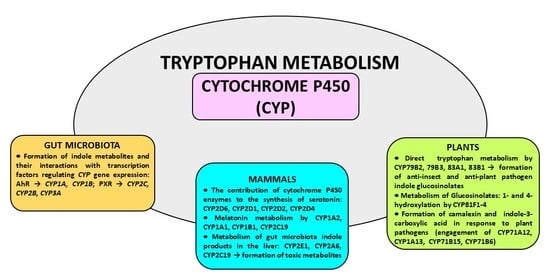
The Engagement of Cytochrome P450 Enzymes in Tryptophan Metabolism
Abstract
1. Introduction
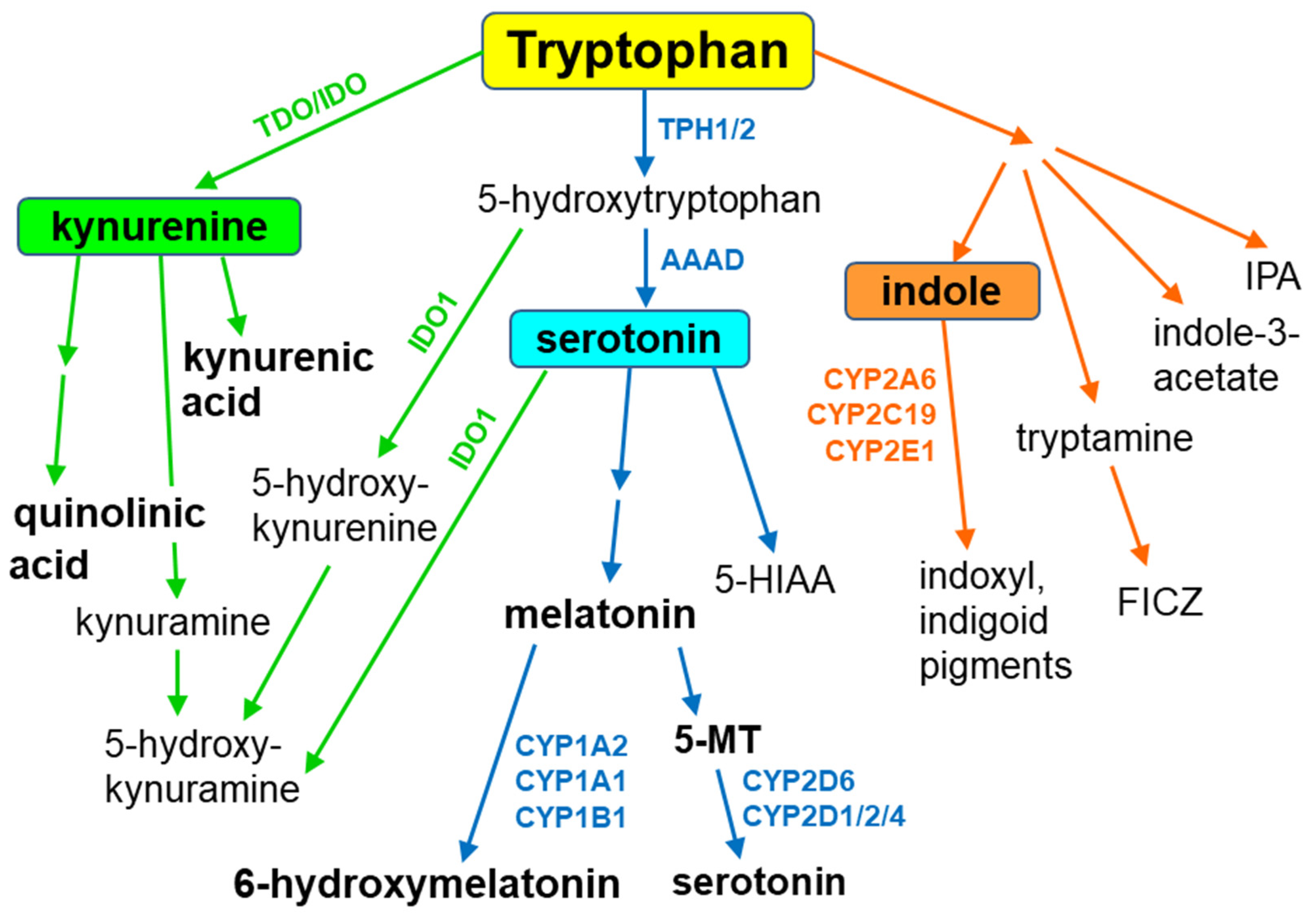
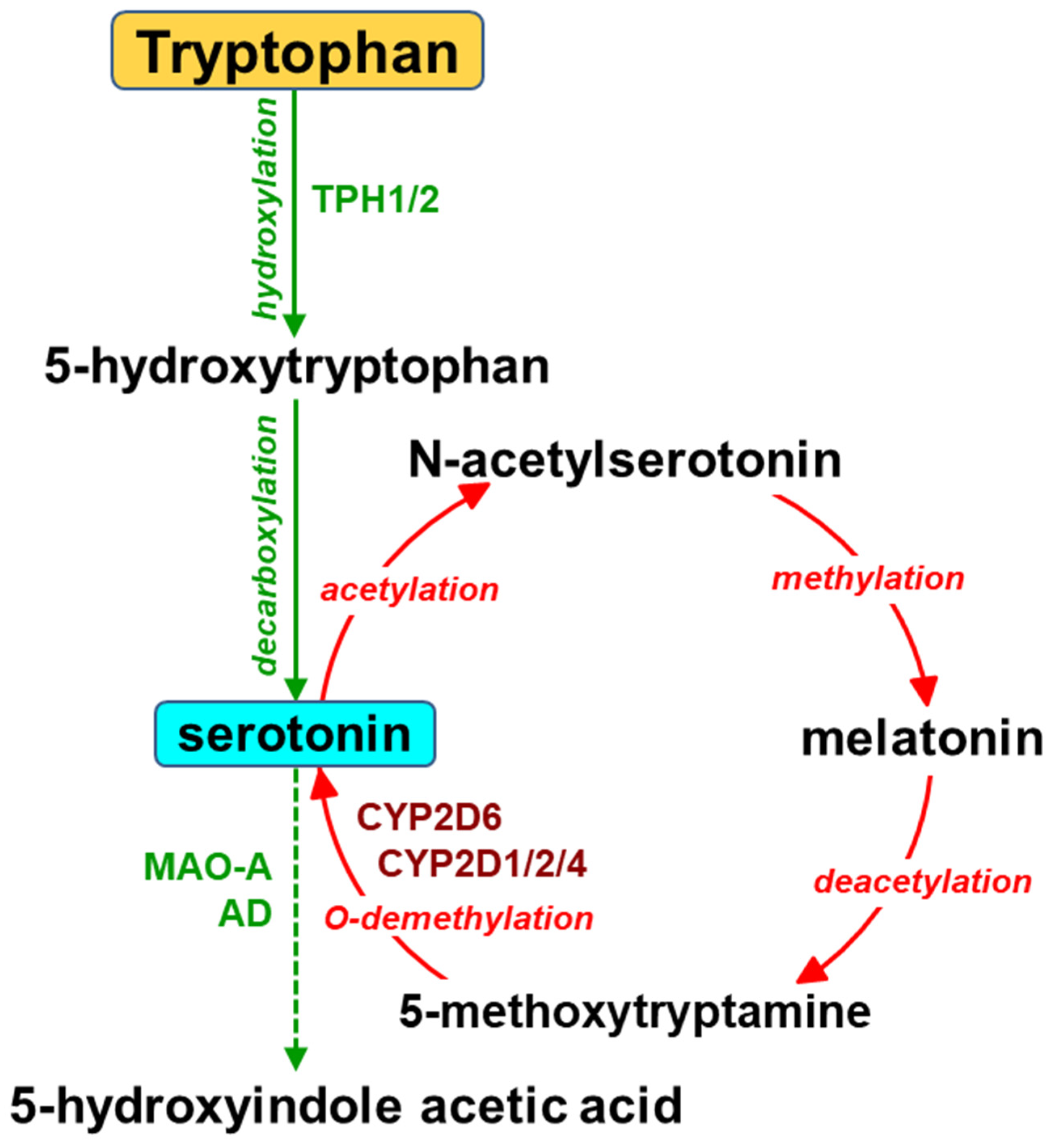
2. The Contribution of Cytochrome P450 to the Synthesis of Serotonin
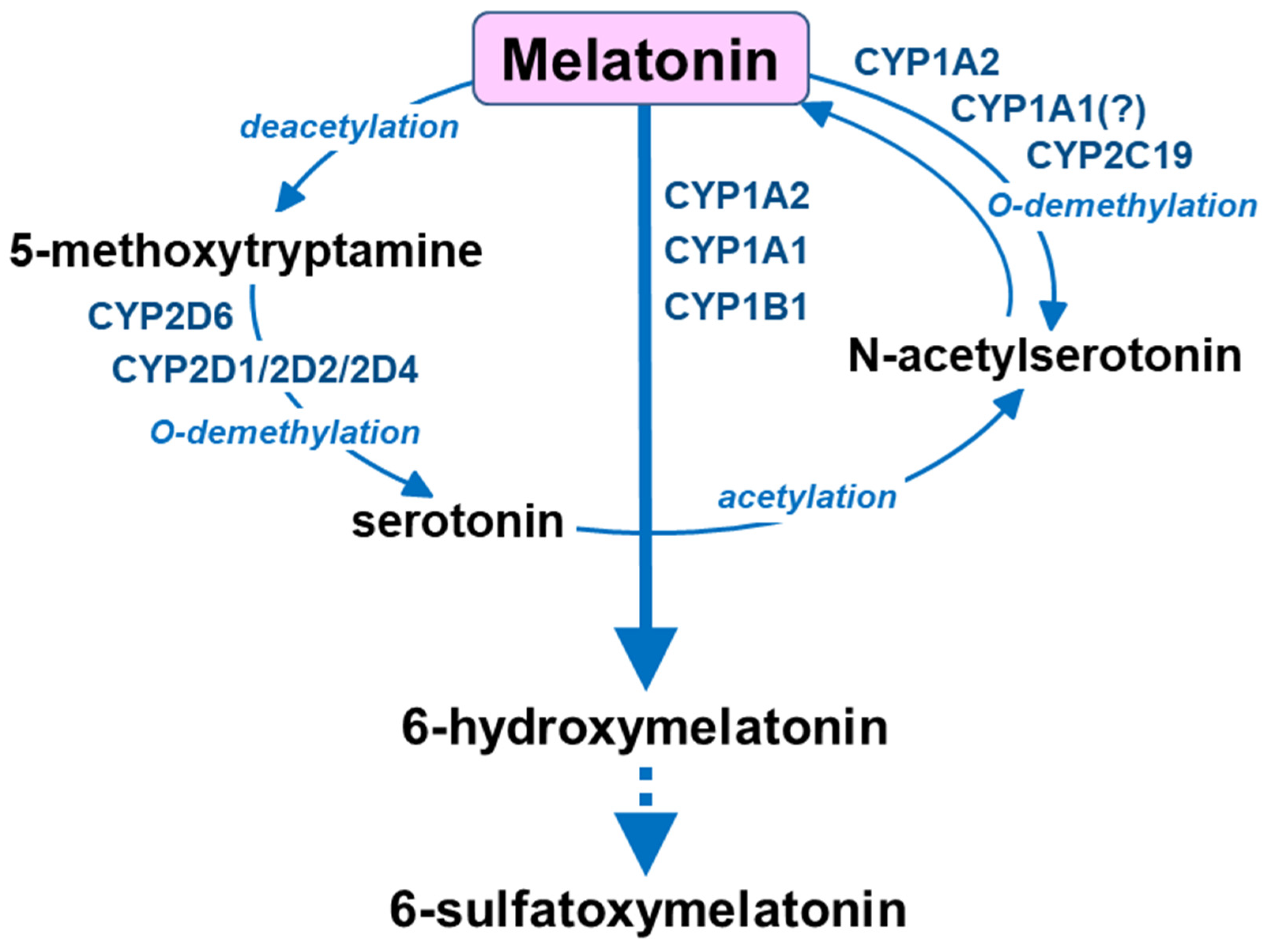
3. Melatonin Metabolism by Cytochrome P450
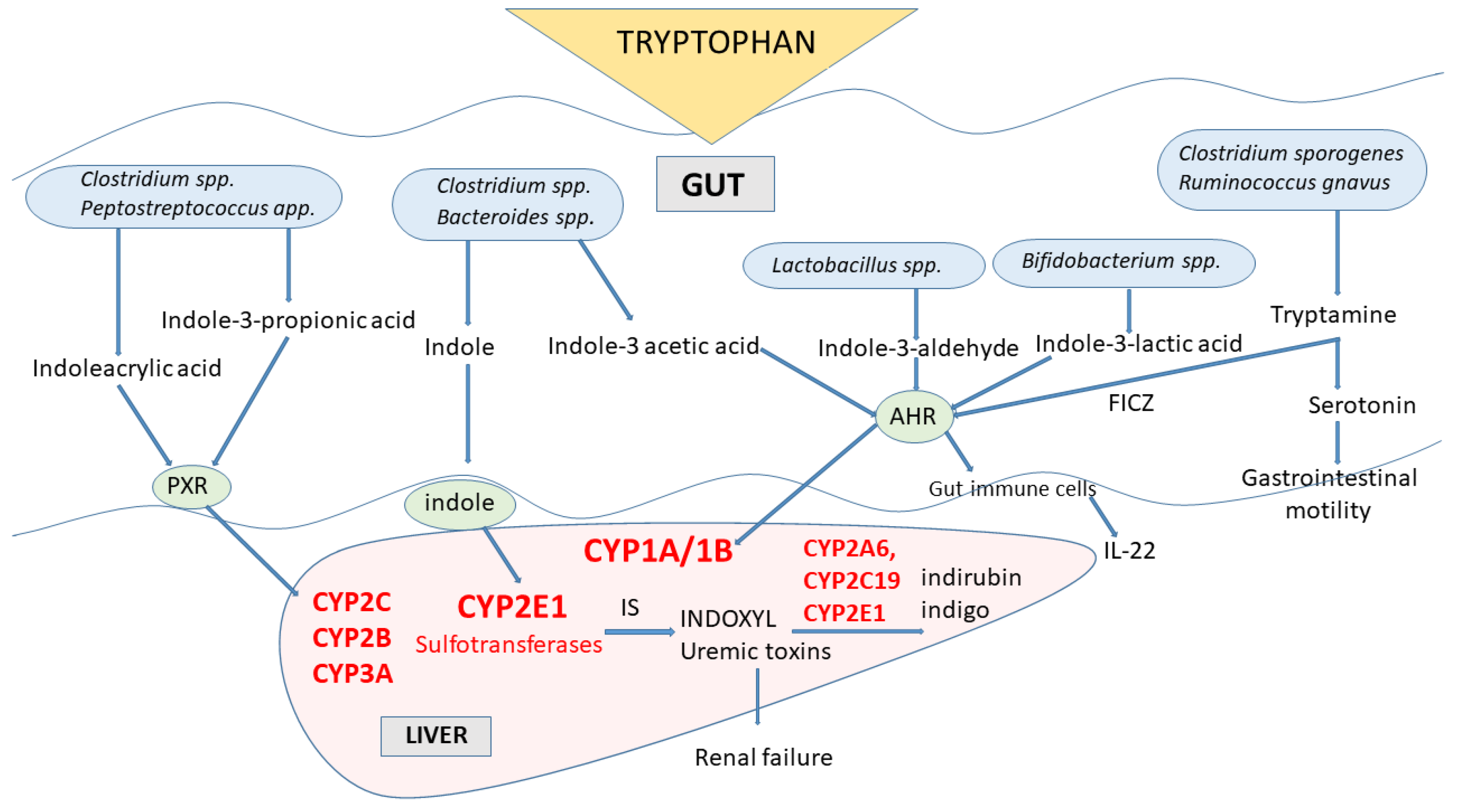
4. Gut Microbiota Indole Products and Their Interaction with Cytochrome P450
Indole Metabolites of Tryptophan-Toxicological Aspects
5. The Involvement of Cytochrome P450 in Tryptophan Metabolism in Plants
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Omura, T. Structural Diversity of Cytochrome P450 Enzyme System. J. Biochem. 2010, 147, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Fujiyama, K.; Hino, T.; Nagano, S. Diverse Reactions Catalyzed by Cytochrome P450 and Biosynthesis of Steroid Hormone. Biophys. Physicobiol. 2022, 19, e190021. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Roles of Cytochrome P450 Enzymes in Pharmacology and Toxicology: Past, Present, and Future. Adv. Pharmacol. 2022, 95, 1–47. [Google Scholar] [CrossRef]
- Nebert, D.W.; Nelson, D.R.; Coon, M.J.; Estabrook, R.W.; Feyereisen, R.; Fujii-Kuriyama, Y.; Gonzalez, F.J.; Guengerich, F.P.; Gunsalus, I.C.; Johnson, E.F. The P450 Superfamily: Update on New Sequences, Gene Mapping, and Recommended Nomenclature. DNA Cell. Biol. 1991, 10, 1–14. [Google Scholar] [CrossRef]
- Crešnar, B.; Petrič, S. Cytochrome P450 Enzymes in the Fungal Kingdom. Biochim. Biophys. Acta 2011, 1814, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant Cytochrome P450 Plasticity and Evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the Occurrence of Cytochrome P450 in Viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Roth, W.; Zadeh, K.; Vekariya, R.; Ge, Y.; Mohamadzadeh, M. Tryptophan Metabolism and Gut-Brain Homeostasis. Int. J. Mol. Sci. 2021, 22, 2973. [Google Scholar] [CrossRef]
- Ravanfar, R.; Sheng, Y.; Gray, H.B.; Winkler, J.R. Tryptophan-96 in Cytochrome P450 BM3 Plays a Key Role in Enzyme Survival. FEBS Lett. 2023, 597, 59–64. [Google Scholar] [CrossRef]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, Tryptophan Metabolism and the Brain-Gut-Microbiome Axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Murray, P.J. Tryptophan and Indole Metabolism in Immune Regulation. Curr. Opin. Immunol. 2021, 70, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-M.; Idle, J.R.; Byrd, L.G.; Krausz, K.W.; Küpfer, A.; Gonzalez, F.J. Regeneration of Serotonin from 5-Methoxytryptamine by Polymorphic Human CYP2D6. Pharmacogenetics 2003, 13, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Haduch, A.; Bromek, E.; Sadakierska-Chudy, A.; Wójcikowski, J.; Daniel, W.A. The Catalytic Competence of Cytochrome P450 in the Synthesis of Serotonin from 5-Methoxytryptamine in the Brain: An in Vitro Study. Pharmacol. Res. 2013, 67, 53–59. [Google Scholar] [CrossRef]
- Haduch, A.; Bromek, E.; Kot, M.; Kamińska, K.; Gołembiowska, K.; Daniel, W.A. The Cytochrome P450 2D-Mediated Formation of Serotonin from 5-Methoxytryptamine in the Brain in Vivo: A Microdialysis Study. J. Neurochem. 2015, 133, 83–92. [Google Scholar] [CrossRef]
