H3 Lysine 4 Methylation Is Required for Full Activation of Genes Involved in α-Ketoglutarate Availability in the Nucleus of Yeast Cells after Diauxic Shift
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Growth Condition
2.2. Chromatin Immuno-Precipitation on DNA Arrays (ChIP on Chip)
2.3. Western Blot Analysis
2.4. Chromatin Immunoprecipitation (ChIP)
2.5. Real Time RT-PCR
2.6. Statistical Analysis
3. Results
3.1. Expression of Jhd2 and Set1 in Different Phases of Yeast Growht
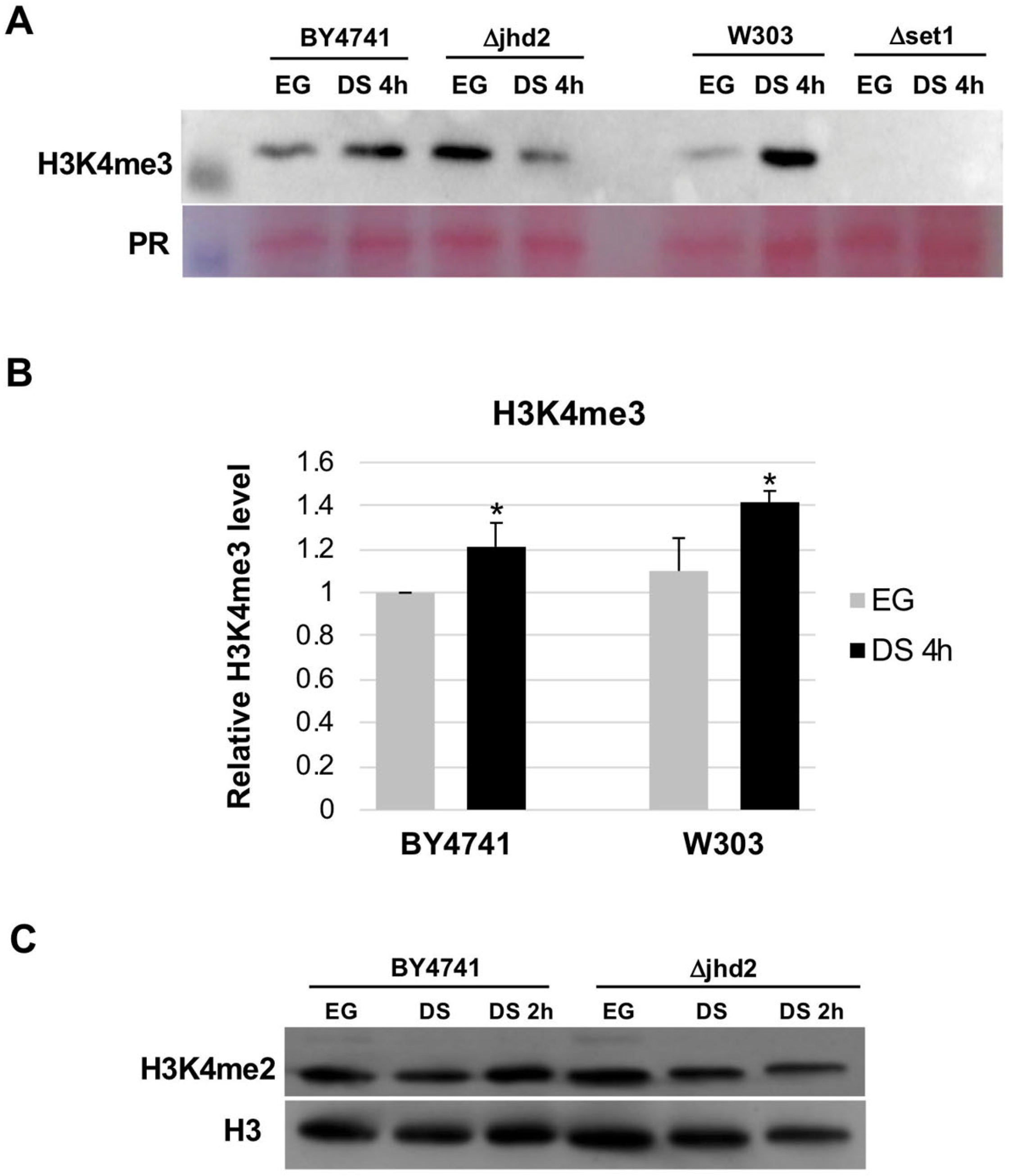
3.2. H3K4 Tri-Methylation Level during DS
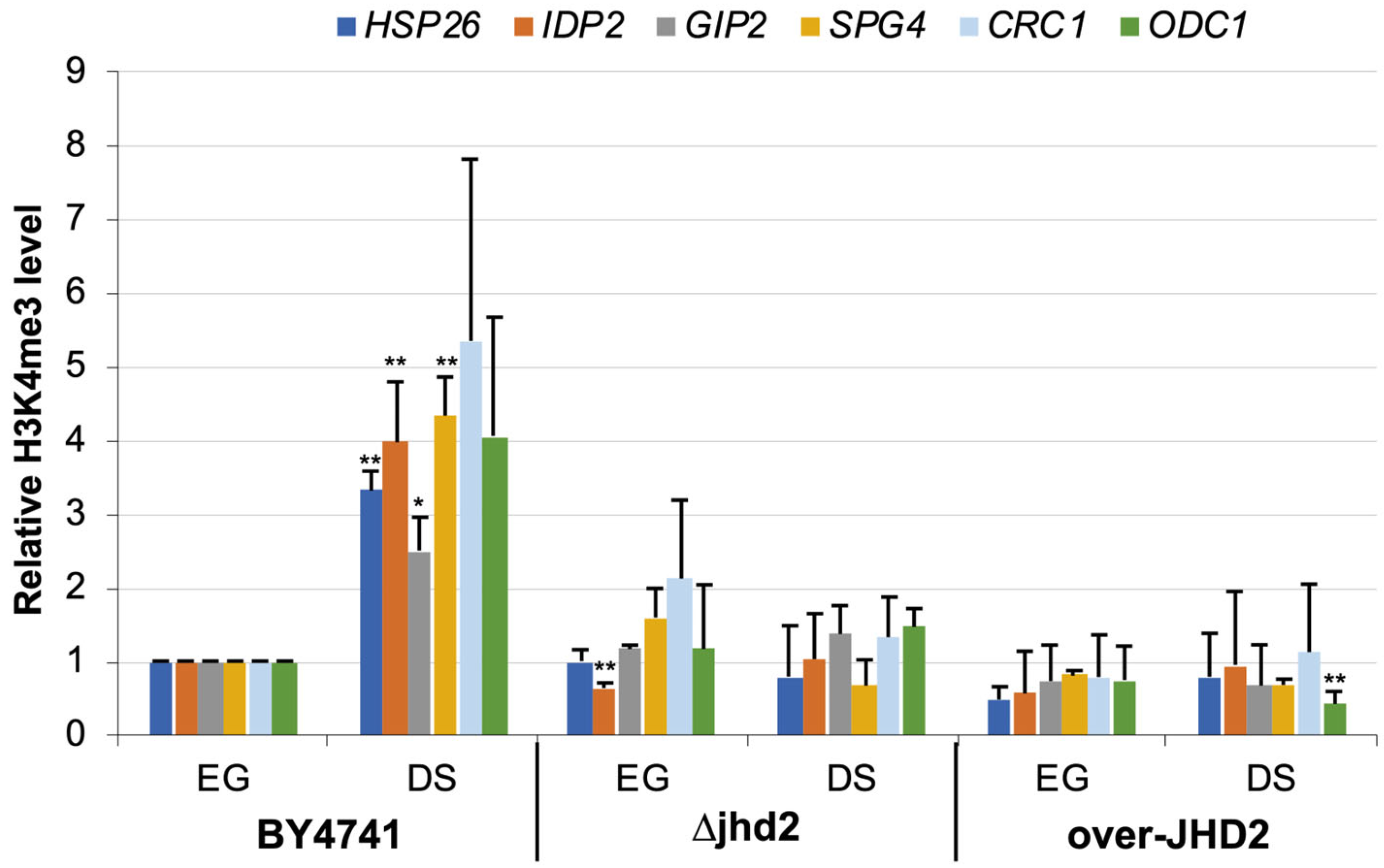
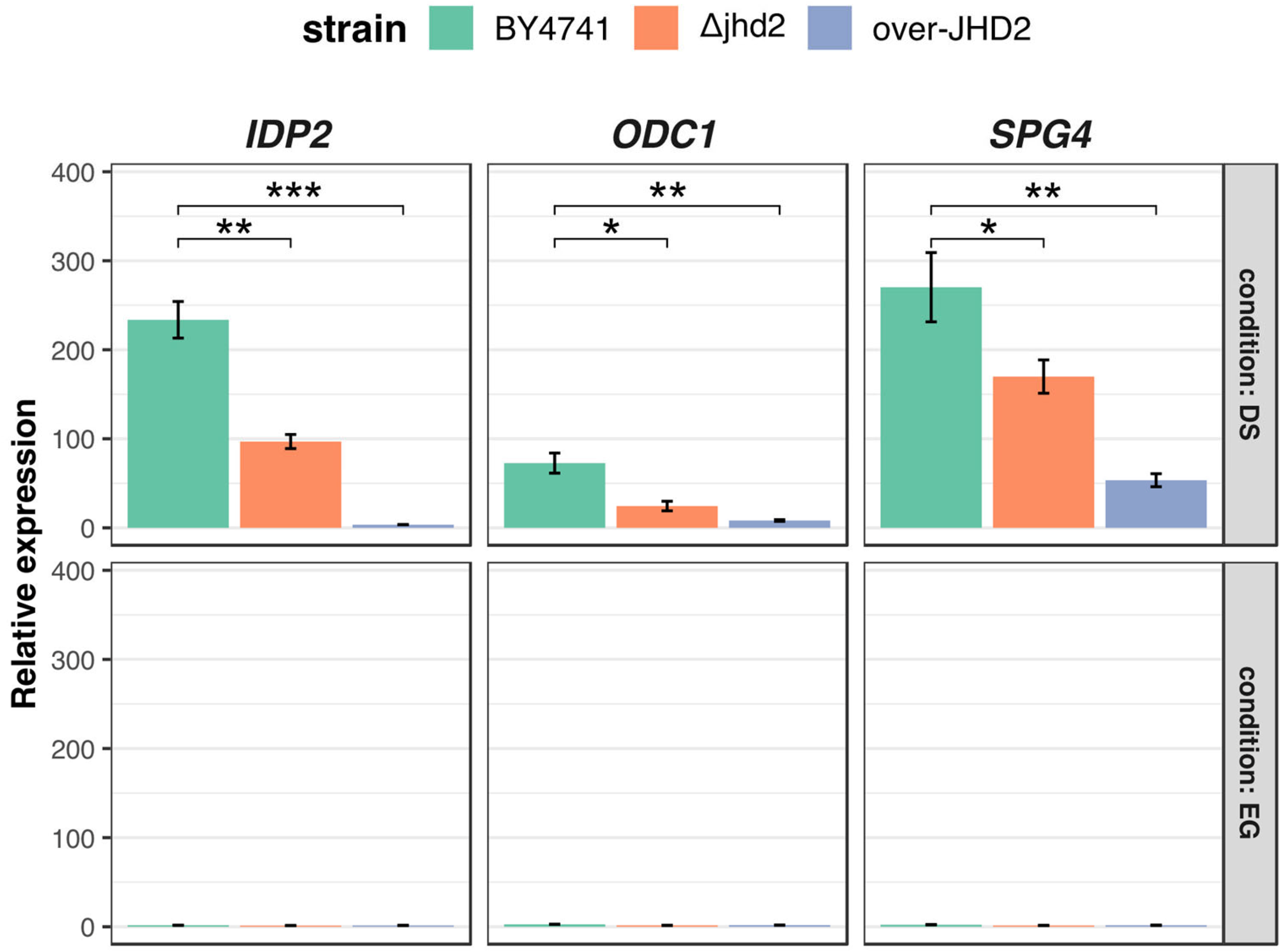
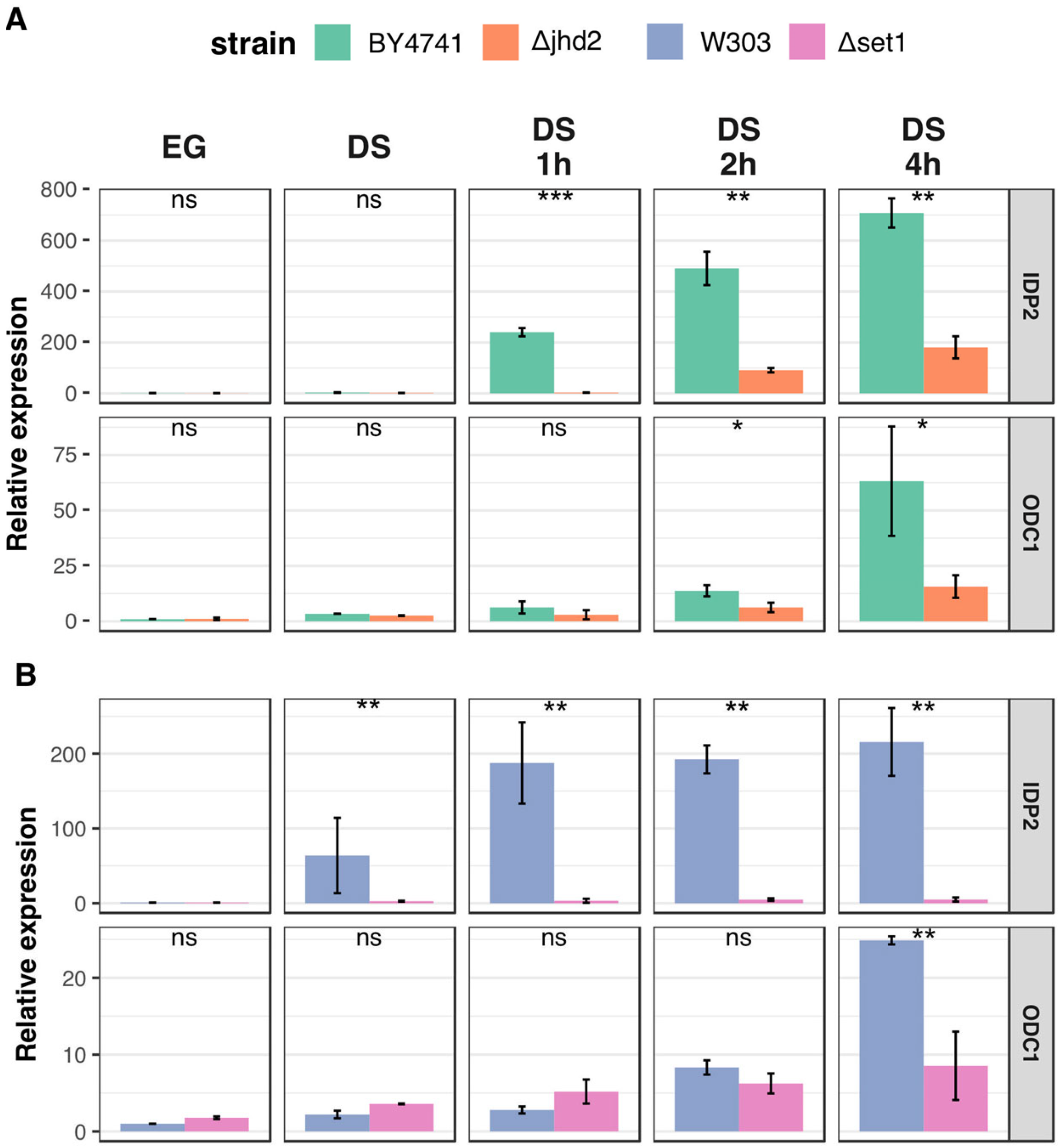
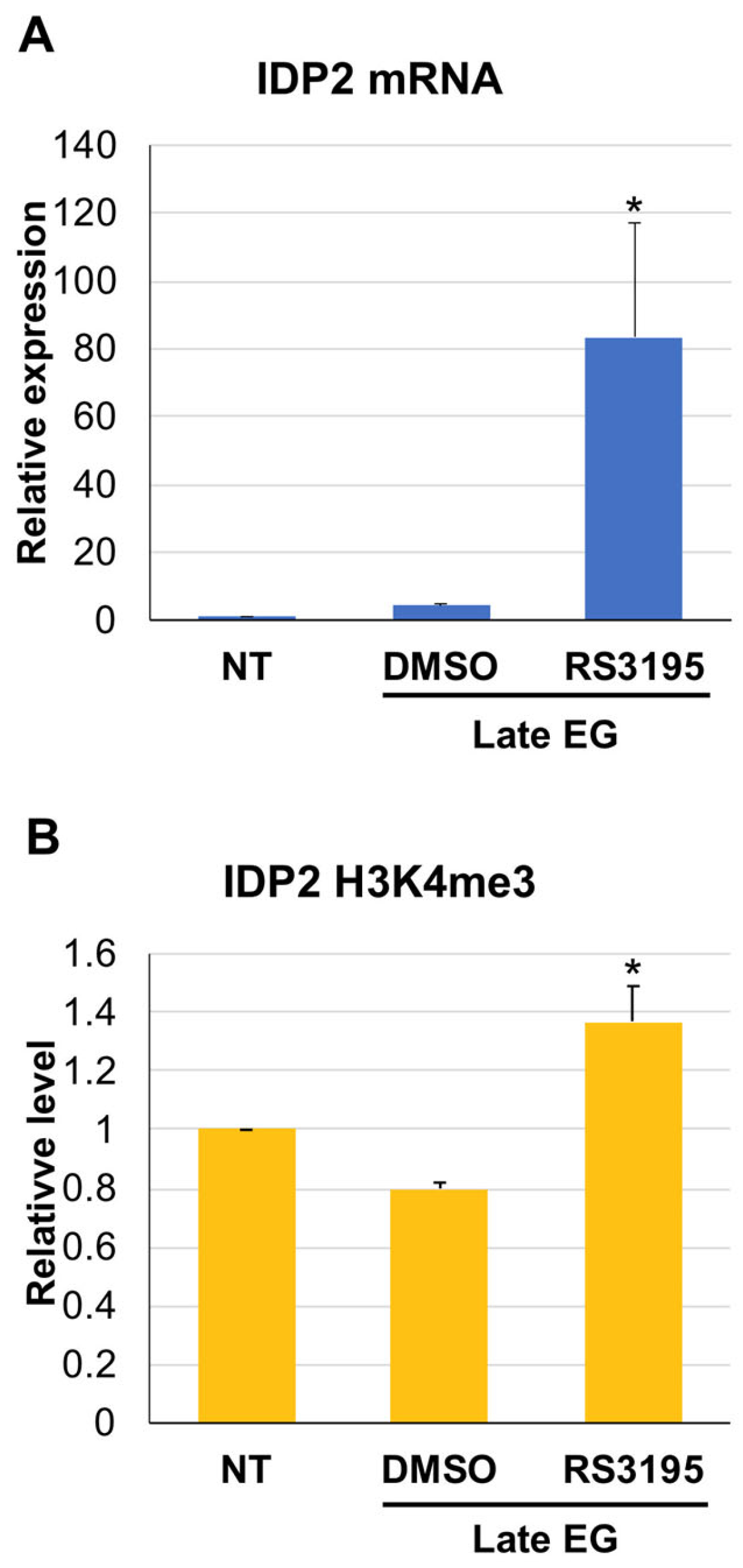
3.3. H3K4 Methylation Stimulates Gene Transcription Induction at DS
3.4. Active Demethylation Is Not Required for DS Genes Repression following Refreshing
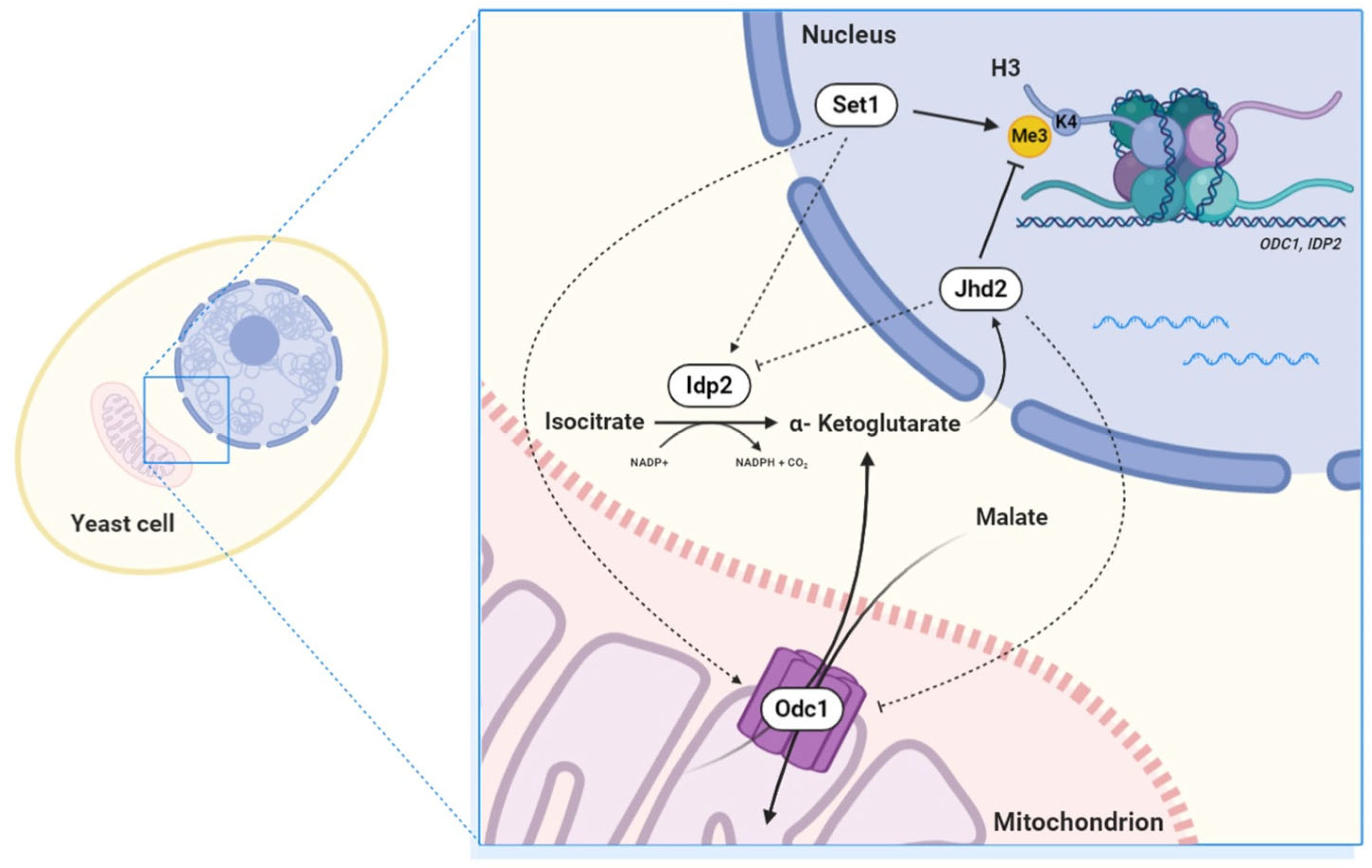
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rando, O.J.; Winston, F. Chromatin and Transcription in Yeast. Genetics 2012, 190, 351–387. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Pokholok, D.K.; Harbison, C.T.; Levine, S.; Cole, M.; Hannett, N.M.; Lee, T.I.; Bell, G.W.; Walker, K.; Rolfe, P.A.; Herbolsheimer, E.; et al. Genome-Wide Map of Nucleosome Acetylation and Methylation in Yeast. Cell 2005, 122, 517–527. [Google Scholar] [CrossRef]
- Pinskaya, M.; Morillon, A. Histone H3 Lysine 4 Di-Methylation: A Novel Mark for Transcriptional Fidelity? Epigenetics 2009, 4, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Dehe, P.M.; Geli, V. The Multiple Faces of Set1. Biochem. Cell Biol. 2006, 84, 536–548. [Google Scholar]
