nang, E.; Cauchie, G.; Hazni, H.; Awang, K.; Bréard, D.; Echavarren, A.M.; Frédérich, M.; et al. Collected mass spectrometry data on monoterpene indole alkaloids from natural product chemistry research. Sci. Data 2019, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Agnès, S.A.; Okpekon, T.; Kouadio, Y.A.; Jagora, A.; Bréard, D.; Costa, E.V.; da Silva, F.M.A.; Koolen, H.H.F.; Le Ray-Richomme, A.M.; Richomme, P.; et al. Implementation of a MS/MS database for isoquinoline alkaloids and other annonaceous metabolites. Sci. Data 2022, 9, 270. [Google Scholar] [CrossRef]
- Kang, K.B.; Jeong, E.; Son, S.; Lee, E.; Lee, S.; Choi, S.Y.; Kim, H.W.; Yang, H.; Shim, S.H. Mass spectrometry data on specialized metabolome of medicinal plants used in East Asian traditional medicine. Sci. Data 2022, 9, 528. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019, 48, D440–D444. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- GNPS. Available online: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c042045869bd47a08dc6adf0c13dc2be (accessed on 9 May 2023).
- Willighagen, E.L.; Mayfield, J.W.; Alvarsson, J.; Berg, A.; Carlsson, L.; Jeliazkova, N.; Kuhn, S.; Pluskal, T.; Rojas-Chertó, M.; Spjuth, O.; et al. The Chemistry Development Kit (CDK) v2.0: Atom typing, depiction, molecular formulas, and substructure searching. J. Cheminform. 2017, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- GNPS. Available online: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c12f1243f858421293acdb3a0addce03 (accessed on 9 May 2023).
- Domon, B.; Costello, C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Popov, R.S.; Ivanchina, N.V.; Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I.; Dolmatov, I.Y.; Stonik, V.A.; Dmitrenok, P.S. Metabolite profiling of triterpene glycosides of the Far Eastern sea cucumber Eupentacta fraudatrix and their distribution in various body components using LC-ESI QTOF-MS. Mar. Drugs 2017, 15, 302. [Google Scholar] [CrossRef]
- Griffiths, W.J. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom. Rev. 2003, 22, 81–152. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.X.; Santos, Á.; Fernandes, C.; Pinto, M.M.M. Liquid chromatography on the different methods for the determination of lipophilicity: An essential analytical tool in medicinal chemistry. Chemosensors 2022, 10, 340. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Martyyas, E.A.; Kalinin, V.I. Triterpene glycosides from the sea cucumber Eupentacta fraudatrix. Structure and biological action of cucumariosides A1, A3, A4, A5, A6, A12 and A15, seven new minor non-sulfated tetraosides and unprecedented 25-keto, 27-norholostane aglycone. Nat. Prod. Commun. 2012, 7, 517–525. [Google Scholar]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Martyyas, E.A.; Kalinin, V.I. Triterpene glycosides from the sea cucumber Eupentacta fraudatrix. Structure and cytotoxic action of cucumariosides A2, A7, A9, A10, A11, A13 and A14, seven new minor non-sulfated tetraosides and an aglycone with an uncommon 18-hydroxy group. Nat. Prod. Commun. 2012, 7, 845–852. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Yurchenko, E.A.; Kalinin, V.I. Structures and cytotoxic properties of cucumariosides H2, H3 and H4 from the sea cucumber Eupentacta fraudatrix. Nat. Prod. Res. 2012, 26, 1765–1774. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Yurchenko, E.A.; Kalinin, V.I. Structure of cucumariosides H5, H6, H7 and H8, triterpene glycosides from the sea cucumber Eupentacta fraudatrix and unprecedented aglycone with 16,22-epoxy-group. Nat. Prod. Commun. 2011, 6, 1075–1082. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Martyyas, E.A.; Kalinin, V.I. Triterpene glycosides from the sea cucumber Eupentacta fraudatrix. Structure and biological action of cucumariosides I1, I3, I4, three new minor disulfated pentaosides. Nat. Prod. Commun. 2013, 8, 1053–1058. [Google Scholar] [CrossRef] [PubMed]