- Hardeland, R. Taxon- and Site-Specific Melatonin Catabolism. Molecules 2017, 22, 2015. [Google Scholar] [CrossRef]
- Haq, S.; Grondin, J.A.; Khan, W.I. Tryptophan-Derived Serotonin-Kynurenine Balance in Immune Activation and Intestinal Inflammation. FASEB J. 2021, 35, e21888. [Google Scholar] [CrossRef]
- Liu, J.-R.; Miao, H.; Deng, D.-Q.; Vaziri, N.D.; Li, P.; Zhao, Y.-Y. Gut Microbiota-Derived Tryptophan Metabolism Mediates Renal Fibrosis by Aryl Hydrocarbon Receptor Signaling Activation. Cell. Mol. Life Sci. 2021, 78, 909–922. [Google Scholar] [CrossRef]
- Gillam, E.M.; Notley, L.M.; Cai, H.; De Voss, J.J.; Guengerich, F.P. Oxidation of Indole by Cytochrome P450 Enzymes. Biochemistry 2000, 39, 13817–13824. [Google Scholar] [CrossRef]
- Yacoub, R.; Wyatt, C.M. Manipulating the Gut Microbiome to Decrease Uremic Toxins. Kidney Int. 2017, 91, 521–523. [Google Scholar] [CrossRef]
- Mosa, A.; Gerber, A.; Neunzig, J.; Bernhardt, R. Products of Gut-Microbial Tryptophan Metabolism Inhibit the Steroid Hormone-Synthesizing Cytochrome P450 11A1. Endocrine 2016, 53, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Adak, S.; Lukowski, A.L.; Schäfer, R.J.B.; Moore, B.S. From Tryptophan to Toxin: Nature’s Convergent Biosynthetic Strategy to Aetokthonotoxin. J. Am. Chem. Soc. 2022, 144, 2861–2866. [Google Scholar] [CrossRef] [PubMed]
- Rannug, A.; Rannug, U. The Tryptophan Derivative 6-Formylindolo[3,2-b]Carbazole, FICZ, a Dynamic Mediator of Endogenous Aryl Hydrocarbon Receptor Signaling, Balances Cell Growth and Differentiation. Crit. Rev. Toxicol. 2018, 48, 555–574. [Google Scholar] [CrossRef] [PubMed]
- Celenza, J.L. Metabolism of Tyrosine and Tryptophan--New Genes for Old Pathways. Curr. Opin. Plant Biol. 2001, 4, 234–240. [Google Scholar] [CrossRef]
- Barry, S.M.; Kers, J.A.; Johnson, E.G.; Song, L.; Aston, P.R.; Patel, B.; Krasnoff, S.B.; Crane, B.R.; Gibson, D.M.; Loria, R.; et al. Cytochrome P450–Catalyzed L-Tryptophan Nitration in Thaxtomin Phytotoxin Biosynthesis. Nat. Chem. Biol. 2012, 8, 814–816. [Google Scholar] [CrossRef]
- Dahlström, A.; Fuxe, K. Localization of Monoamines in the Lower Brain Stem. Experientia 1964, 20, 398–399. [Google Scholar] [CrossRef]
- Törk, I. Anatomy of the Serotonergic System. Ann. N. Y. Acad. Sci. 1990, 600, 9–34; discussion 34–35. [Google Scholar] [CrossRef]
- Di Giovanni, G.; Di Matteo, V.; Pierucci, M.; Esposito, E. Serotonin-Dopamine Interaction: Electrophysiological Evidence. Prog. Brain Res. 2008, 172, 45–71. [Google Scholar] [CrossRef]
- Bromek, E.; Daniel, W.A. The Regulation of Liver Cytochrome P450 Expression and Activity by the Brain Serotonergic System in Different Experimental Models. Expert. Opin. Drug. Metab. Toxicol. 2021, 17, 413–424. [Google Scholar] [CrossRef]
- Keating, D.J.; Spencer, N.J. What Is the Role of Endogenous Gut Serotonin in the Control of Gastrointestinal Motility? Pharmacol. Res. 2019, 140, 50–55. [Google Scholar] [CrossRef]
