Loss of Brain Angiogenesis Inhibitor-3 (BAI3) G-Protein Coupled Receptor in Mice Regulates Adaptive Thermogenesis by Enhancing Energy Expenditure
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal
2.2. Generation of BAI3 Gene-Deletion Mice
2.3. Western Blotting
2.4. Primer Design and PCR
2.5. Quantitative Magnetic Resonance (QMR)
2.6. Cold Exposure, Comprehensive Laboratory Animal Monitoring System, and Thermoneutrality
2.7. Tissue Histology and Analysis
2.8. Quantitative Real-Time PCR
2.9. Statistical Analysis
3. Results
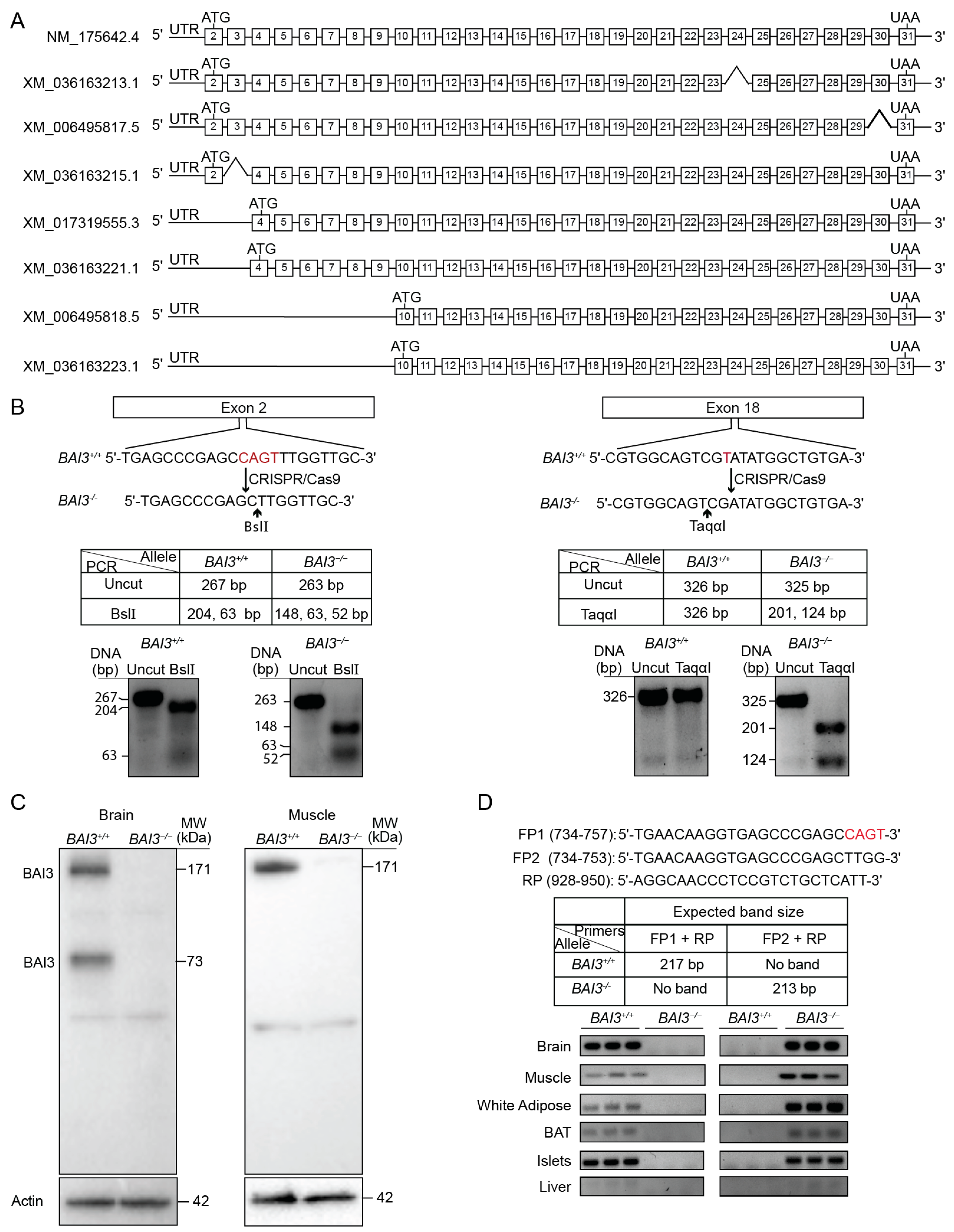
3.1. Generation and Validation of BAI3−/− Mice
3.2. Tissue Expression Profile of BAI3
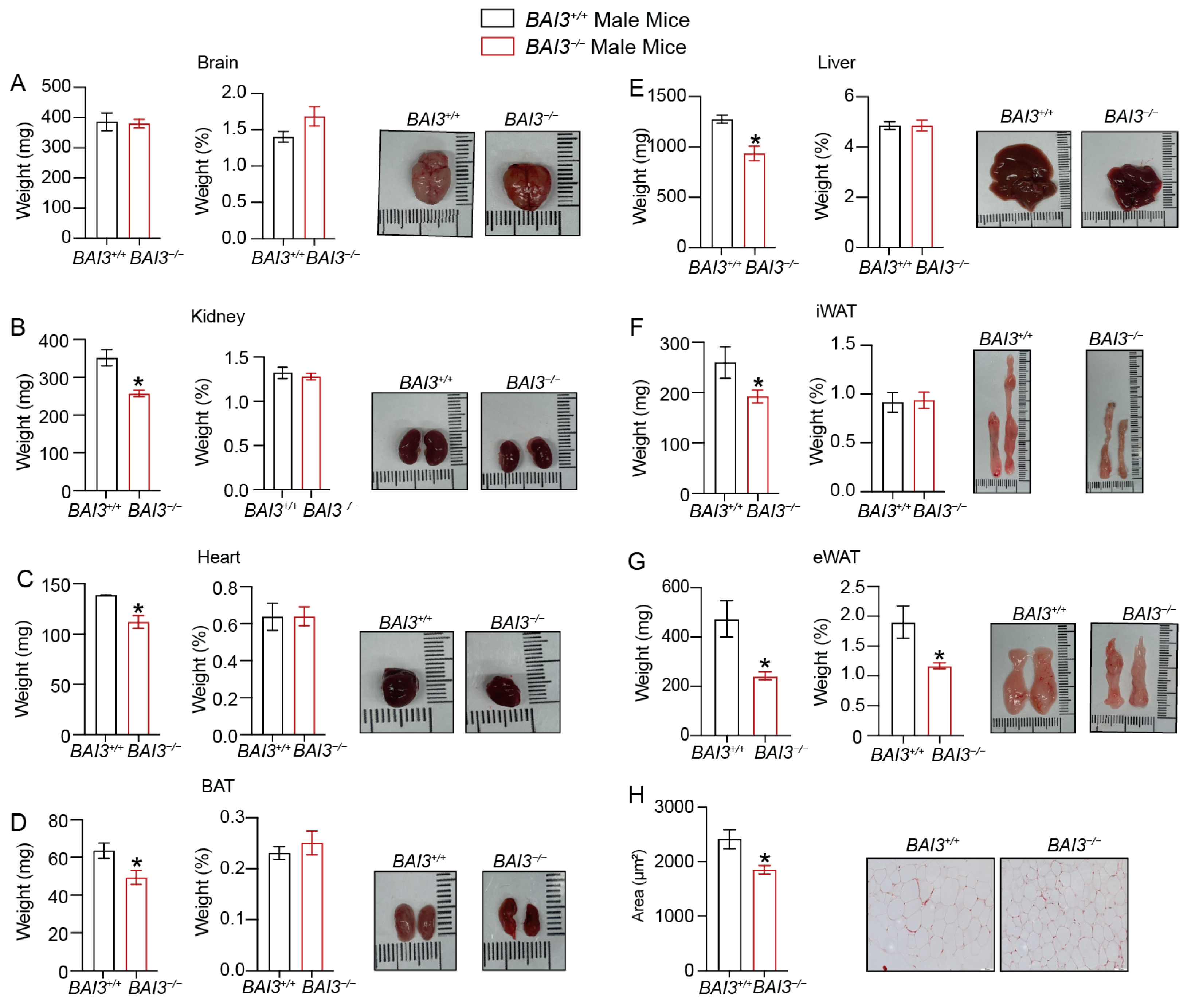
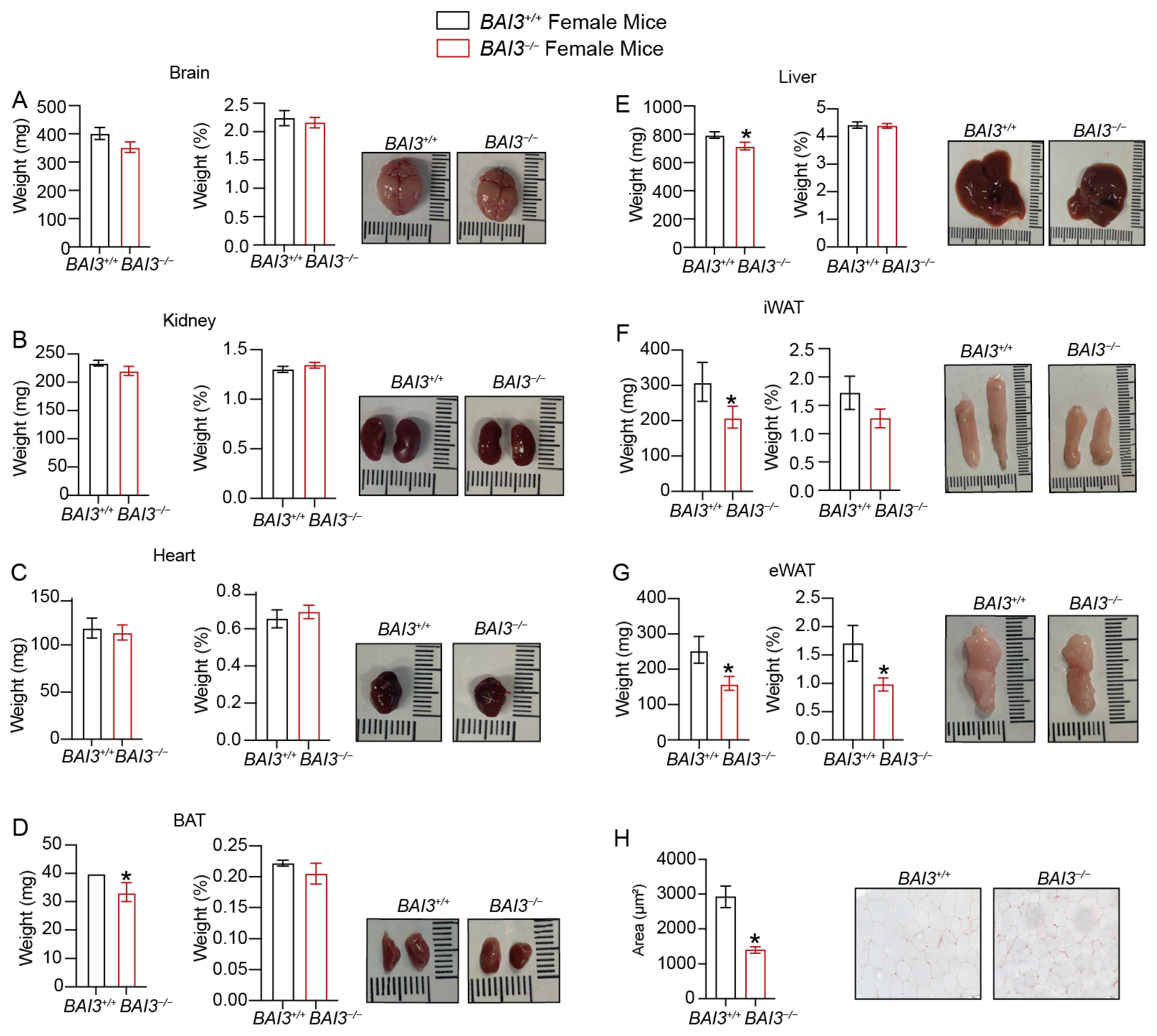
3.3. BAI3−/− Mice Have Reduced Body Weight
3.4. Loss of BAI3 Decreases WAT Tissue Weight
3.5. BAI3−/− Mice Have Enhanced Energy Expenditure
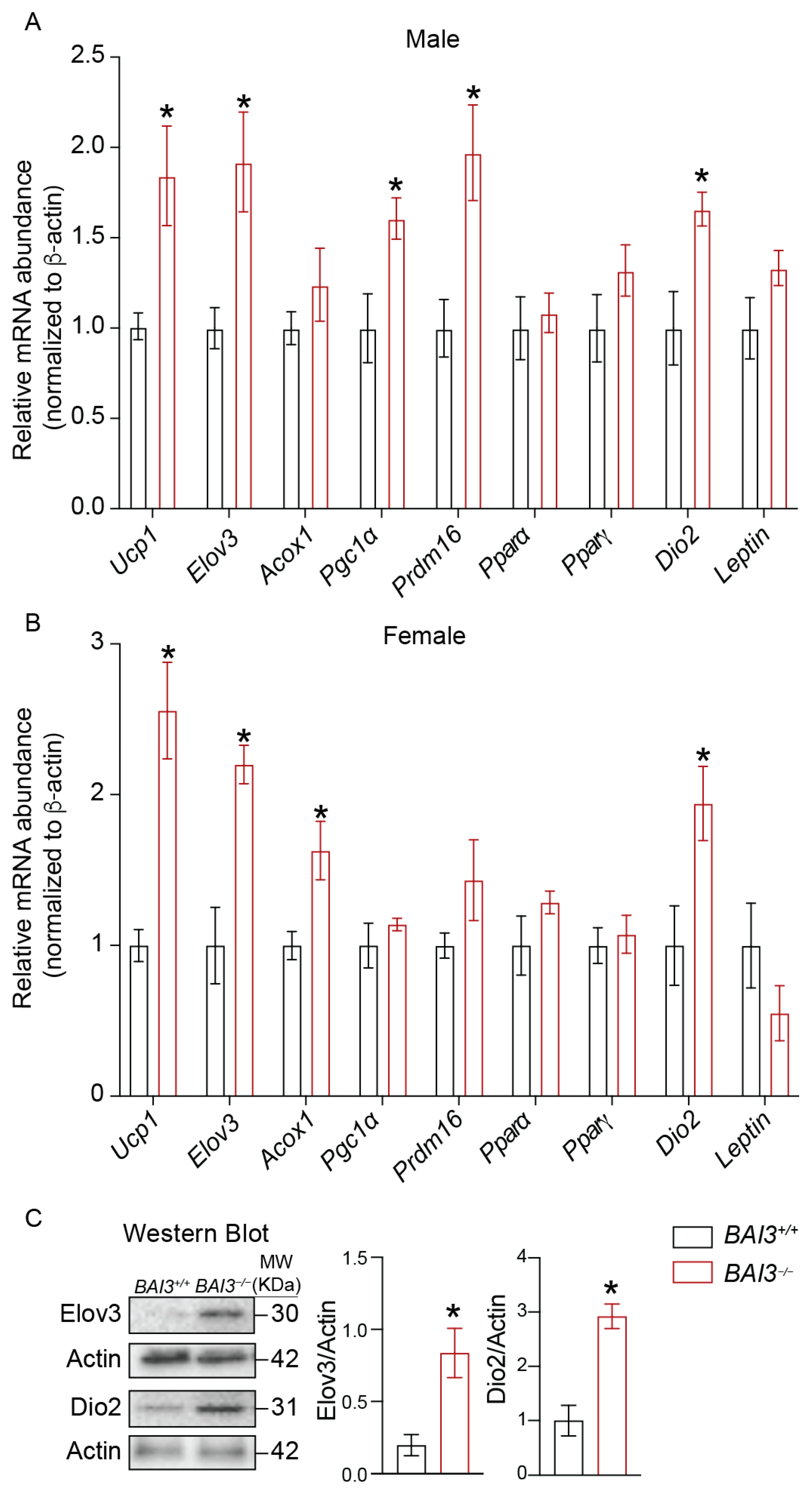
3.6. BAI3−/− Mice Exhibit Increased Expression of Genes Involved in Thermogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romieu, I.; Dossus, L.; Barquera, S.; Blottière, H.M.; Franks, P.W.; Gunter, M.; Hwalla, N.; Hursting, S.D.; Leitzmann, M.; Margetts, B.; et al. Energy balance and obesity: What are the main drivers? Cancer Causes Control. 2017, 28, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.O.; Wyatt, H.R.; Peters, J.C. Energy Balance and Obesity. Circulation 2012, 126, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.C.T.; Delgado, V.; Borlaug, B.A.; Bax, J.J. Diabesity: The combined burden of obesity and diabetes on heart disease and the role of imaging. Nat. Rev. Cardiol. 2021, 18, 291–304. [Google Scholar] [CrossRef]
- Chen, J.; Chen, L.; Sanseau, P.; Freudenberg, J.M.; Rajpal, D.K. Significant obesity-associated gene expression changes occur in the stomach but not intestines in obese mice. Physiol. Rep. 2016, 4, e12793. [Google Scholar] [CrossRef]
- Barbatelli, G.; Murano, I.; Madsen, L.; Hao, Q.; Jimenez, M.; Kristiansen, K.; Giacobino, J.P.; De Matteis, R.; Cinti, S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E1244–E1253. [Google Scholar] [CrossRef]
- Caron, A.; Reynolds, R.P.; Castorena, C.M.; Michael, N.J.; Lee, C.E.; Lee, S.; Berdeaux, R.; Scherer, P.E.; Elmquist, J.K. Adipocyte Gs but not Gi signaling regulates whole-body glucose homeostasis. Mol. Metab. 2019, 27, 11–21. [Google Scholar] [CrossRef]