- Margaritis, T.; Oreal, V.; Brabers, N.; Maestroni, L.; Vitaliano-Prunier, A.; Benschop, J.J.; van Hooff, S.; van Leenen, D.; Dargemont, C.; Géli, V.; et al. Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription. PLoS Genet. 2012, 8, e1002952. [Google Scholar] [CrossRef]
- Zhou, B.O.; Zhou, J.Q. Recent Transcription-Induced Histone H3 Lysine 4 (H3K4) Methylation Inhibits Gene Reactivation. J. Biol. Chem. 2011, 286, 34770–34776. [Google Scholar] [CrossRef]
- Lauberth, S.M.; Nakayama, T.; Wu, X.; Ferris, A.L.; Tang, Z.; Hughes, S.H.; Roeder, R.G. H3K4me3 Interactions with TAF3 Regulate Preinitiation Complex Assembly and Selective Gene Activation. Cell 2013, 152, 1021–1036. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Pokhrel, S.; Palani, S.; Pflueger, C.; Parnell, T.J.; Cairns, B.R.; Bhaskara, S.; Chandrasekharan, M.B. Counteracting H3K4 Methylation Modulators Set1 and Jhd2 Co-Regulate Chromatin Dynamics and Gene Transcription. Nat. Commun. 2016, 7, 11949. [Google Scholar] [CrossRef]
- Cloos, P.A.C.; Christensen, J.; Agger, K.; Helin, K. Erasing the Methyl Mark: Histone Demethylases at the Center of Cellular Differentiation and Disease. Genes Dev. 2008, 22, 1115–1140. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Klose, R.J.; Gardner, K.E.; Zhang, Y. Yeast Jhd2p Is a Histone H3 Lys4 Trimethyl Demethylase. Nat. Struct. Mol. Biol. 2007, 14, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.; Bulloch, E.M.; Yang, L.; Ren, C.; Huang, W. Identification of Yeast Demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 14262–14271. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Soloveychik, M.; Ranger, M.; Schertzberg, M.; Shah, Z.; Raisner, R.; Venkatasubrahmanyan, S.; Tsui, K.; Gebbia, M.; Hughes, T.; et al. Timing of Transcriptional Quiescence during Gametogenesis Is Controlled by Global Histone H3K4 Demethylation. Dev. Cell 2012, 23, 1059–1071. [Google Scholar] [CrossRef]
- Law, M.J.; Ciccaglione, K. Fine-Tuning of Histone H3 Lys4 Methylation during Pseudohyphal Differentiation by the CDK Submodule of RNA Polymerase II. Genetics 2015, 199, 435–453. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.Y.; Ahn, S.H. Yeast Histone H3 Lysine 4 Demethylase Jhd2 Regulates Mitotic RDNA Condensation. BMC Biol. 2014, 12, 75. [Google Scholar] [CrossRef] [PubMed]
- Radman-Livaja, M.; Liu, C.L.; Friedman, N.; Schreiber, S.L.; Rando, O.J. Replication and Active Demethylation Represent Partially Overlapping Mechanisms for Erasure of H3K4me3 in Budding Yeast. PLoS Genet. 2010, 6, e1000837. [Google Scholar] [CrossRef]
- Galdieri, L.; Mehrotra, S.; Yu, S.; Vancura, A. Transcriptional Regulation in Yeast during Diauxic Shift and Stationary Phase. OMICS 2010, 14, 629–638. [Google Scholar] [CrossRef]
- Radonjic, M.; Andrau, J.-C.; Lijnzaad, P.; Kemmeren, P.; Kockelkorn, T.T.J.P.; van Leenen, D.; van Berkum, N.L.; Holstege, F.C.P. Genome-Wide Analyses Reveal RNA Polymerase II Located Upstream of Genes Poised for Rapid Response upon S. cerevisiae Stationary Phase Exit. Mol. Cell 2005, 18, 171–183. [Google Scholar] [CrossRef]
