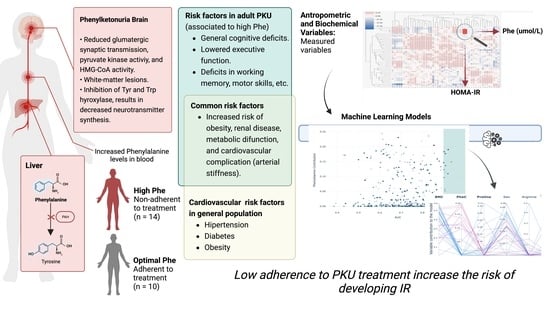
Risk of Developing Insulin Resistance in Adult Subjects with Phenylketonuria: Machine Learning Model Reveals an Association with Phenylalanine Concentrations in Dried Blood Spots
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Participant Description
2.3. Anthropometric Assessment
2.4. Biochemical Analysis
Homeostasis Model Assessment (HOMA)
2.5. Amino Acid and Acylcarnitine Determination by Tandem Mass Spectrometry (MSMS)
2.6. Statistical Analysis
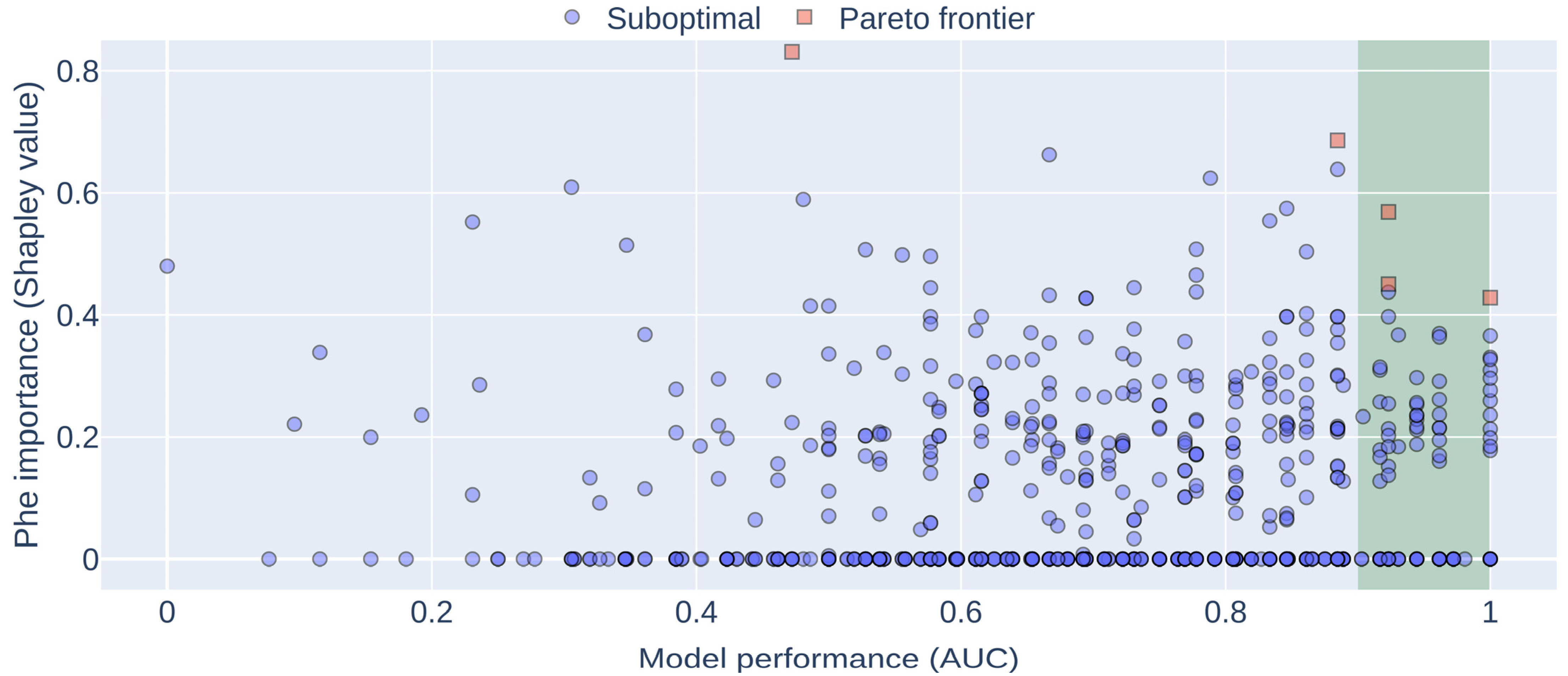
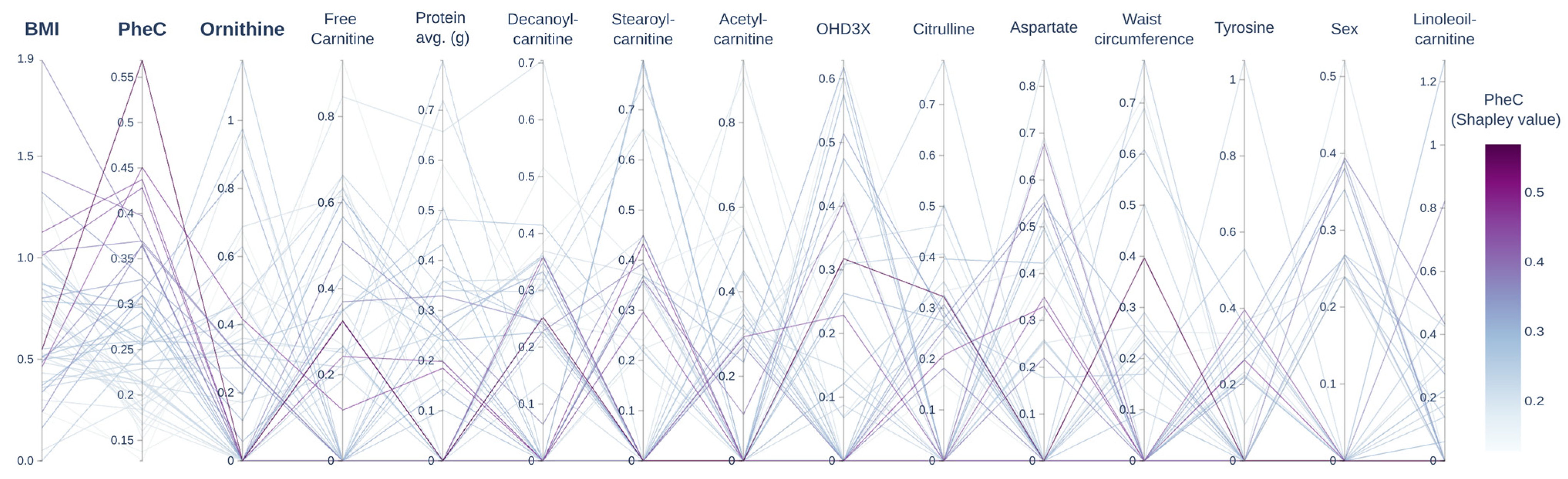
2.7. Machine Learning Model
2.8. Code Availability
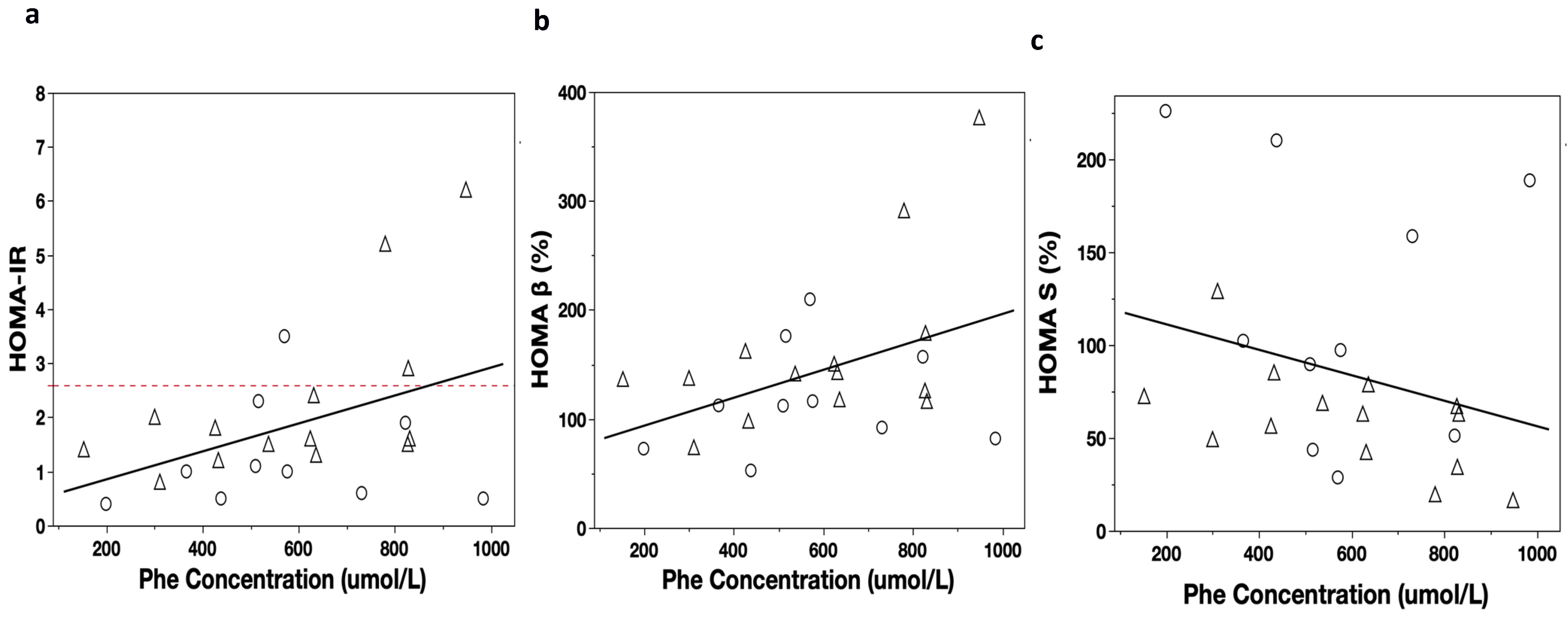
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marta Colombo, C.; Verónica Cornejo, E.; Erna Raimann, B. Errores Innatos en el Metabolismo del Niño, 4th ed.; Universitaria: Santiago, Chile, 2017. [Google Scholar]
- The American College of Medical Genetics and Genomics Therapeutic Committee; Vockley, J.; Andersson, H.C.; Antshel, K.M.; Braverman, N.E.; Burton, B.K.; Frazier, D.M.; Mitchell, J.; Smith, W.E.; Thompson, B.H.; et al. Phenylalanine hydroxylase deficiency: Diagnosis and management guideline. Genet. Med. 2014, 16, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.H.; Cunningham, A.C.; Mofidi, S.; Douglas, T.D.; Frazier, D.M.; Hook, D.G.; Jeffers, L.; McCune, H.; Moseley, K.D.; Ogata, B.; et al. Updated, web-based nutrition management guideline for PKU: An evidence and consensus based approach. Mol. Genet. Metab. 2016, 118, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Castro, G.; Hamilton, V.; Cornejo, V. Chilean Nutrition Management Protocol for Patients With Phenylketonuria. J. Inborn Errors Metab. Screen. 2017, 5, 2326409816689788. [Google Scholar] [CrossRef]
- MacDonald, A.; van Wegberg, A.M.J.; Ahring, K.; Beblo, S.; Bélanger-Quintana, A.; Burlina, A.; Campistol, J.; Coşkun, T.; Feillet, F.; Giżewska, M.; et al. PKU dietary handbook to accompany PKU guidelines. Orphanet J. Rare Dis. 2020, 15, 171. [Google Scholar] [CrossRef]