| Order | Family | Species | Compounds |
|---|---|---|---|
| Dendrochirotida | Cucumariidae | Actinocucumis typica | Typicosides A1, A2, B1, C1, and C2 |
| Colochirus quadrangularis | Quadrangularisosides A, A1, B, B1, B2, C, C1, D, D1–D4, and E | ||
| Colochirus robustus | Colochirosides A1–A3, B1–B3, C, D, and E; Hemoiedemoside B; Lefevreosides B and C; Neothyonidioside | ||
| Cucumaria djakonovi | Cucumarioside A0-1; Frondoside D; Okhotoside A1-1 | ||
| Cucumaria fallax | Fallaxosides C1, C2, D1, D2, D6, and D7 | ||
| Cucumaria japonica | Cucumarioside A2-2 | ||
| Pseudocolochirus violaceus | Violaceusosides C, D, and E; Violaceusides II and A; Holothurinoside A; Liouvilloside A; Philinopside E | ||
| Staurocucumis turqueti | Turquetoside A | ||
| Thyonidium (=Duasmodactyla) kurilensis | Kurilosides A, A1-A3, C1, D, D1, E, F, G, H, I, I1, J, K, and K1; DS-Kurilosides L and M | ||
| Phyllophoridae | Neothynidium (=Massinium) magnum | Magnumosides A3, A4, B3, and C1–C4 | |
| Psolidae | Psolus chitonoides | Chitonoidosides A, A1, B C, D, E, E1, F, G, H, I, J, K, K1, and L | |
| Psolus fabricii | Psolusosides A, B, B1, B2, C1–C3, D1–D5, E, F, G, H, I, J, K, L, M, N, O, P, and Q | ||
| Sclerodactylidae | Cladolabes schmeltzii | Cladolosides A2, B, B1, B2, C, C1, C2, D, D1, D2, E1, E2, F1, F2, G, H1, I1, I2, J1, K1, K2, L1, M, M1, M2, N, O, P, P1–P3, Q, and R; Holotoxin A1 | |
| Eupentacta fraudatrix | Cucumariosides A1–A4, A6, A7, A9–A15, D, H2–H8, and I1–I4 | ||
| Molpadida | Caudinidae | Paracaudina chilensis | Chilensosides A, A1, B, C, D, E, F, and G |
| Valvatida | Solasteridae | Solaster pacificus | Pacificusosides A, B, C, E, G, H, and J; Cucumariosides C1 and C2 |
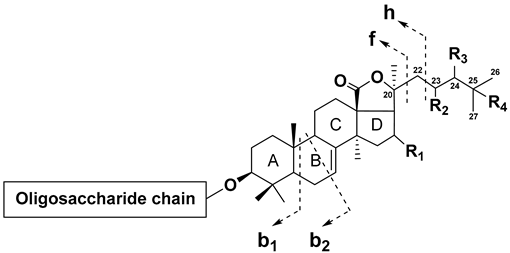 | ||||||||||||
| R1 | R2 | R3 | R4 | Double Bonds | Neutral Losses, Da | Example Compound * | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b1 | b2 | f + C6H8O4 | f + C4H6O4 | f + C3H4O4 | f + CO2 | h | ||||||
| OAc | H | H | H | - | 374 | 362 | 230 | 204 | 190 | 130 | - | Cucumarioside H7 |
| OAc | H | H | H | Δ24 | 372 | 360 | 228 | 202 | 188 | 128 | - | Lefevreoside B |
| OAc | H | H | H | Δ22E,24 | 370 | 358 | 226 | 200 | - | - | - | Typicoside A1 |
| OAc | H | H | H | Δ22Z,24 | 370 | 358 | 226 | 200 | - | - | - | Cucumarioside H5 |
| OAc | H | H | - | Δ25 | 372 | 360 | 228 | 202 | 188 | 128 | 70 | Colochiroside A1 |
| =O | H | H | - | Δ25 | 328 | 316 | - | - | - | 128 | 70 | Philinopside E |
| H | H | H | - | Δ25 | 314 | 302 | - | - | - | 128 | 70 | Colochiroside A2 |
| H | H | H | H | Δ24 | 314 | 302 | - | - | - | - | - | Colochiroside A3 |
| OAc | H | OH | - | Δ25 | 388 | 376 | 244 | 218 | 204 | 144 | 86 | Colochiroside B1 |
| OAc | H | H | OH | Δ23 | 388 | 376 | 244 | 218 | - | - | - | Colochiroside B2 |
| OAc | H | =O | - | Δ25 | 386 | 374 | 242 | 216 | 202 | 142 | 84 | Colochiroside B3 |
| OAc | =O | H | H | - | 388 | 376 | 244 | 218 | 204 | 144 | - | Okhotoside A1-1 |
| OAc | OH | H | H | - | 390 | 378 | 246 | 220 | 206 | - | - | Frondoside D |