- Kershaw, E.E.; Flier, J.S. Adipose Tissue as an Endocrine Organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef]
- Lee, M.-J.; Wu, Y.; Fried, S.K. Adipose tissue heterogeneity: Implication of depot differences in adipose tissue for obesity complications. Mol. Asp. Med. 2013, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, S.; Wang, M.; Cao, Q.; Zhang, Z.; Huang, Q.; Li, J.; Deng, Z.; Liu, T.; Liu, C.-L.; et al. Differential IL18 signaling via IL18 receptor and Na-Cl co-transporter discriminating thermogenesis and glucose metabolism regulation. Nat. Commun. 2022, 13, 7582. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ishibashi, J.; Trefely, S.; Shao, M.; Cowan, A.J.; Sakers, A.; Lim, H.-W.; O’connor, S.; Doan, M.; Cohen, P.; et al. A PRDM16-Driven Metabolic Signal from Adipocytes Regulates Precursor Cell Fate. Cell Metab. 2019, 30, 174–189.e5. [Google Scholar] [CrossRef]
- Caron, A.; Lee, S.; Elmquist, J.K.; Gautron, L. Leptin and brain–adipose crosstalks. Nat. Rev. Neurosci. 2018, 19, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, C.; Gaggini, M.; Carli, F.; Gastaldelli, A. The Subtle Balance between Lipolysis and Lipogenesis: A Critical Point in Metabolic Homeostasis. Nutrients 2015, 7, 9453–9474. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.E.; Ahmadian, M.; Jaworski, K.; Sarkadi-Nagy, E.; Sul, H.S. Regulation of Lipolysis in Adipocytes. Annu. Rev. Nutr. 2007, 27, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Chait, A.; den Hartigh, L.J. Adipose Tissue Distribution, Inflammation and Its Metabolic Consequences, Including Diabetes and Cardiovascular Disease. Front. Cardiovasc. Med. 2020, 7, 22. [Google Scholar] [CrossRef]
- Cannon, B.; Nedergaard, J. Brown Adipose Tissue: Function and Physiological Significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef]
- McNeill, B.T.; Suchacki, K.J.; Stimson, R.H. Mechanisms in endocrinology: Human brown adipose tissue as a therapeutic target: Warming up or cooling down? Eur. J. Endocrinol. 2021, 184, R243–R259. [Google Scholar] [CrossRef]
- Brownstein, A.J.; Veliova, M.; Acin-Perez, R.; Liesa, M.; Shirihai, O.S. ATP-consuming futile cycles as energy dissipating mechanisms to counteract obesity. Rev. Endocr. Metab. Disord. 2021, 23, 121–131. [Google Scholar] [CrossRef]