- Mawe, G.M.; Hurd, M.; Hennig, G.W.; Lavoie, B. Epithelial 5-HT4 Receptors as a Target for Treating Constipation and Intestinal Inflammation. Adv. Exp. Med. Biol. 2022, 1383, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Lesurtel, M.; Graf, R.; Aleil, B.; Walther, D.J.; Tian, Y.; Jochum, W.; Gachet, C.; Bader, M.; Clavien, P.-A. Platelet-Derived Serotonin Mediates Liver Regeneration. Science 2006, 312, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Liu, C.; Shu, B.; Zhai, M.; Deng, C.; He, C.; Luo, M.; Han, T.; Zheng, W.; Zhang, J.; et al. Axis of Serotonin -PERK-YAP in Liver Regeneration. Life Sci. 2018, 209, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Moteki, H.; Ogihara, M. Role of Hepatocyte Growth Regulators in Liver Regeneration. Cells 2023, 12, 208. [Google Scholar] [CrossRef]
- Prozialek, W.C.; Vogel, W.H. Deamination of 5-Methoxytryptamine, Serotonin and Phenylethylamine by Rat MAO in Vitro and in Vivo. Life Sci. 1978, 22, 561–569. [Google Scholar] [CrossRef]
- Kobayashi, S.; Murray, S.; Watson, D.; Sesardic, D.; Davies, D.S.; Boobis, A.R. The Specificity of Inhibition of Debrisoquine 4-Hydroxylase Activity by Quinidine and Quinine in the Rat Is the Inverse of That in Man. Biochem. Pharmacol. 1989, 38, 2795–2799. [Google Scholar] [CrossRef]
- Boobis, A.R.; Sesardic, D.; Murray, B.P.; Edwards, R.J.; Singleton, A.M.; Rich, K.J.; Murray, S.; de la Torre, R.; Segura, J.; Pelkonen, O. Species Variation in the Response of the Cytochrome P-450-Dependent Monooxygenase System to Inducers and Inhibitors. Xenobiotica 1990, 20, 1139–1161. [Google Scholar] [CrossRef]
- Cheng, J.; Zhen, Y.; Miksys, S.; Beyoğlu, D.; Krausz, K.W.; Tyndale, R.F.; Yu, A.; Idle, J.R.; Gonzalez, F.J. Potential Role of CYP2D6 in the Central Nervous System. Xenobiotica 2013, 43, 973–984. [Google Scholar] [CrossRef]
- Patel, S.; Dulluc, J.; Geffard, M. Comparison of Serotonin and 5-Methoxytryptamine Immunoreactivity in Rat Raphe Nuclei. Histochemistry 1986, 85, 259–263. [Google Scholar] [CrossRef]
- Kuban, W.; Daniel, W.A. Cytochrome P450 Expression and Regulation in the Brain. Drug. Metab. Rev. 2021, 53, 1–29. [Google Scholar] [CrossRef]
- Kot, M.; Daniel, W.A. Cytochrome P450 Is Regulated by Noradrenergic and Serotonergic Systems. Pharmacol. Res. 2011, 64, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Miksys, S.; Rao, Y.; Sellers, E.M.; Kwan, M.; Mendis, D.; Tyndale, R.F. Regional and Cellular Distribution of CYP2D Subfamily Members in Rat Brain. Xenobiotica 2000, 30, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Miksys, S.; Rao, Y.; Hoffmann, E.; Mash, D.C.; Tyndale, R.F. Regional and Cellular Expression of CYP2D6 in Human Brain: Higher Levels in Alcoholics: CYP2D6 Is Higher in Brain of Human Alcoholics. J. Neurochem. 2002, 82, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Miksys, S.; Tyndale, R.F. Cytochrome P450-Mediated Drug Metabolism in the Brain. J. Psychiatry Neurosci. 2013, 38, 152–163. [Google Scholar] [CrossRef]
- Mostalac-Preciado, C.R.; de Gortari, P.; López-Rubalcava, C. Antidepressant-like Effects of Mineralocorticoid but Not Glucocorticoid Antagonists in the Lateral Septum: Interactions with the Serotonergic System. Behav. Brain Res. 2011, 223, 88–98. [Google Scholar] [CrossRef]
