Coronary Artery Disease with Elevated Levels of HDL Cholesterol Is Associated with Distinct Lipid Signatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Sample Preparation, Lipid Extraction, and LC-MS/MS Lipidomic Analysis
2.2.1. Sample Preparation
2.2.2. Lipid Extraction
2.2.3. LC-MS/MS Lipidomic Analysis
2.3. Statistical Analysis
3. Results
3.1. Characteristics of Study Participants
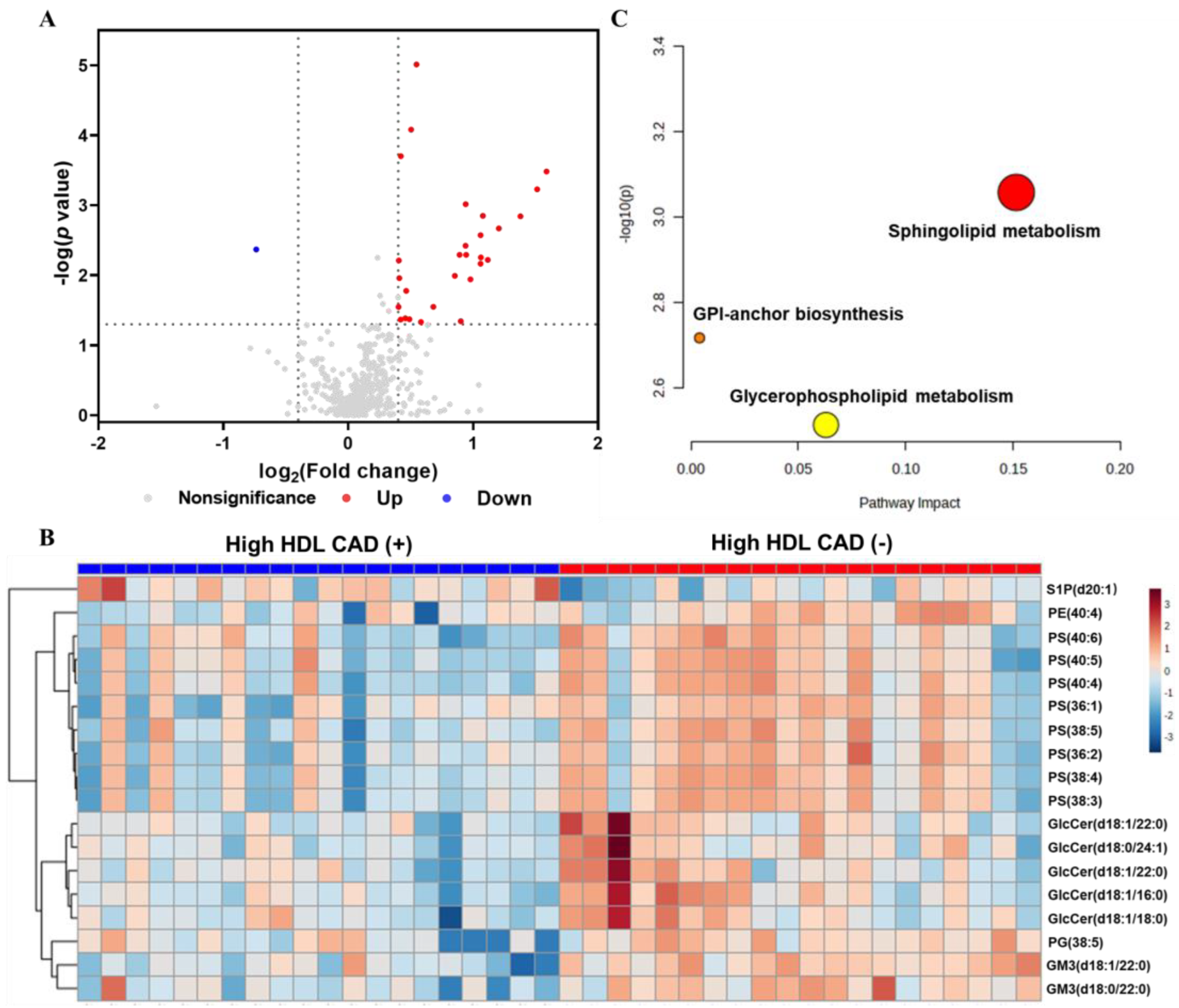
3.2. Comparison of Plasma Lipidomic Profiles between the High HDL CAD (+) and High HDL CAD (−) Groups
3.3. Identification and Hierarchical Clustering Analysis of Differential Lipid Species
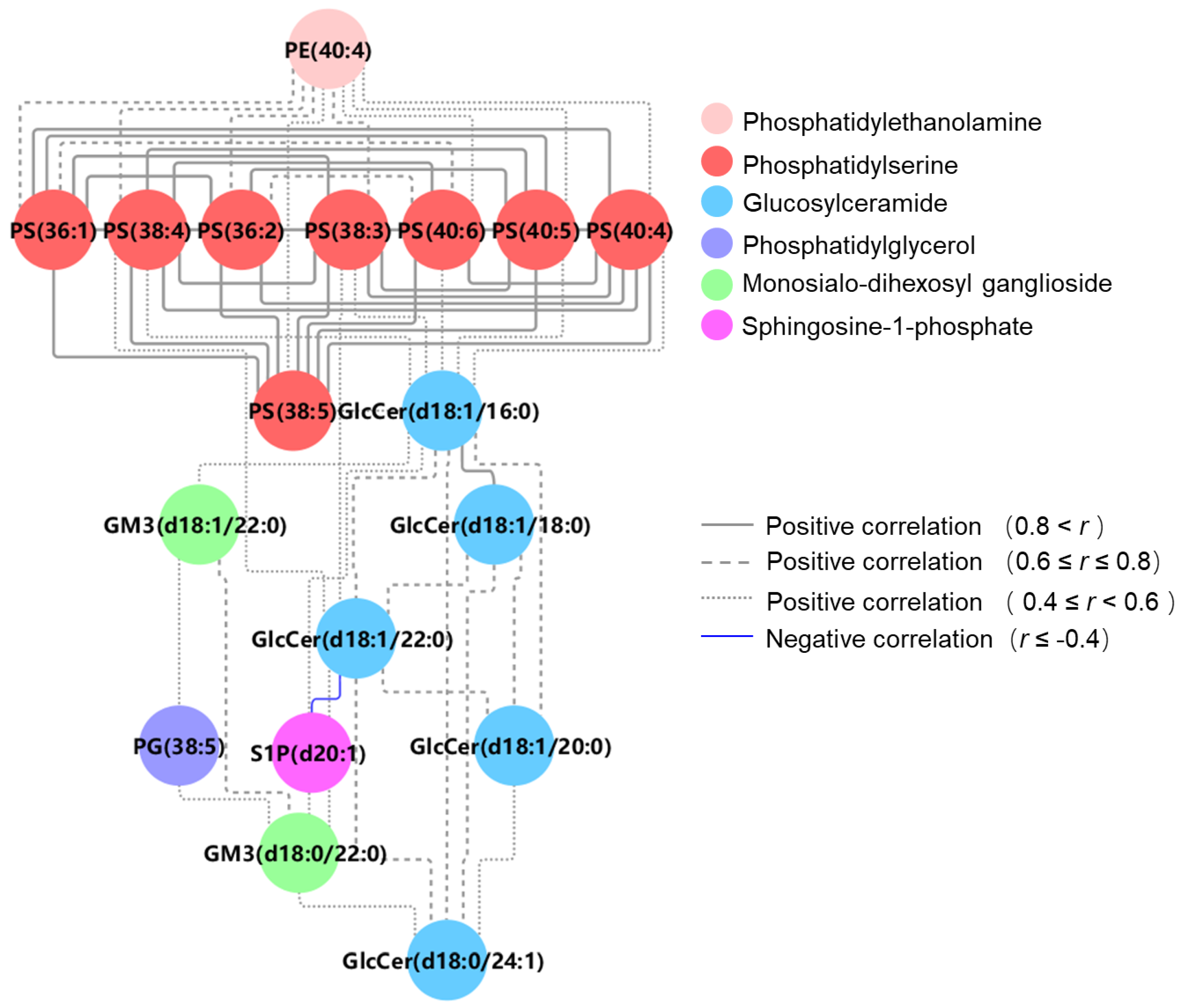
3.4. Differential Lipid Metabolite Pathway and Correlation Analysis
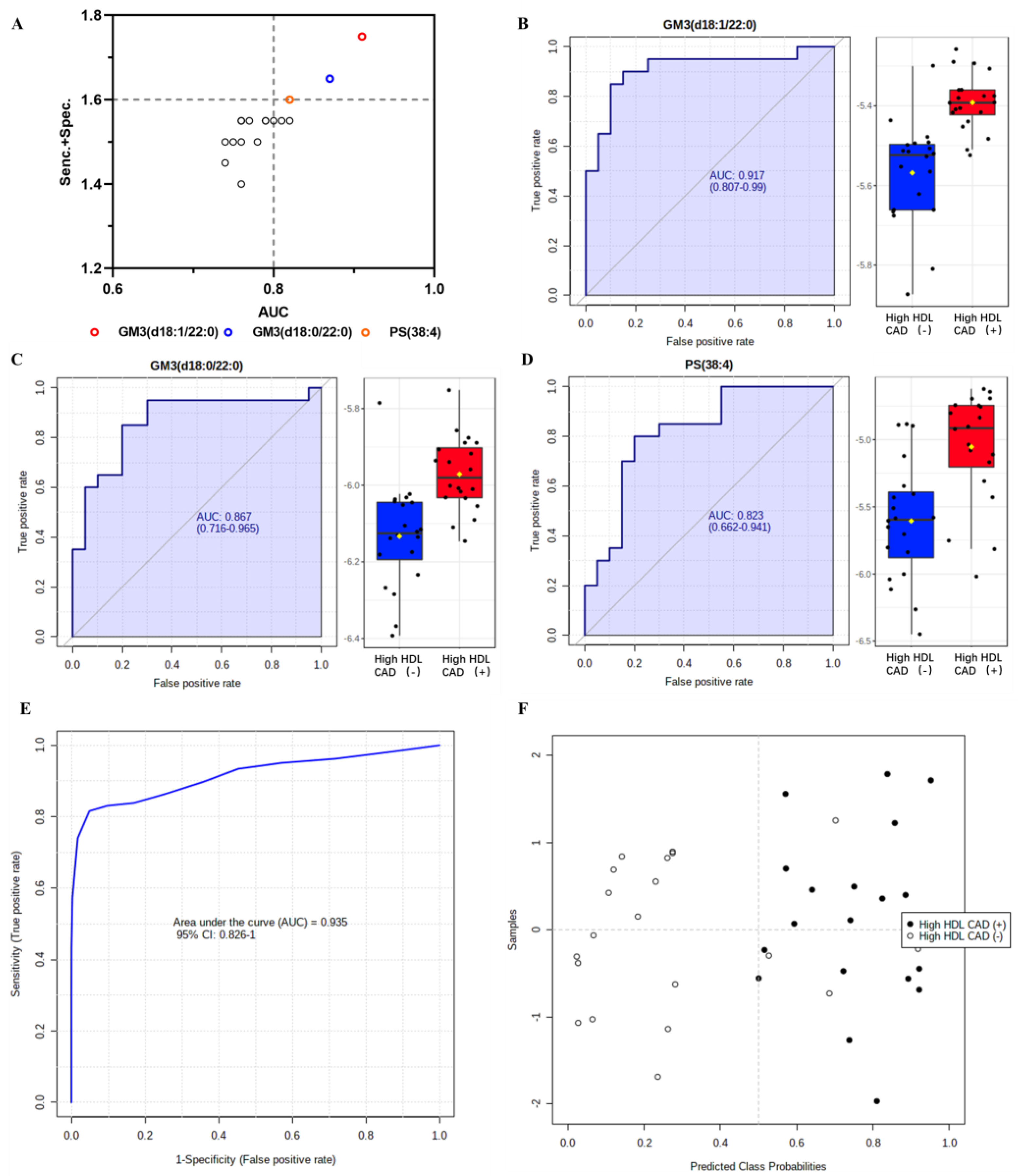
3.5. ROC Analysis and Lipid Biomarker Selection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Malakar, A.K.; Choudhury, D.; Halder, B.; Paul, P.; Uddin, A.; Chakraborty, S. A Review on Coronary Artery Disease, Its Risk Factors, and Therapeutics. J. Cell Physiol. 2019, 234, 16812–16823. [Google Scholar] [CrossRef] [PubMed]
- The Emerging Risk Factors Collaboration Major Lipids, Apolipoproteins, and Risk of Vascular Disease. JAMA 2009, 302, 1993–2000. [CrossRef] [PubMed]
- Barter, P.J.; Caulfield, M.; Eriksson, M.; Grundy, S.M.; Kastelein, J.J.P.; Komajda, M.; Lopez-Sendon, J.; Mosca, L.; Tardif, J.-C.; Waters, D.D.; et al. Effects of Torcetrapib in Patients at High Risk for Coronary Events. N. Engl. J. Med. 2007, 357, 2109–2122. [Google Scholar] [CrossRef]
- Lincoff, A.M.; Nicholls, S.J.; Riesmeyer, J.S.; Barter, P.J.; Brewer, H.B.; Fox, K.A.A.; Gibson, C.M.; Granger, C.; Menon, V.; Montalescot, G.; et al. Evacetrapib and Cardiovascular Outcomes in High-Risk Vascular Disease. N. Engl. J. Med. 2017, 376, 1933–1942. [Google Scholar] [CrossRef]
- DeFaria Yeh, D.; Freeman, M.W.; Meigs, J.B.; Grant, R.W. Risk Factors for Coronary Artery Disease in Patients With Elevated High-Density Lipoprotein Cholesterol. Am. J. Cardiol. 2007, 99, 1–4. [Google Scholar] [CrossRef]
