Multi-Enzymatic Synthesis of Lactobionic Acid Using Wood-Degrading Enzymes Produced by White Rot Fungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Microorganisms
2.2. Culture Conditions and Purification of CDH and LAC
2.3. Enzyme Activity Assay and Protein Determination
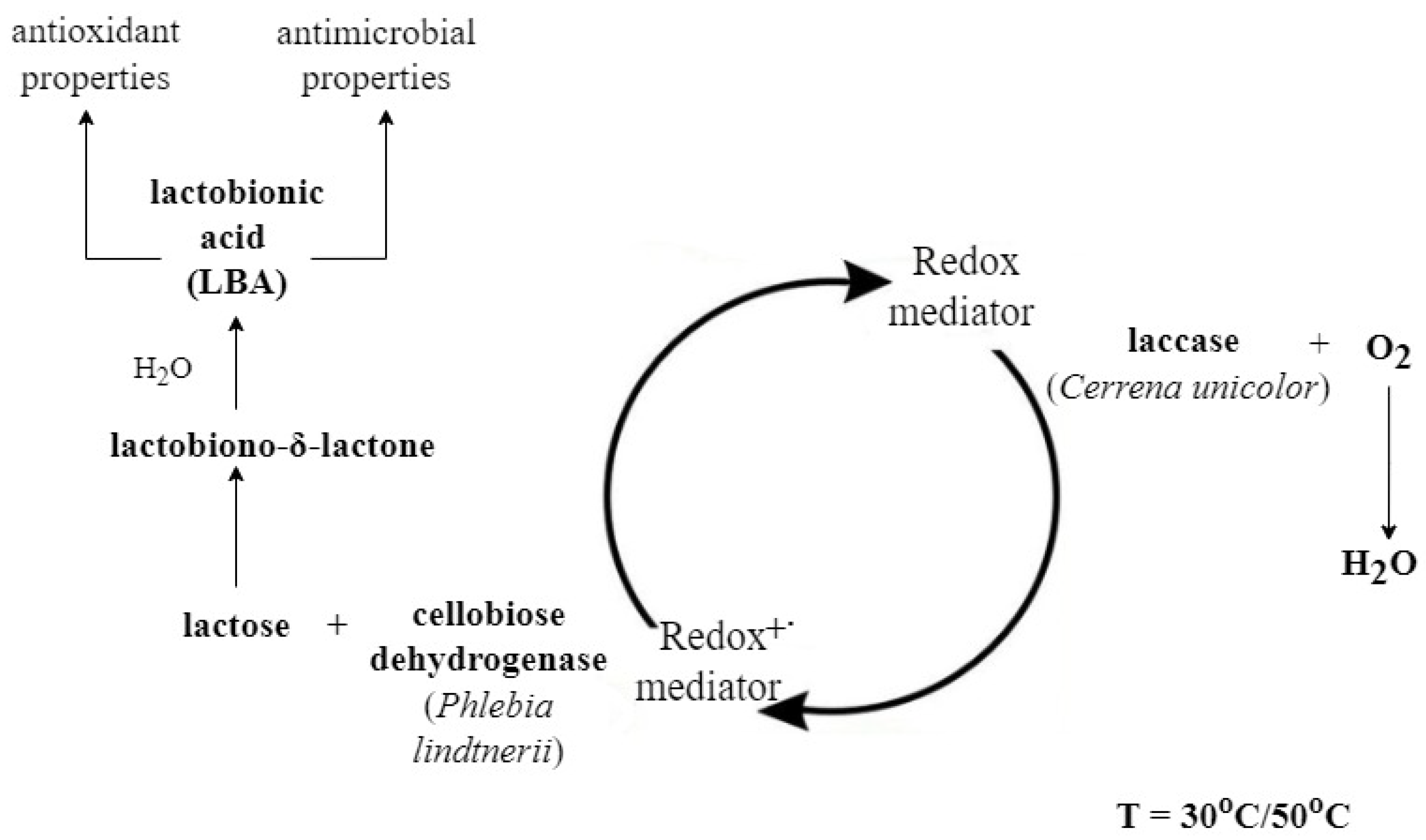
2.4. Enzymatic Oxidation of Lactose and Synthesis of Lactobionic Acid (LBA)
2.4.1. Determination of LBA Using High-Performance Liquid Chromatography
2.4.2. Determination of LBA Using Thin-Layer Chromatography
2.4.3. Determination of LBA using Fourier-Transform Infrared Spectroscopy (FTIR)
2.5. Antibacterial Activity Assay of LBA
2.6. Antioxidant Properties of LBA
2.7. Statistical Analysis
3. Results and Discussion
3.1. Enzymatic Oxidation of Lactose and Synthesis of Lactobionic Acid (LBA)
3.2. Data Analysis for FTIR Spectra
3.3. Antioxidant Properties of LBA
3.4. Antibacterial Properties of LBA
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.; Brahamanage, R.S.; Brooks, S. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Gianinazzi-Pearson, V. Plant cell responses to arbuscular mycorrhizal fungi: Getting to the roots of the symbiosis. Plant Cell 1996, 8, 1871. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Leonowicz, A.; Matuszewska, A.; Luterek, J.; Ziegenhagen, D.; Wojtaś-Wasilewska, M.; Cho, N.-S.; Hofrichter, M.; Rogalski, J. Biodegradation of lignin by white rot fungi. Fungal Genet. Biol. 1999, 27, 175–185. [Google Scholar] [CrossRef]
- Putro, J.N.; Soetaredjo, F.E.; Lin, S.-Y.; Ju, Y.-H.; Ismadji, S. Pretreatment and conversion of lignocellulose biomass into valuable chemicals. RSC Adv. 2016, 6, 46834–46852. [Google Scholar] [CrossRef]
- Goodell, B.; Qian, Y.; Jellison, J. Fungal decay of wood: Soft rot—Brown rot—White rot. ACS Publ. 2008, 982, 9–31. [Google Scholar] [CrossRef]
- Enebak, S.A.; Blanchette, R.A. Canker formation and decay in sugar maple and paper birch infected by Cerrena unicolor. Can. J. For. Res. 1989, 19, 225–231. [Google Scholar] [CrossRef]