- Palmieri, L.; Agrimi, G.; Runswick, M.J.; Fearnley, I.M.; Palmieri, F.; Walker, J.E. Identification in Saccharomyces cerevisiae of Two Isoforms of a Novel Mitochondrial Transporter for 2-Oxoadipate and 2-Oxoglutarate. J. Biol. Chem. 2001, 276, 1916–1922. [Google Scholar] [CrossRef]
- El Moualij, B.; Duyckaerts, C.; Lamotte-Brasseur, J.; Sluse, F.E. Phylogenetic Classification of the Mitochondrial Carrier Family of Saccharomyces cerevisiae. Yeast 1997, 13, 573–581. [Google Scholar] [CrossRef]
- Harismendy, O.; Gendrel, C.-G.; Soularue, P.; Gidrol, X.; Sentenac, A.; Werner, M.; Lefebvre, O. Genome-Wide Location of Yeast RNA Polymerase III Transcription Machinery. EMBO J. 2003, 22, 4738–4747. [Google Scholar] [CrossRef] [PubMed]
- Mannironi, C.; Proietto, M.; Bufalieri, F.; Cundari, E.; Alagia, A.; Danovska, S.; Rinaldi, T.; Famiglini, V.; Coluccia, A.; La Regina, G.; et al. An High-Throughput in Vivo Screening System to Select H3K4-Specific Histone Demethylase Inhibitors. PLoS ONE 2014, 9, e86002. [Google Scholar] [CrossRef]
- Pippa, S.; Mannironi, C.; Licursi, V.; Bombardi, L.; Colotti, G.; Cundari, E.; Mollica, A.; Coluccia, A.; Naccarato, V.; La Regina, G.; et al. Small Molecule Inhibitors of KDM5 Histone Demethylases Increase the Radiosensitivity of Breast Cancer Cells Overexpressing JARID1B. Molecules 2019, 24, 1739. [Google Scholar] [CrossRef] [PubMed]
- Su, X.B.; Pillus, L. Functions for Diverse Metabolic Activities in Heterochromatin. Proc. Natl. Acad. Sci. USA 2016, 113, E1526–E1535. [Google Scholar] [CrossRef]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.S.; et al. IDH1 Mutation Is Sufficient to Establish the Glioma Hypermethylator Phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH Mutation Impairs Histone Demethylation and Results in a Block to Cell Differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef]
