Fatty Acid Metabolites and the Tumor Microenvironment as Potent Regulators of Cancer Stem Cell Signaling
Abstract
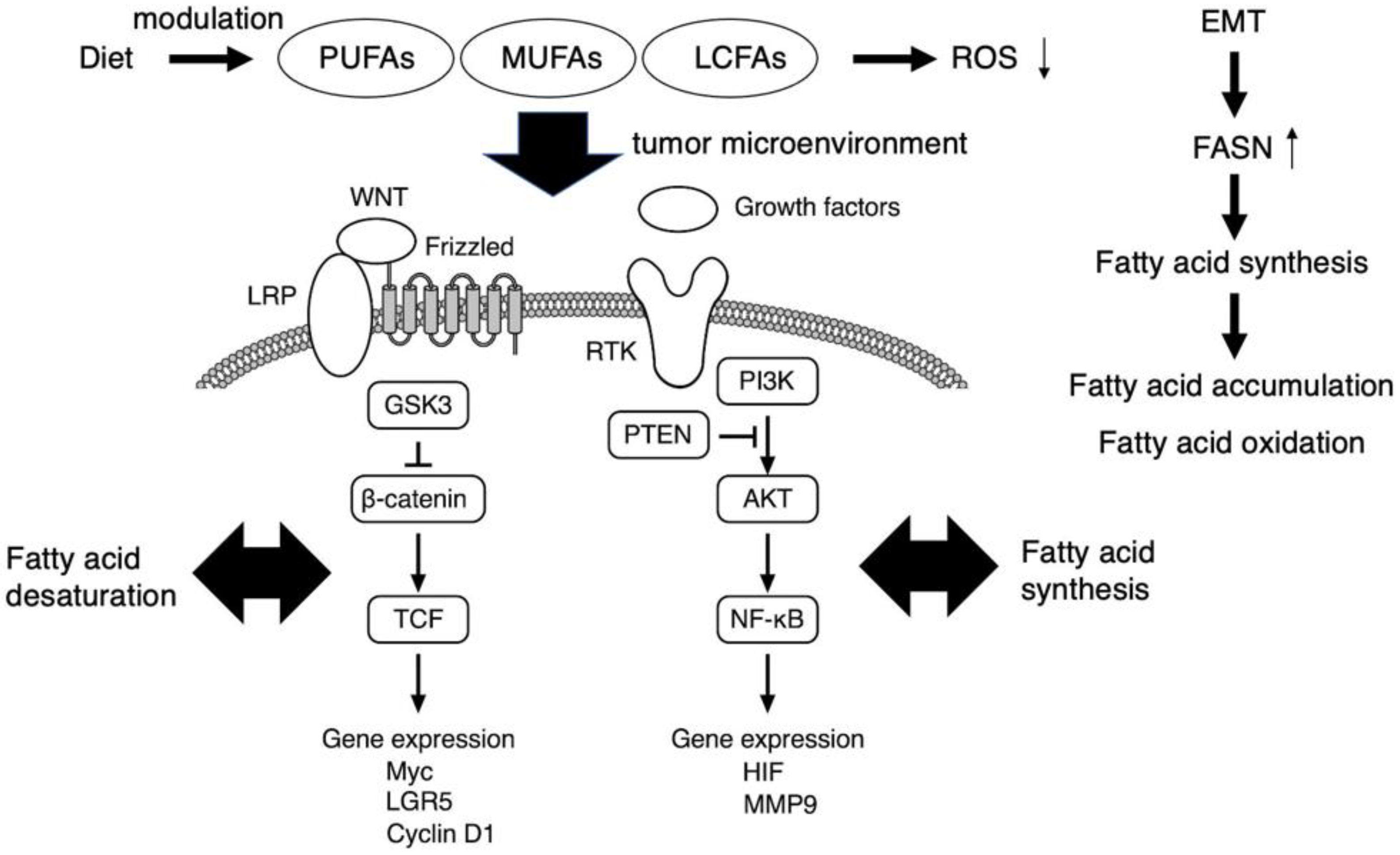
1. Introduction
2. Cancer Cell Signaling and Cancer Stem Cells
3. Metabolic Reprogramming and the Tumor Microenvironment as a Target of EMT-Induced Cancer Stem Cell Phenotypes
4. Therapy Resistance in Cancer Stem Cells
5. Dietary Intervention Impacting Fatty Acid Metabolites and the Tumor Microenvironment as Potent Regulators of Cancer Stem Cell Signaling
6. Integrative Healthcare Approach with Emerging Single-Cell Technologies for Precision Nutrition and Functional Medicine
7. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Gomez, R.L.; Ibragimova, S.; Ramachandran, R.; Philpott, A.; Ali, F.R. Tumoral heterogeneity in neuroblastoma. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188805. [Google Scholar] [CrossRef]
- Rodrigues, F.S.; Ciccarelli, F.D.; Malanchi, I. Reflected stemness as a potential driver of the tumour microenvironment. Trends Cell Biol. 2022, 32, 979–987. [Google Scholar] [CrossRef]
- Antonica, F.; Santomaso, L.; Pernici, D.; Petrucci, L.; Aiello, G.; Cutarelli, A.; Conti, L.; Romanel, A.; Miele, E.; Tebaldi, T.; et al. A slow-cycling/quiescent cells subpopulation is involved in glioma invasiveness. Nat. Commun. 2022, 13, 4767. [Google Scholar]
- Jehanno, C.; Vulin, M.; Richina, V.; Richina, F.; Bentires-Alj, M. Phenotypic plasticity during metastatic colonization. Trends Cell Biol. 2022, 32, 854–867. [Google Scholar] [CrossRef]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Taussig, D.C.; Miraki-Moud, F.; Anjos-Afonso, F.; Pearce, D.J.; Allen, K.; Ridler, C.; Lillington, D.; Oakervee, H.; Cavenagh, J.; Agrawal, S.G.; et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood 2008, 112, 568–575. [Google Scholar]
- Eppert, K.; Takenaka, K.; Lechman, E.R.; Waldron, L.; Nilsson, B.; van Galen, P.; Metzeler, K.H.; Poeppl, A.; Ling, V.; Beyene, J.; et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011, 17, 1086–1093. [Google Scholar] [CrossRef]
- Sarry, J.E.; Murphy, K.; Perry, R.; Sanchez, P.V.; Secreto, A.; Keefer, C.; Swider, C.R.; Strzelecki, A.C.; Cavelier, C.; Récher, C.; et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rγc-deficient mice. J. Clin. Investig. 2011, 121, 384–395. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Meyer, M.J.; Fleming, J.M.; Lin, A.F.; Hussnain, S.A.; Ginsburg, E.; Vonderhaar, B.K. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010, 70, 4624–4633. [Google Scholar] [CrossRef]
- Tang, C.; Ang, B.T.; Pervaiz, S. Cancer stem cell: Target for anti-cancer therapy. FASEB J. 2007, 21, 3777–3785. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; St Clair, R.; Baljevic, M.; White, I.; Jin, D.K.; et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J. Clin. Investig. 2008, 118, 2111–2120. [Google Scholar] [PubMed]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Dalerba, P.; Cho, R.W.; Clarke, M.F. Cancer stem cells: Models and concepts. Annu. Rev. Med. 2007, 58, 267–284. [Google Scholar] [CrossRef]