- Hibi, M.; Hatahira, S.; Nakatani, M.; Yokozeki, K.; Shimizu, S.; Ogawa, J. Extracellular oxidases of Cerrena sp. complementarily functioning in artificial dye decolorization including laccase, manganese peroxidase, and novel versatile peroxidases. Biocatal. Agric. Biotechnol. 2012, 1, 220–225. [Google Scholar] [CrossRef]
- Ledakowicz, J.; Joanna, L.; Stanislaw, L.; Anna, M. Bio-scouring of linen fabrics with laccase complex from Cerrena unicolor. Fibres Text East Eur. 2007, 15, 86–89. [Google Scholar]
- D’Errico, C.; Jørgensen, J.O.; Krogh, K.B.; Spodsberg, N.; Madsen, R.; Monrad, R.N. Enzymatic degradation of lignin-carbohydrate complexes (LCCs): Model studies using a fungal glucuronoyl esterase from Cerrena unicolor. Biotechnol. Bioeng. 2015, 112, 914–922. [Google Scholar] [CrossRef]
- Rogalski, J.; Grzegorz, J. Purification of extracellular laccase from Cerrena unicolor. Prep. Biochem. Biotechnol. 2010, 40, 242–255. [Google Scholar] [CrossRef]
- Matuszewska, A.; Karp, M.; Jaszek, M.; Janusz, G.; Osińska-Jaroszuk, M.; Sulej, J.; Stefaniuk, D.; Tomczak, W.; Giannopoulos, K. Laccase purified from Cerrena unicolor exerts antitumor activity against leukemic cells. Oncol. Lett. 2016, 11, 2009–2018. [Google Scholar] [CrossRef] [PubMed]
- Zmitrovich, I.V.; Ezhov, O.N. Ecology and plectology of Phlebia tremelloidea (Polyporales, Agaricomycetes). Acta Mycol. 2011, 46, 19–25. [Google Scholar] [CrossRef]
- Janusz, G.; Sulej, J.; Jaszek, M.; Osińska-Jaroszuk, M. Effect of different wavelengths of light on laccase, cellobiose dehydrogenase, and proteases produced by Cerrena unicolor, Pycnoporus sanguineus and Phlebia lindtneri. Acta Biochim. Pol. 2016, 63, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Mori, T.; Kamei, I.; Kondo, R. A novel metabolic pathway for biodegradation of DDT by the white rot fungi, Phlebia lindtneri and Phlebia brevispora. Biodegradation 2011, 22, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Mori, T.; Kondo, R. Biotransformation of the organochlorine pesticide trans-chlordane by wood-rot fungi. New Biotechnol. 2011, 29, 107–115. [Google Scholar] [CrossRef]