- Cornejo, V.; Raimann, E.; Cabello, J.F.; Valiente, A.; Becerra, C.; Opazo, M.; Colombo, M. Past, present and future of newborn screening in Chile. J. Inherit. Metab. Dis. 2010, 33, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, V.; Santa María, L.; Fuenzalida, K.; Morales, P.; Desviat, L.R.; Ugarte, M.; Pérez, B.; Cabello, J.F.; Cornejo, V. Characterization of Phenyalanine Hydroxylase Gene Mutations in Chilean PKU Patients. JIMD Rep. 2017, 42, 71–77. [Google Scholar] [CrossRef]
- Burton, B.K.; Leviton, L. Reaching out to the lost generation of adults with early-treated phenylketonuria (PKU). Mol. Genet. Metab. 2010, 101, 146–148. [Google Scholar] [CrossRef]
- Leal-Witt, M.J.; Salazar, M.F.; Peñaloza, F.; Castro, G.; Hamilton, V.; Arias, C.; Peredo, P.; Valiente, A.; De la Parra, A.; Cabello, J.F.; et al. Update on Dietary Compliance, Nutritional Status, and Neuropsychological Functioning in the Chilean Phenylketonuria cohort. J. Inborn Errors Metab. Screen. 2021, 9, e20210003. [Google Scholar] [CrossRef]
- Burlina, A.P.; Lachmann, R.H.; Manara, R.; Cazzorla, C.; Celato, A.; van Spronsen, F.J.; Burlina, A. The neurological and psychological phenotype of adult patients with early-treated phenylketonuria: A systematic review. J. Inherit. Metab. Dis. 2019, 42, 209–219. [Google Scholar] [CrossRef]
- Rocha, J.C.; van Rijn, M.; van Dam, E.; Ahring, K.; Bélanger-Quintana, A.; Dokoupil, K.; Ozel, H.G.; Lammardo, A.M.; Robert, M.; Heidenborg, C.; et al. Weight Management in Phenylketonuria: What Should Be Monitored? Ann. Nutr. Metab. 2016, 68, 60–65. [Google Scholar] [CrossRef]
- Burton, B.K.; Jones, K.B.; Cederbaum, S.; Rohr, F.; Waisbren, S.; Irwin, D.E.; Kim, G.; Lilienstein, J.; Alvarez, I.; Jurecki, E.; et al. Prevalence of comorbid conditions among adult patients diagnosed with phenylketonuria. Mol. Genet. Metab. 2018, 125, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Trefz, K.F.; Muntau, A.C.; Kohlscheen, K.M.; Altevers, J.; Jacob, C.; Braun, S.; Greiner, W.; Jha, A.; Jain, M.; Alvarez, I.; et al. Clinical burden of illness in patients with phenylketonuria (PKU) and associated comorbidities—A retrospective study of German health insurance claims data. Orphanet J. Rare Dis. 2019, 14, 181. [Google Scholar] [CrossRef]