- Gupta, R.; Webb-Myers, R.; Flanagan, S.; Buckland, M.E. Isocitrate Dehydrogenase Mutations in Diffuse Gliomas: Clinical and Aetiological Implications. J. Clin. Pathol. 2011, 64, 835–844. [Google Scholar] [CrossRef]
- Chang, S.; Yim, S.; Park, H. The Cancer Driver Genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: Crosstalk between Histone Demethylation and Hypoxic Reprogramming in Cancer Metabolism. Exp. Mol. Med. 2019, 51, 1–17. [Google Scholar] [CrossRef]
- Carey, B.W.; Finley, L.W.S.; Cross, J.R.; Allis, C.D.; Thompson, C.B. Intracellular α-Ketoglutarate Maintains the Pluripotency of Embryonic Stem Cells. Nature 2015, 518, 413–416. [Google Scholar] [CrossRef]
- Soloveychik, M.; Xu, M.; Zaslaver, O.; Lee, K.; Narula, A.; Jiang, R.; Rosebrock, A.P.; Caudy, A.A.; Meneghini, M.D. Mitochondrial Control through Nutritionally Regulated Global Histone H3 Lysine-4 Demethylation. Sci. Rep 2016, 6, 37942. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wellen, K.E.; Rabinowitz, J.D. Metabolic Control of Methylation and Acetylation. Curr. Opin. Chem. Biol. 2016, 30, 52–60. [Google Scholar] [CrossRef]
- Tran, T.Q.; Lowman, X.H.; Kong, M. Molecular Pathways: Metabolic Control of Histone Methylation and Gene Expression in Cancer. Clin. Cancer Res. 2017, 23, 4004–4009. [Google Scholar] [CrossRef]