- van der Steeg, W.A.; Holme, I.; Boekholdt, S.M.; Larsen, M.L.; Lindahl, C.; Stroes, E.S.G.; Tikkanen, M.J.; Wareham, N.J.; Faergeman, O.; Olsson, A.G.; et al. High-Density Lipoprotein Cholesterol, High-Density Lipoprotein Particle Size, and Apolipoprotein A-I: Significance for Cardiovascular Risk. J. Am. Coll. Cardiol. 2008, 51, 634–642. [Google Scholar] [CrossRef]
- Agerholm-Larsen, B.; Nordestgaard, B.G.; Steffensen, R.; Jensen, G.; Tybjærg-Hansen, A. Elevated HDL Cholesterol Is a Risk Factor for Ischemic Heart Disease in White Women When Caused by a Common Mutation in the Cholesteryl Ester Transfer Protein Gene. Circulation 2000, 101, 1907–1912. [Google Scholar] [CrossRef]
- Andersen, R.V.; Wittrup, H.H.; Tybjærg-Hansen, A.; Steffensen, R.; Schnohr, P.; Nordestgaard, B.G. Hepatic Lipase Mutations, Elevated High-Density Lipoprotein Cholesterol, and Increased Risk of Ischemic Heart Disease: The Copenhagen City Heart Study. J. Am. Coll. Cardiol. 2003, 41, 1972–1982. [Google Scholar] [CrossRef]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Jensen, G.B.; Steffensen, R.; Tybjærg-Hansen, A. Genetic Variation in ABCA1 Predicts Ischemic Heart Disease in the General Population. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 180–186. [Google Scholar] [CrossRef]
- Zanoni, P.; Khetarpal, S.A.; Larach, D.B.; Hancock-Cerutti, W.F.; Millar, J.S.; Cuchel, M.; DerOhannessian, S.; Kontush, A.; Surendran, P.; Saleheen, D.; et al. Rare Variant in Scavenger Receptor BI Raises HDL Cholesterol and Increases Risk of Coronary Heart Disease. Science 2016, 351, 1166–1171. [Google Scholar] [CrossRef]
- Smith, J.D. Dysfunctional HDL as a Diagnostic and Therapeutic Target. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Mathew, A.V.; Yu, H.; Li, L.; He, L.; Gao, W.; Liu, X.; Guo, Y.; Byun, J.; Zhang, J.; et al. Myeloperoxidase Mediated HDL Oxidation and HDL Proteome Changes Do Not Contribute to Dysfunctional HDL in Chinese Subjects with Coronary Artery Disease. PLoS ONE 2018, 13, e0193782. [Google Scholar] [CrossRef]