- Ciullini, I.; Tilli, S.; Scozzafava, A.; Briganti, F. Fungal laccase, cellobiose dehydrogenase, and chemical mediators: Combined actions for the decolorization of different classes of textile dyes. Bioresour. Technol. 2008, 99, 7003–7010. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, P.; Long, L.; Ding, S. Production of lactobionic acid using an immobilized cellobiose dehydrogenase/laccase system on magnetic chitosan spheres. Process Biochem. 2021, 100, 1–9. [Google Scholar] [CrossRef]
- Sulej, J.; Janusz, G.; Mazur, A.; Żuber, K.; Żebracka, A.; Rogalski, J. Cellobiose dehydrogenase from the ligninolytic basidiomycete Phlebia lindtneri. Process Biochem. 2013, 48, 1715–1723. [Google Scholar] [CrossRef]
- Sulej, J.; Osińska-Jaroszuk, M.; Jaszek, M.; Grąz, M.; Kutkowska, J.; Pawlik, A.; Chudzik, A.; Bancerz, R. Antimicrobial and antioxidative potential of free and immobilised cellobiose dehydrogenase isolated from wood degrading fungi. Fungal Biol. 2019, 123, 875–886. [Google Scholar] [CrossRef]
- Sulej, J.; Jaszek, M.; Osińska-Jaroszuk, M.; Matuszewska, A.; Bancerz, R.; Janczarek, M. Natural microbial polysaccharides as effective factors for modification of the catalytic properties of fungal cellobiose dehydrogenase. Arch. Microbiol. 2021, 203, 4433–4448. [Google Scholar] [CrossRef] [PubMed]
- Nyanhongo, G.S.; Thallinger, B.; Guebitz, G.M. Cellobiose dehydrogenase-based biomedical applications. Process Biochem. 2017, 59, 37–45. [Google Scholar] [CrossRef]
- Henriksson, G.; Johansson, G.; Pettersson, G. A critical review of cellobiose dehydrogenases. J. Biotechnol. 2000, 78, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Kunamneni, A.; Ballesteros, A.; Plou, F.J.; Alcalde, M. Fungal laccase—A versatile enzyme for biotechnological applications. Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 1, 233–245. [Google Scholar]
- Baldrian, P. Fungal laccases–occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef] [PubMed]
- Couto, S.R.; Toca-Herrera, J.L. Lacasses in the textile industry. Biotechnol. Mol. Biol. Rev. 2006, 1, 115–120. [Google Scholar] [CrossRef]