- Bromek, E.; Haduch, A.; Daniel, W.A. The Ability of Cytochrome P450 2D Isoforms to Synthesize Dopamine in the Brain: An in Vitro Study. Eur. J. Pharmacol. 2010, 626, 171–178. [Google Scholar] [CrossRef]
- Borroto-Escuela, D.O.; Ambrogini, P.; Chruścicka, B.; Lindskog, M.; Crespo-Ramirez, M.; Hernández-Mondragón, J.C.; Perez de la Mora, M.; Schellekens, H.; Fuxe, K. The Role of Central Serotonin Neurons and 5-HT Heteroreceptor Complexes in the Pathophysiology of Depression: A Historical Perspective and Future Prospects. Int. J. Mol. Sci. 2021, 22, 1927. [Google Scholar] [CrossRef]
- Correia, A.S.; Vale, N. Tryptophan Metabolism in Depression: A Narrative Review with a Focus on Serotonin and Kynurenine Pathways. Int. J. Mol. Sci. 2022, 23, 8493. [Google Scholar] [CrossRef]
- Pourhamzeh, M.; Moravej, F.G.; Arabi, M.; Shahriari, E.; Mehrabi, S.; Ward, R.; Ahadi, R.; Joghataei, M.T. The Roles of Serotonin in Neuropsychiatric Disorders. Cell. Mol. Neurobiol. 2022, 42, 1671–1692. [Google Scholar] [CrossRef]
- Haduch, A.; Pukło, R.; Alenina, N.; Nikiforuk, A.; Popik, P.; Bader, M.; Daniel, W.A. The Effect of Ageing and Cerebral Serotonin Deficit on the Activity of Cytochrome P450 2D (CYP2D) in the Brain and Liver of Male Rats. Neurochem. Int. 2020, 141, 104884. [Google Scholar] [CrossRef]
- Haduch, A.; Danek, P.J.; Kuban, W.; Pukło, R.; Alenina, N.; Gołębiowska, J.; Popik, P.; Bader, M.; Daniel, W.A. Cytochrome P450 2D (CYP2D) Enzyme Dysfunction Associated with Aging and Serotonin Deficiency in the Brain and Liver of Female Dark Agouti Rats. Neurochem. Int. 2022, 152, 105223. [Google Scholar] [CrossRef] [PubMed]
- Warner, M.; Gustafsson, J.A. Effect of Ethanol on Cytochrome P450 in the Rat Brain. Proc. Natl. Acad. Sci. USA 1994, 91, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Miksys, S.; Lee, A.; Mash, D.C.; Tyndale, R.F. Induction of the Drug Metabolizing Enzyme CYP2D in Monkey Brain by Chronic Nicotine Treatment. Neuropharmacology 2008, 55, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.T.; Miksys, S.; Hoffmann, E.; Tyndale, R.F. Ethanol Self-Administration and Nicotine Treatment Increase Brain Levels of CYP2D in African Green Monkeys. Br. J. Pharmacol. 2014, 171, 3077–3088. [Google Scholar] [CrossRef]
- Yue, J.; Miksys, S.; Hoffmann, E.; Tyndale, R.F. Chronic Nicotine Treatment Induces Rat CYP2D in the Brain but Not in the Liver: An Investigation of Induction and Time Course. J. Psychiatry Neurosci. 2008, 33, 54–63. [Google Scholar]
- Haduch, A.; Bromek, E.; Daniel, W.A. The Effect of Psychotropic Drugs on Cytochrome P450 2D (CYP2D) in Rat Brain. Eur. J. Pharmacol. 2011, 651, 51–58. [Google Scholar] [CrossRef]
- Haduch, A.; Rysz, M.; Papp, M.; Daniel, W.A. The Activity of Brain and Liver Cytochrome P450 2D (CYP2D) Is Differently Affected by Antidepressants in the Chronic Mild Stress (CMS) Model of Depression in the Rat. Biochem. Pharmacol. 2018, 156, 398–405. [Google Scholar] [CrossRef]
- Haduch, A.; Daniel, W.A. The Engagement of Brain Cytochrome P450 in the Metabolism of Endogenous Neuroactive Substrates: A Possible Role in Mental Disorders. Drug Metab. Rev. 2018, 50, 415–429. [Google Scholar] [CrossRef]