- Peng, X.-R.; Gennemark, P.; O’mahony, G.; Bartesaghi, S. Unlock the Thermogenic Potential of Adipose Tissue: Pharmacological Modulation and Implications for Treatment of Diabetes and Obesity. Front. Endocrinol. 2015, 6, 174. [Google Scholar] [CrossRef]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef]
- Sugiyama, M.G.; Agellon, L.B. Sex differences in lipid metabolism and metabolic disease risk. Biochem. Cell Biol. 2012, 90, 124–141. [Google Scholar] [CrossRef] [PubMed]
- Woodward, C.J.H.; Emery, P.W. Energy balance in rats given chronic hormone treatment. Br. J. Nutr. 1989, 61, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, M.D.; Krull, J.E.; Douglass, J.D.; Fasnacht, R.; Lara-Lince, F.; Meek, T.H.; Shi, X.; Damian, V.; Nguyen, H.T.; Matsen, M.E.; et al. Sex differences in microglial CX3CR1 signalling determine obesity susceptibility in mice. Nat. Commun. 2017, 8, 14556. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Frank, A.P.; Palmer, B.F.; Rodriguez-Navas, C.; Criollo, A.; Clegg, D.J. A sexually dimorphic hypothalamic response to chronic high-fat diet consumption. Int. J. Obes. 2016, 40, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.L.; Rennie, C.; Tarnopolsky, M.A. Substrate utilization during endurance exercise in men and women after endurance training. Am. J. Physiol. Metab. 2001, 280, E898–E907. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.C. Sexual Dimorphism in the Effects of Exercise on Metabolism of Lipids to Support Resting Metabolism. Front. Endocrinol. 2014, 5, 162. [Google Scholar] [CrossRef]
- Horton, T.J.; Pagliassotti, M.J.; Hobbs, K.; Hill, J.O.; Miotto, P.M.; McGlory, C.; Holloway, T.M.; Phillips, S.M.; Holloway, G.P.; Janssens, S.; et al. Fuel metabolism in men and women during and after long-duration exercise. J. Appl. Physiol. 1985, 85, 1823–1832. [Google Scholar] [CrossRef]
- Frias, J.P.; Macaraeg, G.B.; Ofrecio, J.; Yu, J.G.; Olefsky, J.M.; Kruszynska, Y.T. Decreased Susceptibility to Fatty Acid–Induced Peripheral Tissue Insulin Resistance in Women. Diabetes 2001, 50, 1344–1350. [Google Scholar] [CrossRef]
- Basu, R.; Man, C.D.; Campioni, M.; Basu, A.; Klee, G.; Toffolo, G.; Cobelli, C.; Rizza, R.A. Effects of Age and Sex on Postprandial Glucose Metabolism. Diabetes 2006, 55, 2001–2014. [Google Scholar] [CrossRef]