- Dick, E. Stem cell concepts renew cancer research. Blood 2008, 112, 4793–4807. [Google Scholar] [CrossRef]
- Sellers, Z.P.; Schneider, G.; Bujko, K.; Suszynska, M.; Pedziwiatr, D. Do cancer cell lines have fixed or fluctuating stem cell phenotypes? Studies with the NTera2 cell line. Stem Cell Rev. 2017, 13, 603–610. [Google Scholar] [CrossRef]
- Visvader, J.E.; Clevers, H. Tissue-specific designs of stem cell hierarchies. Nat. Cell Biol. 2016, 18, 349–355. [Google Scholar] [CrossRef]
- Gudiño, V.; Pohl, S.Ö.; Billard, C.V.; Cammareri, P.; Bolado, A.; Aitken, S.; Stevenson, D.; Hall, A.E.; Agostino, M.; Cassidy, J.; et al. RAC1B modulates intestinal tumourigenesis via modulation of WNT and EGFR signalling pathways. Nat. Commun. 2021, 12, 2335. [Google Scholar] [CrossRef]
- Dubrovska, A.; Kim, S.; Salamone, R.J.; Walker, J.R.; Maira, S.M.; García-Echeverría, C.; Schultz, P.G.; Reddy, V.A. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc. Natl. Acad. Sci. USA 2009, 106, 268–273. [Google Scholar] [CrossRef]
- Matsuda, S.; Ichimura, M.; Ogino, M.; Nakano, N.; Minami, A.; Murai, T.; Kitagishi, Y. Effective PI3K modulators for improved therapy against malignant tumors and for neuroprotection of brain damage after tumor therapy. Int. J. Oncol. 2016, 49, 1785–1790. [Google Scholar] [CrossRef]
- Matsuda, S.; Nakagawa, Y.; Kitagishi, Y.; Nakanishi, A.; Murai, T. Reactive oxygen species, superoxide dimutases, and PTEN-p53-AKT-MDM2 signaling loop network in mesenchymal stem/stromal cells regulation. Cells 2018, 7, 36. [Google Scholar] [CrossRef]
- Minematsu, H.; Afify, S.M.; Sugihara, Y.; Hassan, G.; Zahra, M.H.; Seno, A.; Adachi, M.; Seno, M. Cancer stem cells induced by chronic stimulation with prostaglandin E2 exhibited constitutively activated PI3K axis. Sci. Rep. 2022, 12, 15628. [Google Scholar] [CrossRef]
- Brou, C.; Logeat, F.; Gupta, N.; Bessia, C.; LeBail, O.; Doedens, J.R.; Cumano, A.; Roux, P.; Black, R.A.; Israël, A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 2000, 5, 207–216. [Google Scholar] [CrossRef]
- Schroeter, E.H.; Kisslinger, J.A.; Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 1998, 393, 382–386. [Google Scholar] [CrossRef]
- Vieira de Castro, J.; Gonçalves, C.S.; Hormigo, A.; Costa, B.M. Exploiting the Complexities of Glioblastoma Stem Cells: Insights for Cancer Initiation and Therapeutic Targeting. Int. J. Mol. Sci. 2020, 21, 5278. [Google Scholar] [CrossRef] [PubMed]