- Virk, A.P.; Sharma, P.; Capalash, N. Use of laccase in pulp and paper industry. Biotechnol. Prog. 2012, 28, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Minussi, R.C.; Pastore, G.M.; Durán, N. Potential applications of laccase in the food industry. Trends Food Sci. Technol. 2002, 13, 205–216. [Google Scholar] [CrossRef]
- Khatami, S.H.; Vakili, O.; Movahedpour, A.; Ghesmati, Z.; Ghasemi, H.; Taheri-Anganeh, M. Laccase: Various types and applications. Biotechnol. Appl. Biochem. 2022, 69, 2658–2672. [Google Scholar] [CrossRef]
- Qiang, T.; Chen, L.; Zhang, Q.; Liu, X. A sustainable and cleaner speedy tanning system based on condensed tannins catalyzed by laccase. J. Clean. Prod. 2018, 197, 1117–1123. [Google Scholar] [CrossRef]
- Cardoso, T.; Marques, C.; Sotiles, A.R.; Dagostin, J.L.A.; Masson, M.L. Characterization of lactobionic acid evidencing its potential for food industry application. J. Food Process Eng. 2019, 42, e132772019. [Google Scholar] [CrossRef]
- Alonso, S.; Rendueles, M.; Díaz, M. Bio-production of lactobionic acid: Current status, applications and future prospects. Biotechnol. Adv. 2013, 31, 1275–1291. [Google Scholar] [CrossRef]
- Minal, N.; Balakrishnan, S.; Chaudhary, N.N.; Jain, A. Lactobionic acid: Significance and application in food and pharmaceutical. Int. J. Fermented Foods 2017, 6, 25–33. [Google Scholar] [CrossRef]
- Han, H.J.; Eom, G.T. Production of lactobionic acid at high salt concentrations by Acinetobacter halotolerans isolated from seaside soil. Bioprocess Biosyst. Eng. 2022, 45, 1683–1691. [Google Scholar] [CrossRef]
- Lee, S.S.; Oh, Y.-R.; Jeong, B.-Y.; Eom, G.T. Isolation of new lactobionic acid-producing microorganisms and improvement of their production ability by heterologous expression of glucose dehydrogenase from Pseudomonas taetrolens. Enzym. Microb. Technol. 2022, 153, 109954. [Google Scholar] [CrossRef] [PubMed]
- Han, H.J.; Oh, Y.-R.; Eom, G.T. Isolation and characterization of a new superior lactobionic acid-producing bacterium, Enterobacter cloacae KRICT-1, from environmental soil samples. ACS Food Sci. Technol. 2021, 2, 66–74. [Google Scholar] [CrossRef]