- van Genugten, R.E.; Utzschneider, K.M.; Tong, J.; Gerchman, F.; Zraika, S.; Udayasankar, J.; Boyko, E.J.; Fujimoto, W.Y.; Kahn, S.E.; the American Diabetes Association GENNID Study Group. Effects of Sex and Hormone Replacement Therapy Use on the Prevalence of Isolated Impaired Fasting Glucose and Isolated Impaired Glucose Tolerance in Subjects with a Family History of Type 2 Diabetes. Diabetes 2006, 55, 3529–3535. [Google Scholar] [CrossRef]
- Stephenson, J.R.; Purcell, R.H.; Hall, R.A. The BAI subfamily of adhesion GPCRs: Synaptic regulation and beyond. Trends Pharmacol. Sci. 2014, 35, 208–215. [Google Scholar] [CrossRef]
- Bolliger, M.F.; Martinelli, D.C.; Südhof, T.C. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 2534–2539. [Google Scholar] [CrossRef] [PubMed]
- Araç, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Südhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378. [Google Scholar] [CrossRef]
- Hamoud, N.; Tran, V.; Aimi, T.; Kakegawa, W.; Lahaie, S.; Thibault, M.-P.; Pelletier, A.; Wong, G.W.; Kim, I.-S.; Kania, A.; et al. Spatiotemporal regulation of the GPCR activity of BAI3 by C1qL4 and Stabilin-2 controls myoblast fusion. Nat. Commun. 2018, 9, 4470. [Google Scholar] [CrossRef]
- Hamoud, N.; Tran, V.; Croteau, L.-P.; Kania, A.; Côté, J.-F. G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc. Natl. Acad. Sci. USA 2014, 111, 3745–3750. [Google Scholar] [CrossRef]
- Kim, J.H.; Jin, P.; Duan, R.; Chen, E.H. Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Dev. 2015, 32, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.K.; Madsen, L.; Pedersen, L.M.; Hallenborg, P.; Hagland, H.; Viste, K.; Døskeland, S.O.; Kristiansen, K. Cyclic AMP (cAMP)-Mediated Stimulation of Adipocyte Differentiation Requires the Synergistic Action of Epac- and cAMP-Dependent Protein Kinase-Dependent Processes. Mol. Cell Biol. 2008, 28, 3804–3816. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fromme, T.; Schweizer, S.; Schöttl, T.; Klingenspor, M. Taking control over intracellular fatty acid levels is essential for the analysis of thermogenic function in cultured primary brown and brite/beige adipocytes. EMBO Rep. 2014, 15, 1069–1076. [Google Scholar] [CrossRef]