- Couce, M.L.; Sánchez-Pintos, P.; Vitoria, I.; De Castro, M.-J.; Aldámiz-Echevarría, L.; Correcher, P.; Fernández-Marmiesse, A.; Roca, I.; Hermida, A.; Martínez-Olmos, M.; et al. Carbohydrate status in patients with phenylketonuria. Orphanet J. Rare Dis. 2018, 13, 103. [Google Scholar] [CrossRef]
- Mokou, M.; Mischak, H.; Frantzi, M. Statistical determination of cancer biomarkers: Moving forward clinically. Expert Rev. Mol. Diagn. 2023, 23, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Basu, D.; Sinha, R.; Sahu, S.; Malla, J.; Chakravorty, N.; Ghosal, P.S. Identification of severity and passive measurement of oxidative stress biomarkers for β–thalassemia patients: K-means, random forest, XGBoost, decision tree, neural network based novel framework. Adv. Redox Res. 2022, 5, 100034. [Google Scholar] [CrossRef]
- Sharma, A.; Verbeke, W.J.M.I. Improving Diagnosis of Depression With XGBOOST Machine Learning Model and a Large Biomarkers Dutch Dataset (n = 11,081). Front. Big Data 2020, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Caumo, A.; Perseghin, G.; Brunani, A.; Luzi, L. New Insights on the Simultaneous Assessment of Insulin Sensitivity and -Cell Function With the HOMA2 Method. Diabetes Care 2006, 29, 2733–2734. [Google Scholar] [CrossRef]
- Ascaso, J.F.; Pardo, S.; Real, J.T.; Lorente, R.I.; Priego, A.; Carmena, R. Diagnosing Insulin Resistance by Simple Quantitative Methods in Subjects with Normal Glucose Metabolism. Diabetes Care 2003, 26, 3320–3325. [Google Scholar] [CrossRef]
- Burrows, R.; Correa-Burrows, P.; Reyes, M.; Blanco, E.; Albala, C.; Gahagan, S. Healthy Chilean Adolescents with HOMA-IR ≥ 2.6 Have Increased Cardiometabolic Risk: Association with Genetic, Biological, and Environmental Factors. J. Diabetes Res. 2015, 2015, e783296. [Google Scholar] [CrossRef]