- Baminger, U.; Nidetzky, B.; Kulbe, K.D.; Haltrich, D. A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J. Microbiol. Methods 1999, 35, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Leonowicz, A.; Grzywnowicz, K. Quantitative estimation of laccase forms in some white-rot fungi using syringaldazine as a substrate. Enzym. Microb. Technol. 1981, 3, 55–58. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ludwig, R.; Ozga, M.; Zámocky, M.; Peterbauer, C.; Kulbe, K.D.; Haltrich, D. Continuous enzymatic regeneration of electron acceptors used by flavoenzymes: Cellobiose dehydrogenase-catalyzed production of lactobionic acid as an example. Biocatal. Biotransform. 2004, 22, 97–104. [Google Scholar] [CrossRef]
- Baminger, U.; Subramaniam, S.S.; Renganathan, V.; Haltrich, D. Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Appl. Environ. Microbiol. 2001, 67, 1766–1774. [Google Scholar] [CrossRef]
- Kiryu, T.; Kiso, T.; Nakano, H.; Ooe, K.; Kimura, T.; Murakami, H. Involvement of Acetobacter orientalis in the production of lactobionic acid in Caucasian yogurt (“Caspian Sea yogurt”) in Japan. J. Dairy Sci. 2009, 92, 25–34. [Google Scholar] [CrossRef]
- Veiga, A.; Maria da Graça, T.T.; Rossa, L.S.; Mengarda, M.; Stofella, N.C.; Oliveira, L.J.; Gonçalves, A.G.; Murakami, F.S. Colorimetric microdilution assay: Validation of a standard method for determination of MIC, IC50%, and IC90% of antimicrobial compounds. J. Microbiol. Methods 2019, 162, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Yuan, Y.; Zhang, T.; Song, W.; Sheng, Q.; Yue, T. Inhibitory effects of lactobionic acid on Vibrio parahaemolyticus planktonic cells and biofilms. Food Microbiol. 2022, 103, 103963. [Google Scholar] [CrossRef]
- Paduch, R.; Matysik, G.; Wójciak-Kosior, M.; Kandefer-Szerszen, M.; Skalska-Kaminska, A.; Nowak-Kryska, M.; Niedziela, P. Lamium Album Extracts Express Free Radical Scavenging and Cytotoxic Activities. Pol. J. Environ. Stud. 2008, 17, 569–580. [Google Scholar]