- Collins, S. β-adrenoceptor signaling networks in adipocytes for recruiting stored fat and energy expenditure. Front. Endocrinol. 2011, 2, 102. [Google Scholar] [CrossRef]
- Park, H.; He, A.; Lodhi, I.J. Lipid Regulators of Thermogenic Fat Activation. Trends Endocrinol. Metab. 2019, 30, 710–723. [Google Scholar] [CrossRef]
- Jiang, Y.; Berry, D.C.; Graff, J.M. Distinct cellular and molecular mechanisms for β3 adrenergic receptor-induced beige adipocyte formation. Elife 2017, 6, 30329. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, Z.; Zeng, T.; Lim, Y.C.; Wang, Y.; Xie, X.; Yang, S.; Huang, C.; Xu, M.; Tao, L.; et al. cAMP-MicroRNA-203-IFNγ network regulates subcutaneous white fat browning and glucose tolerance. Mol. Metab. 2019, 28, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Ueta, C.B.; Fernandes, G.W.; Capelo, L.P.; Fonseca, T.L.; Maculan, F.D.; Gouveia, C.H.A.; Brum, P.C.A.; Christoffolete, M.; Aoki, M.S.; Lancellotti, C.L.; et al. β1 Adrenergic receptor is key to cold- and diet-induced thermogenesis in mice. J. Endocrinol. 2012, 214, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Quin, J.D.; Fisher, B.M.; Paterson, K.R.; Inoue, A.; Beastall, G.H.; MacCuish, A.C. Acute Response to Recombinant Insulin-like Growth Factor I in a Patient with Mendenhall’s Syndrome. New Engl. J. Med. 1990, 323, 1425–1426. [Google Scholar] [CrossRef]
- Bray, M.S.; Shaw, C.A.; Moore, M.W.S.; Peliciari-Garcia, R.A.; Zanquetta, M.M.; Durgan, D.J.; Jeong, W.J.; Tsai, J.-Y.; Bugger, H.; Zhang, D.; et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am. J. Physiol. Circ. Physiol. 2008, 294, H1036–H1047. [Google Scholar] [CrossRef]
- Cornelissen, G. Cosinor-based rhythmometry. Theor. Biol. Med. Model. 2014, 11, 16. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Sun, K.; Li, X.; Scherer, P.E. Extracellular Matrix (ECM) and Fibrosis in Adipose Tissue: Overview and Perspectives. Compr. Physiol. 2023, 13, 4387–4407. [Google Scholar] [CrossRef]
- Lanoue, V.; Usardi, A.; Sigoillot, S.M.; Talleur, M.; Iyer, K.; Mariani, J.; Isope, P.; Vodjdani, G.; Heintz, N.; Selimi, F. The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Mol. Psychiatry 2013, 18, 943–950. [Google Scholar] [CrossRef]
- Kee, H.J.; Ahn, K.Y.; Choi, K.C.; Song, J.W.; Heo, T.; Jung, S.; Kim, J.-K.; Bae, C.S.; Kim, K.K. Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett. 2004, 569, 307–316. [Google Scholar] [CrossRef]