- Haduch, A.; Bromek, E.; Pukło, R.; Jastrzębska, J.; Danek, P.J.; Daniel, W.A. The Effect of the Selective N-Methyl-D-Aspartate (NMDA) Receptor GluN2B Subunit Antagonist CP-101,606 on Cytochrome P450 2D (CYP2D) Expression and Activity in the Rat Liver and Brain. Int. J. Mol. Sci. 2022, 23, 13746. [Google Scholar] [CrossRef]
- Daniel, W.A.; Bromek, E.; Danek, P.J.; Haduch, A. The Mechanisms of Interactions of Psychotropic Drugs with Liver and Brain Cytochrome P450 and Their Significance for Drug Effect and Drug-Drug Interactions. Biochem. Pharmacol. 2022, 199, 115006. [Google Scholar] [CrossRef]
- Hedlund, E.; Wyss, A.; Kainu, T.; Backlund, M.; Köhler, C.; Pelto-Huikko, M.; Gustafsson, J.A.; Warner, M. Cytochrome P4502D4 in the Brain: Specific Neuronal Regulation by Clozapine and Toluene. Mol. Pharmacol. 1996, 50, 342–350. [Google Scholar] [PubMed]
- Danek, P.J.; Bromek, E.; Haduch, A.; Daniel, W.A. Chronic Treatment with Asenapine Affects Cytochrome P450 2D (CYP2D) in Rat Brain and Liver. Pharmacological Aspects. Neurochem. Int. 2021, 151, 105209. [Google Scholar] [CrossRef] [PubMed]
- Danek, P.J.; Daniel, W.A. Long-Term Treatment with Atypical Antipsychotic Iloperidone Modulates Cytochrome P450 2D (CYP2D) Expression and Activity in the Liver and Brain via Different Mechanisms. Cells 2021, 10, 3472. [Google Scholar] [CrossRef] [PubMed]
- Danek, P.J.; Daniel, W.A. The Atypical Antipsychotic Lurasidone Affects Brain but Not Liver Cytochrome P450 2D (CYP2D) Activity. A Comparison with Other Novel Neuroleptics and Significance for Drug Treatment of Schizophrenia. Cells 2022, 11, 3513. [Google Scholar] [CrossRef] [PubMed]
- Bertilsson, L.; Alm, C.; De Las Carreras, C.; Widen, J.; Edman, G.; Schalling, D. Debrisoquine Hydroxylation Polymorphism and Personality. Lancet 1989, 1, 555. [Google Scholar] [CrossRef] [PubMed]
- González, I.; Peñas-Lledó, E.M.; Pérez, B.; Dorado, P.; Alvarez, M.; LLerena, A. Relation between CYP2D6 Phenotype and Genotype and Personality in Healthy Volunteers. Pharmacogenomics 2008, 9, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Peñas-LLedó, E.M.; LLerena, A. CYP2D6 Variation, Behaviour and Psychopathology: Implications for Pharmacogenomics-Guided Clinical Trials. Br. J. Clin. Pharmacol. 2014, 77, 673–683. [Google Scholar] [CrossRef]
- Hensler, J.G. Serotonergic Modulation of the Limbic System. Neurosci. Biobehav. Rev. 2006, 30, 203–214. [Google Scholar] [CrossRef]
- Shamir, E.; Laudon, M.; Barak, Y.; Anis, Y.; Rotenberg, V.; Elizur, A.; Zisapel, N. Melatonin Improves Sleep Quality of Patients with Chronic Schizophrenia. J. Clin. Psychiatry 2000, 61, 373–377. [Google Scholar] [CrossRef]
- Dolberg, O.T.; Hirschmann, S.; Grunhaus, L. Melatonin for the Treatment of Sleep Disturbances in Major Depressive Disorder. Am. J. Psychiatry 1998, 155, 1119–1121. [Google Scholar] [CrossRef]
- Dalton, E.J.; Rotondi, D.; Levitan, R.D.; Kennedy, S.H.; Brown, G.M. Use of Slow-Release Melatonin in Treatment-Resistant Depression. J. Psychiatry Neurosci. 2000, 25, 48–52. [Google Scholar]