- Filipp, F.V. Crosstalk between Epigenetics and Metabolism-Yin and Yang of Histone Demethylases and Methyltransferases in Cancer. Brief. Funct. Genom. 2017, 16, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, S.; Gong, X.; Yu, Q.; Zhang, Y.; Luo, M.; Zhang, X.; Workman, J.L.; Yu, X.; Li, S. Glycolysis Regulates Gene Expression by Promoting the Crosstalk between H3K4 Trimethylation and H3K14 Acetylation in Saccharomyces cerevisiae. J. Genet. Genom. 2019, 46, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Soares, L.M.; Radman-Livaja, M.; Lin, S.G.; Rando, O.J.; Buratowski, S. Feedback Control of Set1 Protein Levels Is Important for Proper H3K4 Methylation Patterns. Cell Rep. 2014, 6, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, R.; Singh, S.; Arumugam, S.; Roguev, A.; Stewart, A.F. The Set1 Complex Is Dimeric and Acts with Jhd2 Demethylation to Convey Symmetrical H3K4 Trimethylation. Genes Dev. 2019, 33, 550–564. [Google Scholar] [CrossRef]
- Law, M.J.; Finger, M.A. The Saccharomyces cerevisiae Cdk8 Mediator Represses AQY1 Transcription by Inhibiting Set1p-Dependent Histone Methylation. G3 2017, 7, 1001–1010. [Google Scholar] [CrossRef]
| Yeast Strain | Genotype |
|---|---|
| BY4741 | MATα his3∆ leu2∆0 LYS2 met15∆0 ura3∆0 |
| W303 | MATa, his3-11, ade2-1, leu2-3,112, ura3-1, trp1-D2, can1-100 |
| Δjhd2 BY4741 | MATa, his3∆ leu2∆0 LYS2 met15∆0 ura3∆0 YJR119c::kanMX4 |
| Δset1 W303 | MATa, his3∆ leu2∆0 LYS2 met15∆0 ura3∆0 YJHR119w::kanMX4 |
| over-JHD2 | as BY4741 with plasmid pDPM2 |
| Gene | Description | Rank Difference (DS-EG) | p |
|---|---|---|---|
| IDP2 | Isocitrate dehydrogenase (NADP+), cytosolic | 0.78 | <0.001 |