- Hancock-Cerutti, W.; Millar, J.S.; Valentini, S.; Liu, J.; Billheimer, J.T.; Rader, D.J.; Cuchel, M. Assessing HDL Metabolism in Subjects with Elevated Levels of HDL Cholesterol and Coronary Artery Disease. Molecules 2021, 26, 6862. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, C.; Drozdov, I.; Shalhoub, J.; Humphries, J.; Ladroue, C.; Didangelos, A.; Baumert, M.; Allen, M.; Davies, A.H.; Monaco, C.; et al. Comparative Lipidomics Profiling of Human Atherosclerotic Plaques. Circ. Cardiovasc. Genet. 2011, 4, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Wong, G.; Tsorotes, D.; Barlow, C.K.; Weir, J.M.; Christopher, M.J.; MacIntosh, G.L.; Goudey, B.; Stern, L.; Kowalczyk, A.; et al. Plasma Lipidomic Analysis of Stable and Unstable Coronary Artery Disease. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2723–2732. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, S.-H.; Shin, M.-J.; Hwang, G.-S. Alteration in Metabolic Signature and Lipid Metabolism in Patients with Angina Pectoris and Myocardial Infarction. PLoS ONE 2015, 10, e0135228. [Google Scholar] [CrossRef]
- Hadas, Y.; Vincek, A.S.; Youssef, E.; Żak, M.M.; Chepurko, E.; Sultana, N.; Sharkar, M.T.K.; Guo, N.; Komargodski, R.; Kurian, A.A.; et al. Altering Sphingolipid Metabolism Attenuates Cell Death and Inflammatory Response After Myocardial Infarction. Circulation 2020, 141, 916–930. [Google Scholar] [CrossRef]
- Zhu, Q.; Wu, Y.; Mai, J.; Guo, G.; Meng, J.; Fang, X.; Chen, X.; Liu, C.; Zhong, S. Comprehensive Metabolic Profiling of Inflammation Indicated Key Roles of Glycerophospholipid and Arginine Metabolism in Coronary Artery Disease. Front. Immunol. 2022, 13, 829425. [Google Scholar] [CrossRef]
- Tan, S.H.; Koh, H.W.L.; Chua, J.Y.; Burla, B.; Ong, C.C.; Teo, L.S.L.; Yang, X.; Benke, P.I.; Choi, H.; Torta, F.; et al. Variability of the Plasma Lipidome and Subclinical Coronary Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 100–112. [Google Scholar] [CrossRef]