- Singh, M.; Jadhav, H.R. Melatonin: Functions and Ligands. Drug Discov. Today 2014, 19, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Venegas, C.; García, J.A.; Escames, G.; Ortiz, F.; López, A.; Doerrier, C.; García-Corzo, L.; López, L.C.; Reiter, R.J.; Acuña-Castroviejo, D. Extrapineal Melatonin: Analysis of Its Subcellular Distribution and Daily Fluctuations. J. Pineal Res. 2012, 52, 217–227. [Google Scholar] [CrossRef]
- Acuña-Castroviejo, D.; Escames, G.; Venegas, C.; Díaz-Casado, M.E.; Lima-Cabello, E.; López, L.C.; Rosales-Corral, S.; Tan, D.-X.; Reiter, R.J. Extrapineal Melatonin: Sources, Regulation, and Potential Functions. Cell. Mol. Life Sci. 2014, 71, 2997–3025. [Google Scholar] [CrossRef] [PubMed]
- Pandi-Perumal, S.R.; Trakht, I.; Srinivasan, V.; Spence, D.W.; Maestroni, G.J.M.; Zisapel, N.; Cardinali, D.P. Physiological Effects of Melatonin: Role of Melatonin Receptors and Signal Transduction Pathways. Prog. Neurobiol. 2008, 85, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Green, A.R.; Hughes, J.P.; Tordoff, A.F. The Concentration of 5-Methoxytryptamine in Rat Brain and Its Effects on Behaviour Following Its Peripheral Injection. Neuropharmacology 1975, 14, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Mokkawes, T.; Lim, Z.Q.; de Visser, S.P. Mechanism of Melatonin Metabolism by CYP1A1: What Determines the Bifurcation Pathways of Hydroxylation versus Deformylation? J. Phys. Chem. B. 2022, 126, 9591–9606. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Idle, J.R.; Krausz, K.W.; Gonzalez, F.J. Metabolism of Melatonin by Human Cytochromes P450. Drug Metab. Dispos. 2005, 33, 489–494. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin Metabolism in the Central Nervous System. Curr. Neuropharmacol. 2010, 8, 168–181. [Google Scholar] [CrossRef]
- Rogawski, M.A.; Roth, R.H.; Aghajanian, G.K. Melatonin: Deacetylation to 5-Methoxytryptamine by Liver but Not Brain Aryl Acylamidase. J. Neurochem. 1979, 32, 1219–1226. [Google Scholar] [CrossRef]
- Beck, O.; Jonsson, G. In Vivo Formation of 5-Methoxytryptamine from Melatonin in Rat. J. Neurochem. 1981, 36, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Haduch, A.; Bromek, E.; Wojcikowski, J.; Go embiowska, K.; Daniel, W.A. Melatonin Supports CYP2D-Mediated Serotonin Synthesis in the Brain. Drug Metab. Dispos. 2016, 44, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, O.; Katsumata, Y.; Oya, M. Characterization of Eight Biogenic Indoleamines as Substrates for Type A and Type B Monoamine Oxidase. Biochem. Pharmacol. 1981, 30, 1353–1358. [Google Scholar] [PubMed]
- Galzin, A.M.; Langer, S.Z. Potentiation by Deprenyl of the Autoreceptor-Mediated Inhibition of [3H]-5-Hydroxytryptamine Release by 5-Methoxytryptamine. Naunyn Schmiedebergs Arch. Pharmacol. 1986, 333, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, F.; Pévet, P. 5-Methoxytryptamine Is Metabolized by Monoamine Oxidase A in the Pineal Gland and Plasma of Golden Hamsters. Neurosci. Lett. 1991, 123, 172–174. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin, Its Metabolites and Their Interference with Reactive Nitrogen Compounds. Molecules 2021, 26, 4105. [Google Scholar] [CrossRef]