- Katz, A.; Nambi, S.S.; Mather, K.; Baron, A.D.; Follmann, D.A.; Sullivan, G.; Quon, M.J. Quantitative Insulin Sensitivity Check Index: A Simple, Accurate Method for Assessing Insulin Sensitivity In Humans. J. Clin. Endocrinol. Metab. 2000, 85, 2402–2410. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Heese, R.; Mücke, S.; Jakobs, M.; Gerlach, T.; Piatkowski, N. Shapley Values with Uncertain Value Functions. In Proceedings of the Advances in Intelligent Data Analysis XXI: 21st International Symposium on Intelligent Data Analysis, IDA 2023, Louvain-la-Neuve, Belgium, 12–14 April 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 156–168. [Google Scholar]
- Jurecki, E.R.; Cederbaum, S.; Kopesky, J.; Perry, K.; Rohr, F.; Sanchez-Valle, A.; Viau, K.S.; Sheinin, M.Y.; Cohen-Pfeffer, J.L. Adherence to clinic recommendations among patients with phenylketonuria in the United States. Mol. Genet. Metab. 2017, 120, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Green, B.; Browne, R.; Firman, S.; Hill, M.; Rahman, Y.; Kaalund Hansen, K.; Adam, S.; Skeath, R.; Hallam, P.; Herlihy, I.; et al. Nutritional and Metabolic Characteristics of UK Adult Phenylketonuria Patients with Varying Dietary Adherence. Nutrients 2019, 11, 2459. [Google Scholar] [CrossRef] [PubMed]
- van Spronsen, F.J.; van Wegberg, A.M.; Ahring, K.; Bélanger-Quintana, A.; Blau, N.; Bosch, A.M.; Burlina, A.; Campistol, J.; Feillet, F.; Giżewska, M.; et al. Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol. 2017, 5, 743–756. [Google Scholar] [CrossRef]
- Karam, P.E.; Majdalani, M.N.; Daher, R.T.; Barhoumi, A.; Yazbeck, N. Cardiovascular disease biomarkers in patients with inborn errors of protein metabolism: A pilot study. J. Hum. Nutr. Diet. Off. J. Br. Diet. Assoc. 2015, 28, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Azabdaftari, A.; van der Giet, M.; Schuchardt, M.; Hennermann, J.B.; Plöckinger, U.; Querfeld, U. The cardiovascular phenotype of adult patients with phenylketonuria. Orphanet J. Rare Dis. 2019, 14, 213. [Google Scholar] [CrossRef]