- Hancock-Cerutti, W.; Lhomme, M.; Dauteuille, C.; Lecocq, S.; Chapman, M.J.; Rader, D.J.; Kontush, A.; Cuchel, M. Paradoxical Coronary Artery Disease in Humans with Hyperalphalipoproteinemia Is Associated with Distinct Differences in the High-Density Lipoprotein Phosphosphingolipidome. J. Clin. Lipidol. 2017, 11, 1192–1200.e3. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, A.P.; Rodrigues, A.; Risman, M.; McCoy, M.; Trindade, K.; Qu, L.; Cuchel, M.; Billheimer, J.; Rader, D.J. High-Density Lipoprotein (HDL) Phospholipid Content and Cholesterol Efflux Capacity Are Reduced in Patients With Very High HDL Cholesterol and Coronary Disease. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Laaksonen, R. Identifying New Risk Markers and Potential Targets for Coronary Artery Disease: The Value of the Lipidome and Metabolome. Cardiovasc. Drugs. Ther. 2016, 30, 19–32. [Google Scholar] [CrossRef]
- Hilvo, M.; Meikle, P.J.; Pedersen, E.R.; Tell, G.S.; Dhar, I.; Brenner, H.; Schöttker, B.; Lääperi, M.; Kauhanen, D.; Koistinen, K.M.; et al. Development and Validation of a Ceramide- and Phospholipid-Based Cardiovascular Risk Estimation Score for Coronary Artery Disease Patients. Eur. Heart J. 2020, 41, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, J.; Li, S.; Chen, A.; Dai, C.; Liu, M.; Lu, D.; Chen, Z.; Wang, X.; Qian, J.; et al. Prognostic Implication of Lipidomics in Patients with Coronary Total Occlusion Undergoing PCI. Eur. J. Clin. Investig. 2022, 52, e13826. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Tong, L.; Duan, X.; Petznick, A.; Wenk, M.R.; Shui, G. Extensive Characterization of Human Tear Fluid Collected Using Different Techniques Unravels the Presence of Novel Lipid Amphiphiles1[S]. J. Lipid Res. 2014, 55, 289–298. [Google Scholar] [CrossRef]
- Song, J.-W.; Lam, S.M.; Fan, X.; Cao, W.-J.; Wang, S.-Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.-P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Shui, G.; Cheong, W.F.; Jappar, I.A.; Hoi, A.; Xue, Y.; Fernandis, A.Z.; Tan, B.K.-H.; Wenk, M.R. Derivatization-Independent Cholesterol Analysis in Crude Lipid Extracts by Liquid Chromatography/Mass Spectrometry: Applications to a Rabbit Model for Atherosclerosis. J. Chromatogr. A 2011, 1218, 4357–4365. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Laaksonen, R.; Ekroos, K.; Sysi-Aho, M.; Hilvo, M.; Vihervaara, T.; Kauhanen, D.; Suoniemi, M.; Hurme, R.; März, W.; Scharnagl, H.; et al. Plasma Ceramides Predict Cardiovascular Death in Patients with Stable Coronary Artery Disease and Acute Coronary Syndromes beyond LDL-Cholesterol. Eur. Heart J. 2016, 37, 1967–1976. [Google Scholar] [CrossRef]