- Sticco, M.J.; Palomino, P.A.P.; Lukacsovich, D.; Thompson, B.L.; Földy, C.; Ressl, S.; Martinelli, D.C. C1QL3 promotes cell-cell adhesion by mediating complex formation between ADGRB3/BAI3 and neuronal pentraxins. FASEB J. 2021, 35, e21194. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Nguyen, D.C.; Schaid, M.D.; Lei, X.; Balamurugan, A.N.; Wong, G.W.; Kim, J.; Koltes, J.E.; Kimple, M.E.; Bhatnagar, S. Complement 1q-like-3 protein inhibits insulin secretion from pancreatic β-cells via the cell adhesion G protein–coupled receptor BAI3. J. Biol. Chem. 2018, 293, 18086–18098. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Ke, S.; Chen, Y.; Chen, L.; Lu, X.; Ding, F.; Yang, L.; Tang, Y.; Yu, Y. Expression patterns of C1ql4 and its cell-adhesion GPCR Bai3 in the murine testis and functional roles in steroidogenesis. FASEB J. 2019, 33, 4893–4906. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Hensch, T.K.; Ackerman, L.; Barbel, S.; Jan, L.Y.; Jan, Y.N. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 1996, 379, 837–840. [Google Scholar] [CrossRef] [PubMed]
- Sigoillot, S.M.; Iyer, K.; Binda, F.; González-Calvo, I.; Talleur, M.; Vodjdani, G.; Isope, P.; Selimi, F. The Secreted Protein C1QL1 and Its Receptor BAI3 Control the Synaptic Connectivity of Excitatory Inputs Converging on Cerebellar Purkinje Cells. Cell Rep. 2015, 10, 820–832. [Google Scholar] [CrossRef]
- Wang, C.Y.; Liu, Z.; Ng, Y.H.; Südhof, T.C. A Synaptic Circuit Required for Acquisition but Not Recall of Social Transmission of Food Preference. Neuron 2020, 107, 144–157.e4. [Google Scholar] [CrossRef]
- Abmayr, S.M.; Pavlath, G.K. Myoblast fusion: Lessons from flies and mice. Development 2012, 139, 641–656. [Google Scholar] [CrossRef]
- Yang, L.; Colditz, G.A. Prevalence of Overweight and Obesity in the United States, 2007-2012. JAMA Intern. Med. 2015, 175, 1412–1413. [Google Scholar] [CrossRef]
- Lückerath, W. New ways of functional diagnosis? ZWR 1987, 96, 714–716. [Google Scholar]
- Hong, J.; Stubbins, R.E.; Smith, R.R.E.; Harvey, A.; Núñez, N.P. Differential susceptibility to obesity between male, female and ovariectomized female mice. Nutr. J. 2009, 8, 11. [Google Scholar] [CrossRef]