- Van Hecke, W.; Haltrich, D.; Frahm, B.; Brod, H.; Dewulf, J.; Van Langenhove, H.; Ludwig, R. A biocatalytic cascade reaction sensitive to the gas–liquid interface: Modeling and upscaling in a dynamic membrane aeration reactor. J. Mol. Catal. B Enzym. 2011, 68, 154–161. [Google Scholar] [CrossRef]
- Goderska, K. Biosynthesis of lactobionic acid in whey-containing medium by microencapsulated and free bacteria of Pseudomonas taetrolens. Indian J. Microbiol. 2021, 61, 315–323. [Google Scholar] [CrossRef]
- Sarenkova, I.; Orviz, S.S.; Ciprovica, I.; Rendueles, M.; Diaz, M. Lactobionic acid production from acid whey under different fermentative conditions. J. Adv. Agric. Technol. 2021, 8, 35–40. [Google Scholar] [CrossRef]
- Zavyalova, O.; Gajewska, S.; Dąbrowska-Wisłocka, D.; Sionkowska, A. Characteristics of physicochemical and rheological properties of chitosan hydrogels based on selected hydroxy acids. Eng. Biomater. 2021, 161, 2–7. [Google Scholar] [CrossRef]
- Bisinella, R.Z.B.; Ribeiro, J.C.B.; de Oliveira, C.S.; Colman, T.A.D.; Schnitzler, E.; Masson, M.L. Some instrumental methods applied in food chemistry to characterise lactulose and lactobionic acid. Food Chem. 2017, 220, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Rusciano, D.; Pezzino, S.; Olivieri, M.; Cristaldi, M.; Gagliano, C.; Lupo, G.; Anfuso, C.D. Age-related dry eye lactoferrin and lactobionic acid. Ophthalmic Res. 2018, 60, 94–99. [Google Scholar] [CrossRef]
- Tasic-Kostov, M.; Pavlovic, D.; Lukic, M.; Jaksic, I.; Arsic, I.; Savic, S. Lactobionic acid as antioxidant and moisturizing active in alkyl polyglucoside-based topical emulsions: The colloidal structure, stability and efficacy evaluation. Int. J. Cosmet. Sci. 2012, 34, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Wojeicchowski, J.P.; Cardoso, T.; Mafra, M.R.; Mitterer-Daltoé, M.L.; Masson, M.L. Lactobionic acid as a suitable food preservative for yacon juice. Innov. Food Sci. Emerg. Technol. 2020, 64, 102400. [Google Scholar] [CrossRef]
- Audina, M. A review on anti-aging properties of polyhydroxy acid. World J. Pharm. Res. 2021, 10, 137–141. [Google Scholar] [CrossRef]