- Nallasamy, P.; Nimmakayala, R.K.; Parte, S.; Are, A.C.; Batra, S.K.; Ponnusamy, M.P. Tumor microenvironment enriches the stemness features: The architectural event of therapy resistance and metastasis. Mol. Cancer 2022, 21, 225. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.; Damaghi, M.; Marunaka, Y.; Spugnini, E.; Fais, S.; Gillies, R. Causes, Consequences, and Therapy of Tumors Acidosis. Cancer Metastasis Rev. 2019, 38, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Granja, S.; Tavares-Valente, D.; Queiros, O.; Baltazar, F. Value of pH regulators in the diagnosis, prognosis and treatment of cancer. Semin. Cancer Biol. 2017, 43, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Daniel, Y.; Lelou, E.; Aninat, C.; Corlu, A.; Cabillic, F. Interplay between metabolism Rrprogramming and epithelial-to-mesenchymal transition in cancer stem cells. Cancers 2021, 13, 1973. [Google Scholar] [CrossRef]
- Carracedo, A.; Cantley, L.C.; Pandol, P.P. Cancer Metabolism: Fatty Acid, Oxidation in the Limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging biological principles of metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Tian, B.; Du, X.; Zheng, S.; Zhang, Y. The role of tumor microenvironment in regulating the plasticity of osteosarcoma cells. Int. J. Mol. Sci. 2022, 23, 16155. [Google Scholar] [CrossRef]
- Mortezaee, K.; Majidpoor, J.; Kharazinejad, E. Epithelial-mesenchymal transition in cancer stemness and heterogeneity: Updated. Med. Oncol. 2022, 39, 193. [Google Scholar] [CrossRef]
- Garg, M. Emerging roles of epithelial-mesenchymal plasticity in invasion-metastasis cascade and therapy resistance. Cancer Metastasis Rev. 2022, 41, 131–145. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Ledford, H. Cancer theory faces doubts. Nature 2011, 472, 273. [Google Scholar] [CrossRef]
- Cortina, C.; Turon, G.; Stork, D.; Hernando-Momblona, X.; Sevillano, M.; Aguilera, M.; Tosi, S.; Merlos-Suárez, A.; Attolini, C.S.; Sancho, E.; et al. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 2017, 9, 869–879. [Google Scholar] [CrossRef]
- Voog, J.; Jones, D.L. Stem cells and the niche: A dynamic duo. Cell Stem Cell 2010, 6, 103–115. [Google Scholar] [CrossRef]
- Oskarsson, T.; Batlle, E.; Massagué, J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell. 2014, 14, 306–321. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Morrison, S.J.; Spradling, A.C. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell 2008, 32, 598–611. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- de Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.C.; Richiardone, E.; Jorge, J.; Polónia, B.; Xavier, C.P.R.; Salaroglio, I.C.; Riganti, C.; Vasconcelos, M.H.; Corbet, C.; Sarmento-Ribeiro, A.B. Impact of cancer metabolism on therapy resistance—Clinical implications. Drug Resist. Updat. 2021, 59, 100797. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhu, X.; Wu, Y. Effects of glucose metabolism, lipid metabolism, and glutamine metabolism on tumor microenvironment and clinical implications. Biomolecules 2022, 12, 580. [Google Scholar] [CrossRef] [PubMed]