- Tu, C.; Xie, L.; Wang, Z.; Zhang, L.; Wu, H.; Ni, W.; Li, C.; Li, L.; Zeng, Y. Association between Ceramides and Coronary Artery Stenosis in Patients with Coronary Artery Disease. Lipids Health Dis. 2020, 19, 151. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Suzuki, J.; Segawa, K.; Fujii, T. Exposure of Phosphatidylserine on the Cell Surface. Cell Death Differ. 2016, 23, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, D.; Curaj, A.; Staudt, M.; Cordes, F.; Dumitraşcu, A.R.; Rolles, B.; Beckers, C.; Soppert, J.; Rusu, M.; Simsekyilmaz, S.; et al. Phosphatidylserine Supplementation as a Novel Strategy for Reducing Myocardial Infarct Size and Preventing Adverse Left Ventricular Remodeling. Int. J. Mol. Sci. 2021, 22, 4401. [Google Scholar] [CrossRef] [PubMed]
- Kenis, H.; Zandbergen, H.R.; Hofstra, L.; Petrov, A.D.; Dumont, E.A.; Blankenberg, F.D.; Haider, N.; Bitsch, N.; Gijbels, M.; Verjans, J.W.H.; et al. Annexin A5 Uptake in Ischemic Myocardium: Demonstration of Reversible Phosphatidylserine Externalization and Feasibility of Radionuclide Imaging. J. Nucl. Med. 2010, 51, 259–267. [Google Scholar] [CrossRef]
- Mallat, Z.; Hugel, B.; Ohan, J.; Lesèche, G.; Freyssinet, J.-M.; Tedgui, A. Shed Membrane Microparticles With Procoagulant Potential in Human Atherosclerotic Plaques. Circulation 1999, 99, 348–353. [Google Scholar] [CrossRef]
- Wang, X.; Du, H.; Li, X. Artesunate Attenuates Atherosclerosis by Inhibiting Macrophage M1-like Polarization and Improving Metabolism. Int. Immunopharmacol. 2022, 102, 108413. [Google Scholar] [CrossRef]
- Rueda, R. The Role of Complex Lipids in Attaining Metabolic Health. Curr. Cardiovasc. Risk Rep. 2014, 8, 371. [Google Scholar] [CrossRef]