- Casimiro, I.; Stull, N.D.; Tersey, S.A.; Mirmira, R.G. Phenotypic sexual dimorphism in response to dietary fat manipulation in C57BL/6J mice. J. Diabetes Its Complicat. 2021, 35, 107795. [Google Scholar] [CrossRef] [PubMed]
- Ahima, R.S.; Saper, C.B.; Flier, J.S.; Elmquist, J.K. Leptin Regulation of Neuroendocrine Systems. Front. Neuroendocr. 2000, 21, 263–307. [Google Scholar] [CrossRef] [PubMed]
- Plum, L.; Belgardt, B.F.; Brüning, J.C. Central insulin action in energy and glucose homeostasis. J. Clin. Investig. 2006, 116, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-H.; Song, N.-J.; Choi, J.H.; Yun, U.J.; Park, K.W. Mechanisms underlying UCP1 dependent and independent adipocyte thermogenesis. Obes. Rev. 2019, 20, 241–251. [Google Scholar] [CrossRef]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]



| Cyclophilin F | CAG ACG CCA CTG TCG CTT T |
| Cyclophilin R | TGT CTT TGG AAC TTT GTC TG |
| Ucp1 F | AGG CTT CCA GTA CCA TTA GGT |
| Ucp1R | CTG AGT GAG GCA AAG CTG ATT T |
| Dio2 F | GGC TGA CTT CCT GTT GGT AT |
| Dio2 R | TTG GTT CCG GTG CTT CTT |
| Elov3 F | TAC ATC TGG AGG CAG GAG AA |
| Elov3 R | GGT GGA AGA AGT GAG CGA ATA G |
| Prdm16 F | CAC AAG TCC TAC ACG CAG TT |
| Prdm16 R | TTG TTG AGG GAG GAG GTA GT |
| Pparα F | CGA AGA CAA AGA GGC AGA GG |
| Pparα R | TGA TGT CAC AGA ACG GCT TC |
| Pgc1α F | ATG TGT CGC CTT CTT GCT CT |
| Pgc1α R | ATC TAC TGC CTG GGG ACC TT |
| Leptin F | GTG GCT TTG GTC CTA TCT GTC |
| Leptin R | CGT GTG TGA AAT GTC ATT GAT CC |
| Pparγ1 F | GCG GCT GAG AAA TCA CGT TC |
| Pparγ1 R | GAA TAT CAG TGG TTC ACC GCT TC |
| Acox1 F | GTC TCC GTC ATG AAT CCC GA |
| Acox1 R | TGC GAT GCC AAA TTC CCT CA |
| Male | Mesor | Amplitude | Phase | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BAI3+/+ | BAI3−/− | p-Value | BAI3+/+ | BAI3−/− | p-Value | BAI3+/+ | BAI3−/− | p-Value | |
| RER | 0.876 ± 0.001 | 0.883 ± 0.002 | <0.01 | 0.072 ± 0.002 | 0.083 ± 0.002 | <0.01 | 17.24 ± 0.084 | 16.73 ± 0.099 | <0.01 |
| Energy expenditure | 0.0183 ± 0.00015 | 0.0225 ± 0.00016 | 6.45 × 10−19 | 0.003458 ± 0.00021 | 0.004 ± 0.00022 | 0.07 | 17.89 ± 0.236 | 17.58 ± 0.213 | 0.33 |
| Food intake | 0.054785 ± 0.002 | 0.043344 ± 0.0014 | 7.57 × 10−5 | 0.032303 ± 0.0028 | 0.027483 ± 0.0019 | 0.18 | 17.22 ± 0.33 | 16.17 ± 0.27 | 0.03 |
| Activity | 527.513 ± 24.02 | 460.992 ± 19.14 | 0.04 | 392.07 ± 3 4 | 406.235 ± 27.09 | 0.74 | 17.22 ± 0.33 | 17.76 ± 0.25 | 0.212 |