| MLS1 | Malate synthase 1, functions in glyoxylate cycle, has near identity to Dal7p | 0.57 | <0.01 |
| ECM13 | Protein possibly involved in cell wall structure or biosynthesis | 0.55 | <0.01 |
| GND2 | 6-Phosphogluconate dehydrogenase, decarboxylating, converts 6-phosphogluconate + NADP to ribulose-5-phosphate + NADPH + CO2 | 0.49 | <0.001 |
| PXA1 | Protein required for long-chain fatty acid transport across the peroxisomal membrane, member of the ATP-binding cassette (ABC) superfamily, has similarity to a human gene associated with adrenoleukodystrophy | 0.49 | <0.001 |
| YPT53 | GTP-binding protein involved in endocytosis and transport of proteins to the vacuole, member of the rab family in the ras superfamily | 0.48 | <0.01 |
| ODC1 | 2-Oxodicarboxylate transporter, has specificity for 2-oxoadipate and 2-oxoglutarate, member of the mitochondrial carrier (MCF) family of membrane transporters | 0.48 | <0.05 |
| ATH1 | Vacuolar acid trehalase, converts alpha, alpha-trehalose to glucose | 0.47 | <0.001 |
| GPX1 | Glutathione peroxidase, involved in cellular protection against lipid and non-lipid hydroperoxides | 0.45 | <0.05 |
| MTH1 | Repressor of hexose transport genes | 0.45 | <0.05 |
| ADY2 | Protein required for proper ascus formation, has strong similarity to Ydr384p and Ynr002p | 0.44 | <0.05 |
| HSP26 | Heat shock protein of 26 kDa, expressed during entry to stationary phase and induced by osmostress, may be required for resistance to ethanol and acetaldehyde | 0.42 | <0.001 |
| LEE1 | Protein containing two CCCH-type zinc finger domains, which bind DNA or RNA | 0.4 | <0.001 |