- Rodrigues, C.; Pinto, A.; Faria, A.; Teixeira, D.; van Wegberg, A.M.J.; Ahring, K.; Feillet, F.; Calhau, C.; MacDonald, A.; Moreira-Rosário, A.; et al. Is the Phenylalanine-Restricted Diet a Risk Factor for Overweight or Obesity in Patients with Phenylketonuria (PKU)? A Systematic Review and Meta-Analysis. Nutrients 2021, 13, 3443. [Google Scholar] [CrossRef]
- Stewart, M.R.; Hemli, S.; Kolodny, E.H.; Miller, A.L.; Pallotta, J.A. Carbohydrate Metabolism in Phenylketonuria. Pediatr. Res. 1980, 14, 849–853. [Google Scholar] [CrossRef]
- Kanufre, V.C.; Soares, R.D.L.; Alves, M.R.A.; Aguiar, M.J.B.; Starling, A.L.P.; Norton, R.C. Metabolic syndrome in children and adolescents with phenylketonuria. J. Pediatr. 2015, 91, 98–103. [Google Scholar] [CrossRef]
- Petersen, M.C.; Shulman, G.I. Mechanisms of Insulin Action and Insulin Resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef]
- Zhou, Q.; Sun, W.-W.; Chen, J.-C.; Zhang, H.-L.; Liu, J.; Lin, Y.; Lin, P.-C.; Wu, B.-X.; An, Y.-P.; Huang, L.; et al. Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRβ. Nat. Commun. 2022, 13, 4291. [Google Scholar] [CrossRef]
- Muniyappa, R.; Lee, S.; Chen, H.; Quon, M.J. Current approaches for assessing insulin sensitivity and resistance in vivo: Advantages, limitations, and appropriate usage. Am. J. Physiol.-Endocrinol. Metab. 2008, 294, E15–E26. [Google Scholar] [CrossRef] [PubMed]
- Abdesselam, A.; Zidoum, H.; Zadjali, F.; Hedjam, R.; Al-Ansari, A.; Bayoumi, R.; Al-Yahyaee, S.; Hassan, M.; Albarwani, S. Estimate of the HOMA-IR Cut-off Value for Identifying Subjects at Risk of Insulin Resistance Using a Machine Learning Approach. Sultan Qaboos Univ. Med. J. 2021, 21, 604–612. [Google Scholar] [CrossRef]
- Friedemann, C.; Heneghan, C.; Mahtani, K.; Thompson, M.; Perera, R.; Ward, A.M. Cardiovascular disease risk in healthy children and its association with body mass index: Systematic review and meta-analysis. BMJ 2012, 345, e4759. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, C.; Yao, J.; Jiang, Q.; Chen, N.; Huang, H.; Liang, J.; Li, L.; Lin, L. Overweight, obesity, and their associations with insulin resistance and β-cell function among Chinese: A cross-sectional study in China. Metabolism 2010, 59, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Sener, A.; Best, L.C.; Yates, A.P.; Kadiata, M.M.; Olivares, E.; Louchami, K.; Jijakli, H.; Ladrière, L.; Malaisse, W.J. Stimulus-secretion coupling of arginine-induced insulin release. Endocrine 2000, 13, 329–340. [Google Scholar] [CrossRef]
- Nowak, C.; Hetty, S.; Salihovic, S.; Castillejo-Lopez, C.; Ganna, A.; Cook, N.L.; Broeckling, C.D.; Prenni, J.E.; Shen, X.; Giedraitis, V.; et al. Glucose challenge metabolomics implicates medium-chain acylcarnitines in insulin resistance. Sci. Rep. 2018, 8, 8691. [Google Scholar] [CrossRef]
- Bene, J.; Hadzsiev, K.; Melegh, B. Role of carnitine and its derivatives in the development and management of type 2 diabetes. Nutr. Diabetes 2018, 8, 8. [Google Scholar] [CrossRef]
| G1 Group (n = 10) | G2 Group (n = 14) | G3 Group (n = 24) | p-Value | |
|---|---|---|---|---|
| Age (years) | 24 (19–27) | 24 (18–26) | 23 (19–26) | NS ** |
| Sex (F/M) | 5:5 | 5:9 | 11:13 | NS *** |
| Weight (kg) b | 65 (58–94) | 75 (59–81) | 68 (61–83) | NS ** |
| Height (m) a | 163 ± 9 (157–170) | 163 ± 8 (158–168) | 165 ± 9 (161–168) | NS * |
| BMI (kg/m2) a | 26 ± 5 (23–30) | 28 ± 7 (24–32) | 26 ± 5 (24–28) | NS * |
| Waist circumference (cm) b | 82 (76–97) | 92 (78–103) | 81 (75–91) | NS ** |
| G1 Group (n = 10) | G2 Group (n = 14) | G3 Group (n = 24) | p-Value | |
|---|---|---|---|---|
| Glycemia (mg/dL) a | 85.7 ± 5.5 (81.8–89.7) | 90.7 ± 4.9 (87.8–93.4) | 92.2 ± 8.5 (88.6–95.8) | <0.05 ɛ |
| Insulin (µIU/mL) b | 7.9 (4.1–15.9) | 12.4 (10.6–19.4) | 9.6 (7.1–14.7) | <0.05 ɣ |
| HOMA-IR b | 1.0 (0.5–2.0) | 1.6 (1.4–2.5) | 1.3 (0.9–1.9) | <0.05 ɣǂ |
| HOMA-β (%) b | 112.2 (79.7 –61.7) | 139.1 (117.0–166.0) | 107.5 (90.3–127.5) | <0.05 ǂ |
| HOMA-S (%) a | 119.5 ± 71.6 (68.3–170.7) | 60 ± 28.6 (43.6–76.7) | 82.8 ± 37.5 (66.9–98.7) | <0.05 ɣɛ |
| QUICKI a | 0.36 ± 0.04 (0.33–0.39) | 0.32 ± 0.02 (0.31–0.33) | 0.34 ± 0.03 (0.33–0.35) | <0.05 ɣ |