- Loveless, R.; Bloomquist, R.; Teng, Y. Pyroptosis at the forefront of anticancer immunity. J. Exp. Clin. Cancer Res. 2021, 40, 264. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Nagase, N.; Tsuji, A.; Taniguchi, K.; Kitagishi, Y.; Matsuda, S. Comprehension of the relationship between autophagy and reactive oxygen species for superior cancer therapy with histone deacetylase inhibitors. Oxygen 2021, 1, 22–31. [Google Scholar] [CrossRef]
- Rezayatmand, H.; Razmkhah, M.; Razeghian-Jahromi, I. Drug resistance in cancer therapy: The Pandora’s Box of cancer stem cells. Stem Cell Res. Ther. 2022, 13, 181. [Google Scholar] [CrossRef]
- Nasiri, F.; Kazemi, M.; Mirarefin, S.M.J.; Mahboubi Kancha, M.; Ahmadi Najafabadi, M.; Salem, F.; Dashti Shokoohi, S.; Evazi Bakhshi, S.; Safarzadeh Kozani, P.; Safarzadeh Kozani, P. CAR-T cell therapy in triple-negative breast cancer: Hunting the invisible devil. Front. Immunol. 2022, 13, 1018786. [Google Scholar] [CrossRef]
- Wang, Q.-S.; Wang, Y.; Lv, H.-Y.; Han, Q.-W.; Fan, H.; Guo, B.; Wang, L.L.; Han, W.D. Treatment of CD33-directed chimeric antigen receptor-modified T cells in one patient with relapsed and refractory acute myeloid leukemia. Mol. Ther. 2015, 23, 184–191. [Google Scholar] [CrossRef]
- Patsoukis, N.; Bardhan, K.; Chatterjee, P.; Sari, D.; Liu, B.; Bell, L.N.; Karoly, E.D.; Freeman, G.J.; Petkova, V.; Seth, P.; et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 2015, 6, 6692. [Google Scholar] [CrossRef]
- Jagust, P.; de Luxán-Delgado, B.; Parejo-Alonso, B.; Sancho, P. Metabolism-based therapeutic strategies targeting cancer stem cells. Front. Pharmacol. 2019, 10, 203. [Google Scholar] [CrossRef]
- Tanase, C.; Enciu, A.M.; Codrici, E.; Popescu, I.D.; Dudau, M.; Dobri, A.M.; Pop, S.; Mihai, S.; Gheorghișan-Gălățeanu, A.A.; Hinescu, M.E. Fatty acids, CD36, thrombospondin-1, and CD47 in glioblastoma: Together and/or separately? Int. J. Mol. Sci. 2022, 23, 604. [Google Scholar] [CrossRef]
- Guerrero-Rodríguez, S.L.; Mata-Cruz, C.; Pérez-Tapia, S.M.; Velasco-Velázquez, M.A. Role of CD36 in cancer progression, stemness, and targeting. Front. Cell Dev. Biol. 2022, 10, 1079076. [Google Scholar] [CrossRef]
- Ma, X.; Xiao, L.; Liu, L.; Ye, L.; Su, P.; Bi, E.; Wang, Q.; Yang, M.; Qian, J.; Yi, Q. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metab. 2021, 33, 1001–1012.e5. [Google Scholar] [CrossRef]
- Manzo, T.; Prentice, B.M.; Anderson, K.G.; Raman, A.; Schalck, A.; Codreanu, G.S.; Nava Lauson, C.B.; Tiberti, S.; Raimondi, A.; Jones, M.A.; et al. Accumulation of long-chain fatty acids in the tumor microenvironment drives dysfunction in intrapancreatic CD8+ T cells. J. Exp. Med. 2020, 217, e20191920. [Google Scholar] [CrossRef]
- Mandal, C.C. The novel use of lipids as diagnostic tools and therapeutics in cancer: Recent insights and challenges. Curr. Drug Targets 2022, 23, 542–543. [Google Scholar] [CrossRef]