| Compound | Retention Time, min | m/z of Precursor Ion | Match Score | ||||
|---|---|---|---|---|---|---|---|
| Standard Compound | Features Detected in E. fraudatrix | Δ (min) | Calculated m/z | Measured m/z | Δ (ppm) | ||
| Cucumarioside H8 | 6.3 | 6.3 | 0.0 | 1281.5216 | 1281.5135 | 6.3 | 0.930 |
| Cucumarioside I3 | 6.7 | 6.6 | 0.1 | 702.2487 | 702.2474 | 1.7 | 0.752 |
| Cucumarioside I4 | 7.4 | 7.4 | 0.0 | 630.2093 | 630.2104 | −1.7 | 0.777 |
| Cucumarioside H2 | 7.7 | 7.6 | 0.1 | 1325.5478 | 1325.5474 | 0.3 | 0.905 |
| Colochiroside B1 | 8.1 | 8.1 | 0.0 | 1193.5055 | 1193.4989 | 5.6 | 0.801 |
| Pacificusoside A | 8.1 | 8.2 | 0.1 | 1201.5284 | 1201.5260 | 2.0 | 0.911 |
| Colochiroside B2 | 8.3 | 8.3 | 0.0 | 1193.5055 | 1193.5030 | 2.1 | 0.936 |
| Cucumarioside H3 | 8.8 | 8.8 | 0.0 | 1181.4691 | 1181.4680 | 1.0 | 0.954 |
| Quadrangularisoside A | 9.0 | 9.0 | 0.0 | 1209.5004 | 1209.4946 | 4.8 | 0.907 |
| Cucumarioside A7 | 9.7 | 9.8 | 0.1 | 1113.5487 | 1113.5455 | 2.9 | 0.826 |
| Cucumarioside A11 | 10.0 | 10.0 | 0.0 | 1113.5487 | 1113.5454 | 3.0 | 0.917 |
| Pacificusoside J | 10.1 | 10.1 | 0.0 | 1131.5229 | 1131.5194 | 3.1 | 0.951 |
| Pacificusoside B | 10.5 | 10.6 | 0.1 | 1101.5123 | 1101.5099 | 2.2 | 0.947 |
| Magnumoside B3 | 10.6 | 10.8 | 0.2 | 1135.5000 | 1135.4929 | 6.3 | 0.889 |
| Cucumarioside H4 | 11.7 | 11.5 | 0.2 | 1353.5791 | 1353.5759 | 2.3 | 0.941 |
| Typicoside C2 | 12.0 | 12.0 | 0.0 | 643.2354 | 643.2363 | −1.5 | 0.887 |
| Cucumarioside H5 | 12.4 | 12.6 | 0.2 | 1307.5372 | 1307.5366 | 0.5 | 0.916 |
| Colochiroside A1 | 13.2 | 12.9 | 0.3 | 1193.5051 | 1193.5055 | −0.3 | 0.825 |
| Cucumarioside H6 | 13.6 | 13.4 | 0.2 | 1309.5529 | 1309.5569 | −3.1 | 0.892 |
| Typicoside A1 | 13.7 | 13.7 | 0.0 | 1175.4950 | 1175.4933 | 1.4 | 0.800 |
| Cucumarioside D | 14.2 | 14.3 | 0.1 | 1257.5910 | 1257.5961 | −4.1 | 0.763 |
| Pacificusoside G | 14.5 | 14.6 | 0.1 | 1081.5225 | 1081.5248 | −2.1 | 0.820 |
| Cucumarioside C1 | 14.7 | 14.8 | 0.1 | 1227.5804 | 1227.5812 | −0.6 | 0.944 |
| Pacificusoside E | 14.9 | 15.0 | 0.1 | 1081.5225 | 1081.5195 | 2.7 | 0.975 |
| Cucumarioside C2 | 15.1 | 15.2 | 0.1 | 1227.5804 | 1227.5812 | −0.6 | 0.976 |
| Cucumarioside A1 | 17.1 | 17.2 | 0.1 | 1097.5538 | 1097.5474 | 5.9 | 0.860 |
| Cucumarioside A15 | 18.7 | 18.7 | 0.0 | 1099.5694 | 1099.5643 | 4.7 | 0.970 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popov, R.S.; Ivanchina, N.V.; Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I.; Malyarenko, T.V.; Stonik, V.A.; Dmitrenok, P.S. A Mass Spectrometry Database for Sea Cucumber Triterpene Glycosides. Metabolites 2023, 13, 783. https://doi.org/10.3390/metabo13070783
Popov RS, Ivanchina NV, Silchenko AS, Avilov SA, Kalinin VI, Malyarenko TV, Stonik VA, Dmitrenok PS. A Mass Spectrometry Database for Sea Cucumber Triterpene Glycosides. Metabolites. 2023; 13(7):783. https://doi.org/10.3390/metabo13070783
Chicago/Turabian StylePopov, Roman S., Natalia V. Ivanchina, Alexandra S. Silchenko, Sergey A. Avilov, Vladimir I. Kalinin, Timofey V. Malyarenko, Valentin A. Stonik, and Pavel S. Dmitrenok. 2023. "A Mass Spectrometry Database for Sea Cucumber Triterpene Glycosides" Metabolites 13, no. 7: 783. https://doi.org/10.3390/metabo13070783
APA StylePopov, R. S., Ivanchina, N. V., Silchenko, A. S., Avilov, S. A., Kalinin, V. I., Malyarenko, T. V., Stonik, V. A., & Dmitrenok, P. S. (2023). A Mass Spectrometry Database for Sea Cucumber Triterpene Glycosides. Metabolites, 13(7), 783. https://doi.org/10.3390/metabo13070783