| Female | Mesor | Amplitude | Phase | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BAI3+/+ | BAI3−/− | p-Value | BAI3+/+ | BAI3−/− | p-Value | BAI3+/+ | BAI3−/− | p-Value | |
| RER | 0.897 ± 0.002 | 0.88 ± 0.002 | 1.05 × 10−6 | 0.056 ± 0.003 | 0.068 ± 0.002 | <0.01 | 15.52 ± 0.17 | 16.83 ± 0.13 | 4.4 × 10−7 |
| Energy expenditure | 0.0228 ± 0.00015 | 0.0262 ± 0.00017 | 2.65 × 10−15 | 0.004022 ± 0.00021 | 0.004706 ± 0.00025 | 0.05 | 17.35 ± 0.20 | 17.80 ± 0.20 | 0.13 |
| Food intake | 0.058294 ± 0.0022 | 0.053863 ± 0.0013 | 0.07 | 0.025098 ± 0.0031 | 0.032829 ± 0.0019 | 0.03 | 15.70 ± 0.47 | 16.31 ± 0.22 | 0.20 |
| Activity | 480.247 ± 20 | 503.837 ± 15 | 0.34 | 366.279 ± 29 | 417.875 ± 21 | 0.14 | 18.18 ± 0.30 | 17.78 ± 0.19 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsharif, H.; Latimer, M.N.; Perez, K.C.; Alexander, J.; Rahman, M.M.; Challa, A.K.; Kim, J.-A.; Ramanadham, S.; Young, M.; Bhatnagar, S. Loss of Brain Angiogenesis Inhibitor-3 (BAI3) G-Protein Coupled Receptor in Mice Regulates Adaptive Thermogenesis by Enhancing Energy Expenditure. Metabolites 2023, 13, 711. https://doi.org/10.3390/metabo13060711
Alsharif H, Latimer MN, Perez KC, Alexander J, Rahman MM, Challa AK, Kim J-A, Ramanadham S, Young M, Bhatnagar S. Loss of Brain Angiogenesis Inhibitor-3 (BAI3) G-Protein Coupled Receptor in Mice Regulates Adaptive Thermogenesis by Enhancing Energy Expenditure. Metabolites. 2023; 13(6):711. https://doi.org/10.3390/metabo13060711
Chicago/Turabian StyleAlsharif, Haifa, Mary N. Latimer, Katherine C. Perez, Justin Alexander, Md Mostafizur Rahman, Anil K. Challa, Jeong-A. Kim, Sasanka Ramanadham, Martin Young, and Sushant Bhatnagar. 2023. "Loss of Brain Angiogenesis Inhibitor-3 (BAI3) G-Protein Coupled Receptor in Mice Regulates Adaptive Thermogenesis by Enhancing Energy Expenditure" Metabolites 13, no. 6: 711. https://doi.org/10.3390/metabo13060711
APA StyleAlsharif, H., Latimer, M. N., Perez, K. C., Alexander, J., Rahman, M. M., Challa, A. K., Kim, J.-A., Ramanadham, S., Young, M., & Bhatnagar, S. (2023). Loss of Brain Angiogenesis Inhibitor-3 (BAI3) G-Protein Coupled Receptor in Mice Regulates Adaptive Thermogenesis by Enhancing Energy Expenditure. Metabolites, 13(6), 711. https://doi.org/10.3390/metabo13060711