- Bietrix, F.; Lombardo, E.; van Roomen, C.P.A.A.; Ottenhoff, R.; Vos, M.; Rensen, P.C.N.; Verhoeven, A.J.; Aerts, J.M.; Groen, A.K. Inhibition of Glycosphingolipid Synthesis Induces a Profound Reduction of Plasma Cholesterol and Inhibits Atherosclerosis Development in APOE*3 Leiden and Low-Density Lipoprotein Receptor−/− Mice. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 931–937. [Google Scholar] [CrossRef]
- Chatterjee, S.B.; Dey, S.; Shi, W.Y.; Thomas, K.; Hutchins, G.M. Accumulation of Glycosphingolipids in Human Atherosclerotic Plaque and Unaffected Aorta Tissues. Glycobiology 1997, 7, 57–65. [Google Scholar] [CrossRef]
- Edsfeldt, A.; Dunér, P.; Ståhlman, M.; Mollet, I.G.; Asciutto, G.; Grufman, H.; Nitulescu, M.; Persson, A.F.; Fisher, R.M.; Melander, O.; et al. Sphingolipids Contribute to Human Atherosclerotic Plaque Inflammation. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1132–1140. [Google Scholar] [CrossRef]
- Mihanfar, A.; Nejabati, H.R.; Fattahi, A.; Latifi, Z.; Pezeshkian, M.; Afrasiabi, A.; Safaie, N.; Jodati, A.R.; Nouri, M. The Role of Sphingosine 1 Phosphate in Coronary Artery Disease and Ischemia Reperfusion Injury. J. Cell Physiol. 2019, 234, 2083–2094. [Google Scholar] [CrossRef] [PubMed]
- Potì, F.; Simoni, M.; Nofer, J.-R. Atheroprotective Role of High-Density Lipoprotein (HDL)-Associated Sphingosine-1-Phosphate (S1P). Cardiovasc. Res. 2014, 103, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Sattler, K.J.E.; Elbasan, Ş.; Keul, P.; Elter-Schulz, M.; Bode, C.; Gräler, M.H.; Bröcker-Preuss, M.; Budde, T.; Erbel, R.; Heusch, G.; et al. Sphingosine 1-Phosphate Levels in Plasma and HDL Are Altered in Coronary Artery Disease. Basic Res. Cardiol. 2010, 105, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wen, S.; Jiang, M.; Zhu, Y.; Ding, L.; Shi, H.; Dong, P.; Yang, J.; Yang, Y. Atherosclerotic Dyslipidemia Revealed by Plasma Lipidomics on ApoE−/− Mice Fed a High-Fat Diet. Atherosclerosis 2017, 262, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Dang, V.T.; Huang, A.; Zhong, L.H.; Shi, Y.; Werstuck, G.H. Comprehensive Plasma Metabolomic Analyses of Atherosclerotic Progression Reveal Alterations in Glycerophospholipid and Sphingolipid Metabolism in Apolipoprotein E-Deficient Mice. Sci. Rep. 2016, 6, 35037. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Zhu, Q.; Lai, W.; Ma, Q.; Liu, C.; Chen, X.; Zhang, Y.; Wang, Z.; Chen, H.; Yan, H.; et al. Insights into the Prognosis of Lipidomic Dysregulation for Death Risk in Patients with Coronary Artery Disease. Clin. Transl. Med. 2020, 10, e189. [Google Scholar] [CrossRef]
- Poss, A.M.; Maschek, J.A.; Cox, J.E.; Hauner, B.J.; Hopkins, P.N.; Hunt, S.C.; Holland, W.L.; Summers, S.A.; Playdon, M.C. Machine Learning Reveals Serum Sphingolipids as Cholesterol-Independent Biomarkers of Coronary Artery Disease. J. Clin. Investig. 2020, 130, 1363–1376. [Google Scholar] [CrossRef]

| Characteristics | High HDL CAD (−) (n = 20) | High HDL CAD (+) (n = 20) | p Value |
|---|---|---|---|
| Age (years) | 59 (54, 64) | 62 (59, 66) | 0.086 |
| Male (%) | 50 | 65 | 0.523 |
| Waist circumference (cm) | 87.10 ± 8.74 | 89.82 ± 9.97 | 0.364 |
| BMI (kg/m2) | 22.97 ± 2.60 | 23.95 ± 4.03 | 0.372 |
| Smokers (%) | 40 | 70 | 0.055 |
| Hypertension history (%) | 50 | 50 | 1.000 |
| HDL-C (mmol/L) | 1.51 (1.40, 1.69) | 1.53 (1.32, 1.58) | 0.277 |
| TC (mmol/L) | 4.84 ± 0.78 | 5.18 ± 1.06 | 0.253 |
| LDL-C (mmol/L) | 2.60 ± 0.75 | 2.99 ± 0.88 | 0.151 |
| TG (mmol/L) | 1.06 (0.79, 1.34) | 1.23 (0.82,1.85) | 0.253 |
| Apo A1 (mg/L) | 1979.80 ± 293.12 | 1929.35 ± 307.83 | 0.599 |
| Apo B (mg/L) | 765.90 ± 233.79 | 942.35 ± 271.61 | 0.034 a |
| Lp(a) (mg/L) | 90.00 (41.00, 247.75) | 231.00 (72.25, 422.00) | 0.046 a |
| Fasting glucose(mmol/L) | 5.11 ± 0.65 | 5.13 ± 0.84 | 0.933 |
| HbA1C (%) | 5.49 ± 0.33 | 5.67 ± 0.41 | 0.123 |
| hs-CRP (mg/L) | 8.73 ± 6.72 | 5.07 ± 3.61 | 0.040 a |
| Lipid Species | Adjusted p Value a | VIP b | FC c | AUC (95% CI) d | Sensitivity d | Specificity d | Sens. + Spec. |
|---|---|---|---|---|---|---|---|
| Sphingolipid | |||||||
| GM3 (d18:1/22:0) | 0.010 | 2.57 | 1.46 | 0.917 (0.807–0.990) | 0.90 | 0.85 | 1.75 |
| GM3 (d18:0/22:0) | 0.009 | 2.30 | 1.42 | 0.870 (0.730–0.964) | 0.85 | 0.80 | 1.65 |
| GlcCer (d18:1/16:0) | 0.026 | 2.78 | 2.08 | 0.779 (0.621–0.914) | 0.80 | 0.70 | 1.50 |
| GlcCer (d18:0/24:1) | 0.015 | 2.22 | 1.85 | 0.765 (0.591–0.899 | 0.80 | 0.75 | 1.55 |