| Total cholesterol (mg/dL) a | 137.1 ± 27.6 (117.3–156.8) | 139.6 ± 27.5 (123.7–155.5) | 154.1 ± 31.1 (140.9–167.3) | NS * |
| HDL cholesterol (mg/dL) a | 50.8 ± 12.8 (41.5–59.9) | 44.3 ± 8.8 (39.2–49.4) | 49.9 ± 9.3 (45.9–53.9) | NS * |
| LDL cholesterol (mg/dL) b | 65.6 (48.7–82.4) | 68.3 (62.2–83.8) | 77 (60.2–99.4) | NS ** |
| Triglycerides (mg/dL) b | 81.5 (54–115) | 93.5 (60–164) | 92.5 (79–122) | NS ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leal-Witt, M.J.; Rojas-Agurto, E.; Muñoz-González, M.; Peñaloza, F.; Arias, C.; Fuenzalida, K.; Bunout, D.; Cornejo, V.; Acevedo, A. Risk of Developing Insulin Resistance in Adult Subjects with Phenylketonuria: Machine Learning Model Reveals an Association with Phenylalanine Concentrations in Dried Blood Spots. Metabolites 2023, 13, 677. https://doi.org/10.3390/metabo13060677
Leal-Witt MJ, Rojas-Agurto E, Muñoz-González M, Peñaloza F, Arias C, Fuenzalida K, Bunout D, Cornejo V, Acevedo A. Risk of Developing Insulin Resistance in Adult Subjects with Phenylketonuria: Machine Learning Model Reveals an Association with Phenylalanine Concentrations in Dried Blood Spots. Metabolites. 2023; 13(6):677. https://doi.org/10.3390/metabo13060677
Chicago/Turabian StyleLeal-Witt, María Jesús, Eugenia Rojas-Agurto, Manuel Muñoz-González, Felipe Peñaloza, Carolina Arias, Karen Fuenzalida, Daniel Bunout, Verónica Cornejo, and Alejandro Acevedo. 2023. "Risk of Developing Insulin Resistance in Adult Subjects with Phenylketonuria: Machine Learning Model Reveals an Association with Phenylalanine Concentrations in Dried Blood Spots" Metabolites 13, no. 6: 677. https://doi.org/10.3390/metabo13060677
APA StyleLeal-Witt, M. J., Rojas-Agurto, E., Muñoz-González, M., Peñaloza, F., Arias, C., Fuenzalida, K., Bunout, D., Cornejo, V., & Acevedo, A. (2023). Risk of Developing Insulin Resistance in Adult Subjects with Phenylketonuria: Machine Learning Model Reveals an Association with Phenylalanine Concentrations in Dried Blood Spots. Metabolites, 13(6), 677. https://doi.org/10.3390/metabo13060677