- Mayne, S.T.; Playdon, M.C.; Rock, C.L. Diet, nutrition, and cancer: Past, present and future. Nat. Rev. Clin. Oncol. 2016, 13, 504–515. [Google Scholar] [CrossRef]
- Ikeda, Y.; Taniguchi, Y.; Sawamura, H.; Yoshikawa, S.; Tsuji, A.; Matsuda, S. Presumed roles of APRO family proteins in cancer invasiveness. Cancers 2022, 14, 4931. [Google Scholar] [CrossRef]
- Ikeda, Y.; Nagase, N.; Tsuji, A.; Taniguchi, K.; Kitagishi, Y.; Matsuda, S. Secretome-microRNA and anti-proliferative APRO family proteins as cancer prevention and stem cell research strategies. Biocell 2022, 46, 1163–1167. [Google Scholar] [CrossRef]
- Mana, M.D.; Kuo, E.Y.; Yilmaz, Ö.H. Dietary regulation of adult stem cells. Curr. Stem Cell Rep. 2017, 3, 1–8. [Google Scholar] [CrossRef]
- Beyaz, S.; Mana, S.D.; Roper, J.; Kedrin, D.; Saadatpour, A.; Hong, S.J.; Bauer-Rowe, K.E.; Xifaras, M.E.; Akkad, A.; Arias, E.; et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016, 531, 53–58. [Google Scholar] [CrossRef]
- Silver, D.J.; Roversi, G.A.; Bithi, N.; Wang, S.Z.; Troike, K.M.; Neumann, C.K.; Ahuja, G.K.; Reizes, O.; Brown, J.M.; Hine, C.; et al. Severe consequences of a high-lipid diet include hydrogen sulfide dysfunction and enhanced aggression in glioblastoma. J. Clin. Investig. 2021, 131, e138276. [Google Scholar] [CrossRef] [PubMed]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martín, M.; Castellanos, A.; Attolini, C.S.; Berenguer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Parris, A.B.; Howard, E.W.; Shi, Y.; Yang, S.; Jiang, Y.; Kong, L.; Yang, X. Caloric restriction inhibits mammary tumorigenesis in MMTV-ErbB2 transgenic mice through the suppression of ER and ErbB2 pathways and inhibition of epithelial cell stemness in premalignant mammary tissues. Carcinogenesis 2018, 39, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; Forbes-Hernandez, T.Y.; Iglesias, R.C.; Ruiz, R.; Elexpuru Zabaleta, M.; Dominguez, I.; Cianciosi, D.; Quiles, J.L.; Giampieri, F.; Battino, M. Effects of caloric restriction on immunosurveillance, microbiota and cancer cell phenotype: Possible implications for cancer treatment. Semin. Cancer Biol. 2021, 273, 45–57. [Google Scholar] [CrossRef]
- Sawamura, H.; Taniguchi, K.; Ikeda, Y.; Tsuji, A.; Kitagishi, Y.; Matsuda, S. Roles of gut dysbiosis, anti-proliferative proteins, and post-transcriptional regulation in carcinogenesis. J. Transl. Genet. Genom. 2022, 6, 157–168. [Google Scholar] [CrossRef]
- Sawamura, H.; Taniguchi, K.; Ikeda, Y.; Tsuji, A.; Kitagishi, Y.; Matsuda, S. Gut microbiota potentiates the effect of immune checkpoint therapy against cancers. Recent Prog. Nutr. 2022, 2, 1. [Google Scholar] [CrossRef]
- Hey, T.; Tansley, S.; Tolle, K. The Fourth Paradigm: Data-Intensive Scientific Discovery; Microsoft Research: Redmond, WA, USA, 2009. [Google Scholar]
- Sampedro, J.J.; Morales-Soriano, R.; Barceló, C. Challenges in precision medicine in pancreatic cancer: A focus in cancer stem cells and microbiota. Front. Oncol. 2022, 12, 995357. [Google Scholar]