| GlcCer (d18:1/22:0) | 0.037 | 2.39 | 2.08 | 0.751 (0.586–0.900) | 0.85 | 0.65 | 1.50 |
| GlcCer (d18:1/18:0) | 0.034 | 2.33 | 1.81 | 0.739 (0.571–0.889) | 0.90 | 0.60 | 1.50 |
| GlcCer (d18:1/20:0) | 0.020 | 2.27 | 1.97 | 0.735 (0.555–0.866) | 0.95 | 0.50 | 1.45 |
| S1P (d20:1) | 0.015 | 2.34 | 0.60 | 0.760 (0.615–0.889) | 0.75 | 0.65 | 1.40 |
| Glycerophospholipid | |||||||
| PS (38:4) | 0.008 | 4.16 | 3.01 | 0.823 (0.662–0.941) | 0.80 | 0.80 | 1.60 |
| PS (38:3) | 0.008 | 3.97 | 2.85 | 0.818 (0.666–0.944) | 0.85 | 0.70 | 1.55 |
| PS (36:1) | 0.012 | 3.04 | 1.92 | 0.809 (0.661–0.925) | 0.85 | 0.70 | 1.55 |
| PS (36:2) | 0.014 | 3.05 | 2.11 | 0.796 (0.666–0.909) | 0.75 | 0.80 | 1.55 |
| PS (40:4) | 0.010 | 3.72 | 2.60 | 0.792 (0.636–0.929) | 0.80 | 0.75 | 1.55 |
| PS (38:5) | 0.034 | 3.42 | 2.31 | 0.785 (0.645–0.922) | 0.75 | 0.80 | 1.55 |
| PS (40:6) | 0.010 | 3.33 | 2.09 | 0.758 (0.579–0.887) | 0.70 | 0.85 | 1.55 |
| PS (40:5) | 0.019 | 3.00 | 2.17 | 0.755 (0.560–0.906) | 0.70 | 0.85 | 1.55 |
| PE (40:4) | 0.024 | 2.82 | 1.92 | 0.758 (0.599–0.891) | 0.90 | 0.60 | 1.50 |
| PG (38:5) | 0.037 | 3.49 | 1.92 | 0.769 (0.604–0.899) | 0.65 | 0.90 | 1.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, W.; Yu, H.; Wang, G. Coronary Artery Disease with Elevated Levels of HDL Cholesterol Is Associated with Distinct Lipid Signatures. Metabolites 2023, 13, 695. https://doi.org/10.3390/metabo13060695
Xia W, Yu H, Wang G. Coronary Artery Disease with Elevated Levels of HDL Cholesterol Is Associated with Distinct Lipid Signatures. Metabolites. 2023; 13(6):695. https://doi.org/10.3390/metabo13060695
Chicago/Turabian StyleXia, Wanying, Haiyi Yu, and Guisong Wang. 2023. "Coronary Artery Disease with Elevated Levels of HDL Cholesterol Is Associated with Distinct Lipid Signatures" Metabolites 13, no. 6: 695. https://doi.org/10.3390/metabo13060695
APA StyleXia, W., Yu, H., & Wang, G. (2023). Coronary Artery Disease with Elevated Levels of HDL Cholesterol Is Associated with Distinct Lipid Signatures. Metabolites, 13(6), 695. https://doi.org/10.3390/metabo13060695