- Tordjman, S.; Chokron, S.; Delorme, R.; Charrier, A.; Bellissant, E.; Jaafari, N.; Fougerou, C. Melatonin: Pharmacology, Functions and Therapeutic Benefits. Curr. Neuropharmacol. 2017, 15, 434–443. [Google Scholar] [CrossRef]
- Liu, J.; Clough, S.J.; Hutchinson, A.J.; Adamah-Biassi, E.B.; Popovska-Gorevski, M.; Dubocovich, M.L. MT1 and MT2 Melatonin Receptors: A Therapeutic Perspective. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 361–383. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial Tryptophan Catabolites in Health and Disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Pascussi, J.-M.; Gerbal-Chaloin, S.; Duret, C.; Daujat-Chavanieu, M.; Vilarem, M.-J.; Maurel, P. The Tangle of Nuclear Receptors That Controls Xenobiotic Metabolism and Transport: Crosstalk and Consequences. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 1–32. [Google Scholar] [CrossRef]
- Ricardo, M.G.; Schwark, M.; Llanes, D.; Niedermeyer, T.H.J.; Westermann, B. Total Synthesis of Aetokthonotoxin, the Cyanobacterial Neurotoxin Causing Vacuolar Myelinopathy. Chemistry 2021, 27, 12032–12035. [Google Scholar] [CrossRef] [PubMed]
- Kawajiri, K.; Fujii-Kuriyama, Y. Cytochrome P450 Gene Regulation and Physiological Functions Mediated by the Aryl Hydrocarbon Receptor. Arch. Biochem. Biophys. 2007, 464, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Ma, N.; He, T.; Johnston, L.J.; Ma, X. Tryptophan (Trp) Modulates Gut Homeostasis via Aryl Hydrocarbon Receptor (AhR). Crit. Rev. Food Sci. Nutr. 2020, 60, 1760–1768. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Quintanilla, D.; Boos, D.; Khorram, O. Further characterization of tryptophan metabolism and its dysregulation in fibroids. F S Sci. 2022, 3, 392–400. [Google Scholar] [CrossRef]
- Liu, P.; Zhou, Y.; Dong, X.; Zheng, B.; Liang, B.; Liang, R.; Liu, Z.; Li, L.; Gong, P. ZNF165 Is Involved in the Regulation of Immune Microenvironment and Promoting the Proliferation and Migration of Hepatocellular Carcinoma by AhR/CYP1A1. J. Immunol. Res. 2022, 2022, 4446805. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Panwar, R.; Mittal, P.; Hassan, M.I.; Singh, I.K. Plant Cytochrome P450s: Role in Stress Tolerance and Potential Applications for Human Welfare. Int. J. Biol. Macromol. 2021, 184, 874–886. [Google Scholar] [CrossRef] [PubMed]
- Minerdi, D.; Savoi, S.; Sabbatini, P. Role of Cytochrome P450 Enzyme in Plant Microorganisms’ Communication: A Focus on Grapevine. Int. J. Mol. Sci. 2023, 24, 4695. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of Glucosinolates--Gene Discovery and Beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Hull, A.K.; Vij, R.; Celenza, J.L. Arabidopsis Cytochrome P450s That Catalyze the First Step of Tryptophan-Dependent Indole-3-Acetic Acid Biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2379–2384. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Hansen, C.H.; Wittstock, U.; Halkier, B.A. Cytochrome P450 CYP79B2 from Arabidopsis Catalyzes the Conversion of Tryptophan to Indole-3-Acetaldoxime, a Precursor of Indole Glucosinolates and Indole-3-Acetic Acid. J. Biol. Chem. 2000, 275, 33712–33717. [Google Scholar] [CrossRef]