- Li, L.; Xun, C.; Yu, C.H. Role of microRNA-regulated cancer stem cells in recurrent hepatocellular carcinoma. World J. Hepatol. 2022, 14, 1985–1996. [Google Scholar] [CrossRef]
- Rajabi, A.; Kayedi, M.; Rahimi, S.; Dashti, F.; Mirazimi, S.M.A.; Homayoonfal, M.; Mahdian, S.M.A.; Hamblin, M.R.; Tamtaji, O.R.; Afrasiabi, A.; et al. Non-coding RNAs and glioma: Focus on cancer stem cells. Mol. Ther. Oncolytics 2022, 27, 100–123. [Google Scholar] [CrossRef]
- Tsang, S.V.; Rainusso, N.; Liu, M.; Nomura, M.; Patel, T.D.; Nakahata, K.; Kim, H.R.; Huang, S.; Rajapakshe, K.; Coarfa, C.; et al. LncRNA PVT-1 promotes osteosarcoma cancer stem-like properties through direct interaction with TRIM28 and TSC2 ubiquitination. Oncogene 2022, 41, 5373–5384. [Google Scholar]
- Hall, A.E.; Pohl, S.Ö.; Cammareri, P.; Aitken, S.; Younger, N.T.; Raponi, M.; Billard, C.V.; Carrancio, A.B.; Bastem, A.; Freile, P.; et al. RNA splicing is a key mediator of tumour cell plasticity and a therapeutic vulnerability in colorectal cancer. Nat. Commun. 2022, 13, 2791. [Google Scholar] [CrossRef]
- Öther-Gee Pohl, S.; Myant, K.B. Alternative RNA splicing in tumour heterogeneity, plasticity and therapy. Dis. Model Mech. 2022, 15, dmm049233. [Google Scholar] [CrossRef]
- Steponaitis, G.; Stakaitis, R.; Valiulyte, I.; Krusnauskas, R.; Dragunaite, R.; Urbanavičiūtė, R.; Tamasauskas, A.; Skiriute, D. Transcriptome-wide analysis of glioma stem cell specific m6A modifications in long-non-coding RNAs. Sci. Rep. 2022, 12, 5431. [Google Scholar] [CrossRef]
- Costa-Pinheiro, P.; Ramalho-Carvalho, J.; Vieira, F.Q.; Torres-Ferreira, J.; Oliveira, J.; Gonçalves, C.S.; Costa, B.M.; Henrique, R.; Jerónimo, C. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clin. Epigenetics 2015, 7, 42. [Google Scholar] [CrossRef]
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Kardousha, A.; Islam, Z.; Qureshi, R.; Alam, T.; Kolatkar, P.R.; Alajez, N.M. Long non-coding RNA and RNA-binding protein interactions in cancer: Experimental and machine learning approaches. Semin. Cancer Biol. 2022, 86 Pt 3, 325–345. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef]
- Peterson, V.M.; Zhang, K.X.; Kumar, N.; Wong, J.; Li, L.; Wilson, D.C.; Moore, R.; McClanahan, T.K.; Sadekova, S.; Klappenbach, J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017, 35, 936–939. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, R.; Park, J.E.; Teichmann, S.A.; James, K.R. Immunology in the era of single-cell technologies. Annu. Rev. Immunol. 2020, 38, 727–757. [Google Scholar] [CrossRef]
- Efremova, M.; Teichmann, S.A. Computational methods for single-cell omics across modalities. Nat. Methods 2020, 17, 14–17. [Google Scholar] [CrossRef]