| TRX3 | Mitochondrial thioredoxin, has similarity to cytoplasmic thioredoxins Trx1p and Trx2p | 0.38 | <0.05 |
| CRC1 | Mitochondrial carnitine carrier, member of the mitochondrial carrier family (MCF) of membrane transporters | 0.38 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Nisio, E.; Danovska, S.; Condemi, L.; Cirigliano, A.; Rinaldi, T.; Licursi, V.; Negri, R. H3 Lysine 4 Methylation Is Required for Full Activation of Genes Involved in α-Ketoglutarate Availability in the Nucleus of Yeast Cells after Diauxic Shift. Metabolites 2023, 13, 507. https://doi.org/10.3390/metabo13040507
Di Nisio E, Danovska S, Condemi L, Cirigliano A, Rinaldi T, Licursi V, Negri R. H3 Lysine 4 Methylation Is Required for Full Activation of Genes Involved in α-Ketoglutarate Availability in the Nucleus of Yeast Cells after Diauxic Shift. Metabolites. 2023; 13(4):507. https://doi.org/10.3390/metabo13040507
Chicago/Turabian StyleDi Nisio, Elena, Svetlana Danovska, Livia Condemi, Angela Cirigliano, Teresa Rinaldi, Valerio Licursi, and Rodolfo Negri. 2023. "H3 Lysine 4 Methylation Is Required for Full Activation of Genes Involved in α-Ketoglutarate Availability in the Nucleus of Yeast Cells after Diauxic Shift" Metabolites 13, no. 4: 507. https://doi.org/10.3390/metabo13040507
APA StyleDi Nisio, E., Danovska, S., Condemi, L., Cirigliano, A., Rinaldi, T., Licursi, V., & Negri, R. (2023). H3 Lysine 4 Methylation Is Required for Full Activation of Genes Involved in α-Ketoglutarate Availability in the Nucleus of Yeast Cells after Diauxic Shift. Metabolites, 13(4), 507. https://doi.org/10.3390/metabo13040507