- Bak, S.; Tax, F.E.; Feldmann, K.A.; Galbraith, D.W.; Feyereisen, R. CYP83B1, a Cytochrome P450 at the Metabolic Branch Point in Auxin and Indole Glucosinolate Biosynthesis in Arabidopsis. Plant Cell 2001, 13, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Naur, P.; Petersen, B.L.; Mikkelsen, M.D.; Bak, S.; Rasmussen, H.; Olsen, C.E.; Halkier, B.A. CYP83A1 and CYP83B1, Two Nonredundant Cytochrome P450 Enzymes Metabolizing Oximes in the Biosynthesis of Glucosinolates in Arabidopsis. Plant Physiol. 2003, 133, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Pfalz, M.; Mukhaimar, M.; Perreau, F.; Kirk, J.; Hansen, C.I.C.; Olsen, C.E.; Agerbirk, N.; Kroymann, J. Methyl Transfer in Glucosinolate Biosynthesis Mediated by Indole Glucosinolate O-Methyltransferase 5. Plant Physiol. 2016, 172, 2190–2203. [Google Scholar] [CrossRef]
- Pastorczyk, M.; Kosaka, A.; Piślewska-Bednarek, M.; López, G.; Frerigmann, H.; Kułak, K.; Glawischnig, E.; Molina, A.; Takano, Y.; Bednarek, P. The Role of CYP71A12 Monooxygenase in Pathogen-Triggered Tryptophan Metabolism and Arabidopsis Immunity. New Phytol. 2020, 225, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Piślewska-Bednarek, M.; Sánchez-Vallet, A.; Molina, A.; Glawischnig, E.; Gigolashvili, T.; Bednarek, P. Regulation of Pathogen-Triggered Tryptophan Metabolism in Arabidopsis Thaliana by MYB Transcription Factors and Indole Glucosinolate Conversion Products. Mol. Plant 2016, 9, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Dodani, S.C.; Cahn, J.K.B.; Heinisch, T.; Brinkmann-Chen, S.; McIntosh, J.A.; Arnold, F.H. Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE. Chembiochem 2014, 15, 2259–2267. [Google Scholar] [CrossRef] [PubMed]
- Louka, S.; Barry, S.M.; Heyes, D.J.; Mubarak, M.Q.E.; Ali, H.S.; Alkhalaf, L.M.; Munro, A.W.; Scrutton, N.S.; Challis, G.L.; de Visser, S.P. Catalytic Mechanism of Aromatic Nitration by Cytochrome P450 TxtE: Involvement of a Ferric-Peroxynitrite Intermediate. J. Am. Chem. Soc. 2020, 142, 15764–15779. [Google Scholar] [CrossRef]
- Saroay, R.; Roiban, G.-D.; Alkhalaf, L.M.; Challis, G.L. Expanding the Substrate Scope of Nitrating Cytochrome P450 TxtE by Active Site Engineering of a Reductase Fusion. Chembiochem 2021, 22, 2262–2265. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haduch, A.; Bromek, E.; Kuban, W.; Daniel, W.A. The Engagement of Cytochrome P450 Enzymes in Tryptophan Metabolism. Metabolites 2023, 13, 629. https://doi.org/10.3390/metabo13050629
Haduch A, Bromek E, Kuban W, Daniel WA. The Engagement of Cytochrome P450 Enzymes in Tryptophan Metabolism. Metabolites. 2023; 13(5):629. https://doi.org/10.3390/metabo13050629
Chicago/Turabian StyleHaduch, Anna, Ewa Bromek, Wojciech Kuban, and Władysława Anna Daniel. 2023. "The Engagement of Cytochrome P450 Enzymes in Tryptophan Metabolism" Metabolites 13, no. 5: 629. https://doi.org/10.3390/metabo13050629
APA StyleHaduch, A., Bromek, E., Kuban, W., & Daniel, W. A. (2023). The Engagement of Cytochrome P450 Enzymes in Tryptophan Metabolism. Metabolites, 13(5), 629. https://doi.org/10.3390/metabo13050629