- Lee, A.C.; Lee, Y.; Choi, A.; Lee, H.B.; Shin, K.; Lee, H.; Kim, J.Y.; Ryu, H.S.; Kim, H.S.; Ryu, S.Y.; et al. Spatial epitranscriptomics reveals A-to-I editome specific to cancer stem cell microniches. Nat. Commun. 2022, 13, 2540. [Google Scholar] [CrossRef] [PubMed]
- Ben-Chetrit, N.; Niu, X.; Swett, A.D.; Sotelo, J.; Jiao, M.S.; Stewart, C.M.; Potenski, C.; Mielinis, P.; Roelli, P.; Stoeckius, M.; et al. Integration of whole transcriptome spatial profiling with protein markers. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Ikeda, Y.; Tsuji, A.; Matsuda, S. Potential diets to improve mitochondrial activity in amyotrophic lateral sclerosis. Diseases 2022, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Yoshikawa, S.; Ikeda, Y.; Taniguchi, K.; Sawamura, H.; Tsuji, A.; Matsuda, S. Encouraging tactics with genetically modified probiotics to improve immunity for the prevention of immune-related diseases including cardio-metabolic disorders. Biomolecules 2023, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Human Cell Atlas. Available online: https://www.humancellatlas.org (accessed on 28 February 2023).
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef]
- Moselhy, J.; Srinivasan, S.; Ankem, M.K.; Damodaran, C. Natural products that target cancer stem cells. Anticancer Res. 2015, 35, 5773–5788. [Google Scholar]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
| Name | Other Name | Type | Function in Normal Cells |
|---|---|---|---|
| CD44 | Pgp1 | type I transmembrane | receptor for hyaluronan [11] |
| CD133 | prominin-1 | pentaspan transmembrane | costimulatory factor of T cells [11] |
| CD24 | heat stable antigen | GPI-anchored | costimulatory factor of T cells [12] |
| CD326 | EpCAM | GPI-anchored | cell adhesion [16] |
| LGR5 | GPR49 | GPCR | receptor for R-spondin [4] |
| Name | Type | Function in Normal Cells |
|---|---|---|
| PI3K | protein kinase | cell survival and proliferation |
| PTEN | protein phosphatase | tumor suppressor |
| WNT | secreted glycoprotein | Frizzled binding |
| Notch | type I transmembrane | development and differentiation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murai, T.; Matsuda, S. Fatty Acid Metabolites and the Tumor Microenvironment as Potent Regulators of Cancer Stem Cell Signaling. Metabolites 2023, 13, 709. https://doi.org/10.3390/metabo13060709
Murai T, Matsuda S. Fatty Acid Metabolites and the Tumor Microenvironment as Potent Regulators of Cancer Stem Cell Signaling. Metabolites. 2023; 13(6):709. https://doi.org/10.3390/metabo13060709
Chicago/Turabian StyleMurai, Toshiyuki, and Satoru Matsuda. 2023. "Fatty Acid Metabolites and the Tumor Microenvironment as Potent Regulators of Cancer Stem Cell Signaling" Metabolites 13, no. 6: 709. https://doi.org/10.3390/metabo13060709
APA StyleMurai, T., & Matsuda, S. (2023). Fatty Acid Metabolites and the Tumor Microenvironment as Potent Regulators of Cancer Stem Cell Signaling. Metabolites, 13(6), 709. https://doi.org/10.3390/metabo13060709