- Cao, J.; Fu, H.; Gao, L.; Zheng, Y. Antibacterial activity and mechanism of lactobionic acid against Staphylococcus aureus. Folia Microbiol. 2019, 64, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zheng, Y. iTRAQ-based quantitative proteomic analysis of the antimicrobial mechanism of lactobionic acid against Staphylococcus aureus. Food Funct. 2021, 12, 1349–1360. [Google Scholar] [CrossRef]
- Hou, W.; Kang, S.; Chang, J.; Tian, X.; Shi, C. Correlation Analysis between GlpQ-Regulated Degradation of Wall Teichoic Acid and Biofilm Formation Triggered by Lactobionic Acid in Staphylococcus aureus. Foods 2022, 11, 3438. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kong, F.; Liang, X.; Li, M.; Yang, N.; Cao, X.; Yang, M.; Tao, D.; Yue, X.; Zheng, Y. Label-free quantitative proteomics reveals the multitargeted antibacterial mechanisms of lactobionic acid against methicillin-resistant Staphylococcus aureus (MRSA) using SWATH-MS technology. J. Agric. Food Chem. 2019, 67, 12322–12332. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhong, Q. Lactobionic acid enhances the synergistic effect of nisin and thymol against Listeria monocytogenes Scott A in tryptic soy broth and milk. Int. J. Food Microbiol. 2017, 260, 36–41. [Google Scholar] [CrossRef]
- Coroli, A.; Romano, R.; Saccani, A.; Raddadi, N.; Mele, E.; Mascia, L. An in-vitro evaluation of the characteristics of zein-based films for the release of lactobionic acid and the effects of oleic acid. Polymers 2021, 13, 1826. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kong, F.; Shi, X.; Han, H.; Li, M.; Guan, B.; Yang, M.; Cao, X.; Tao, D.; Zheng, Y. Antibacterial activity and mechanism of lactobionic acid against Pseudomonas fluorescens and Methicillin-resistant Staphylococcus aureus and its application on whole milk. Food Control. 2020, 108, 106876. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, X.; Chen, J.; Liu, H.; Liu, Y. Antibacterial mechanism of lactobionic acid against Shewanella baltica and Shewanella putrefaciens and its application on refrigerated shrimp. Food Biosci. 2023, 51, 102291. [Google Scholar] [CrossRef]
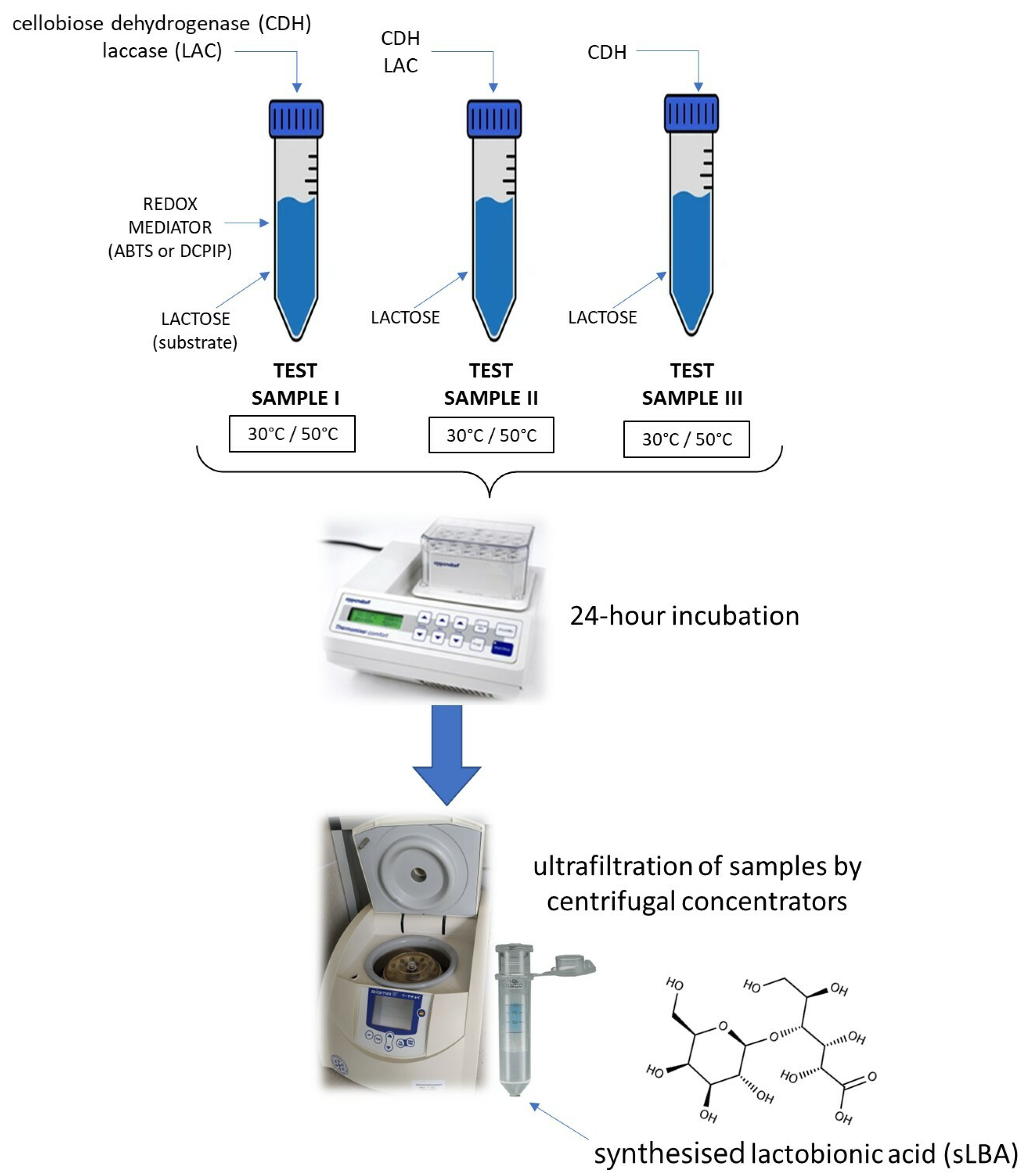
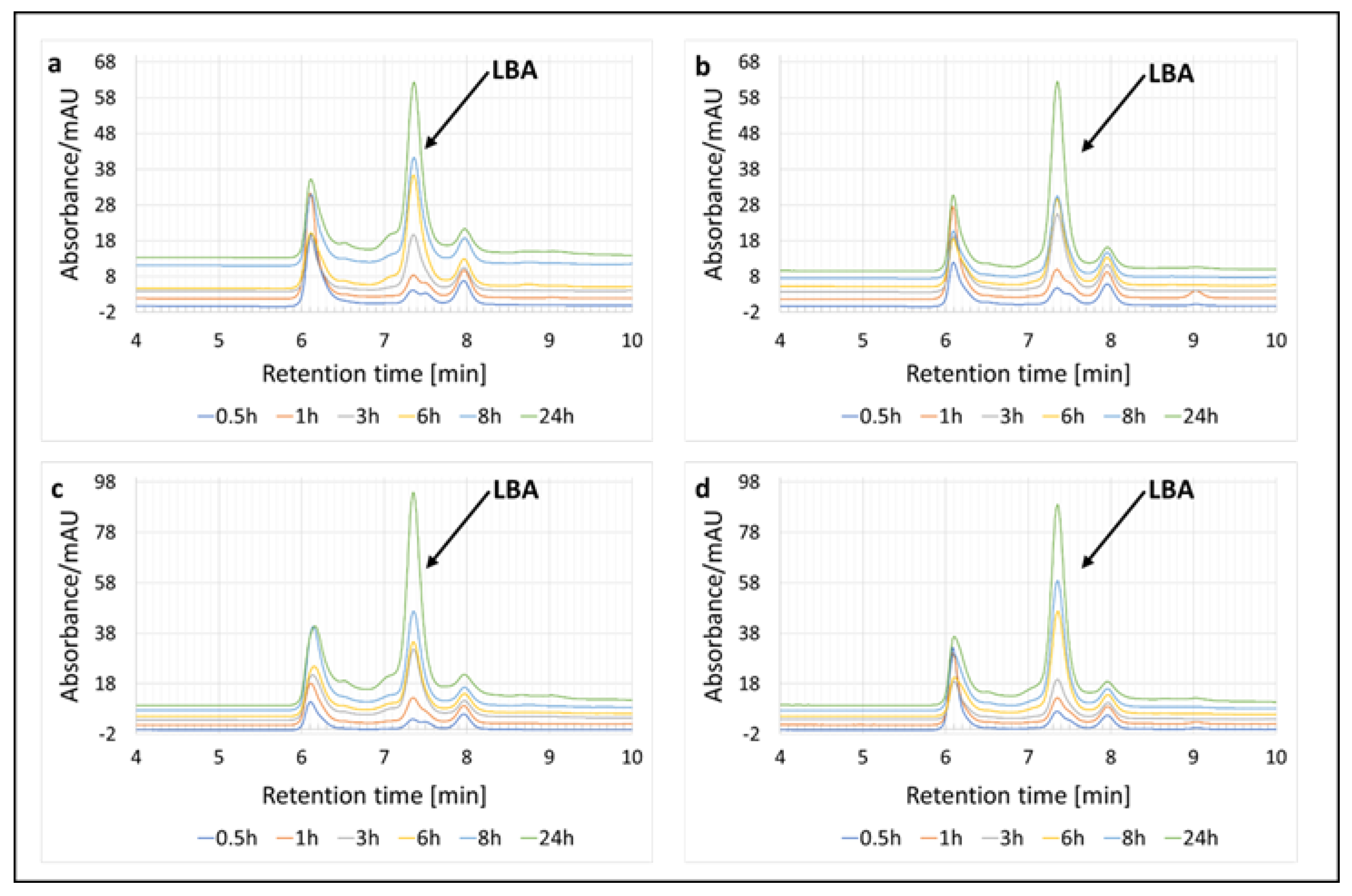
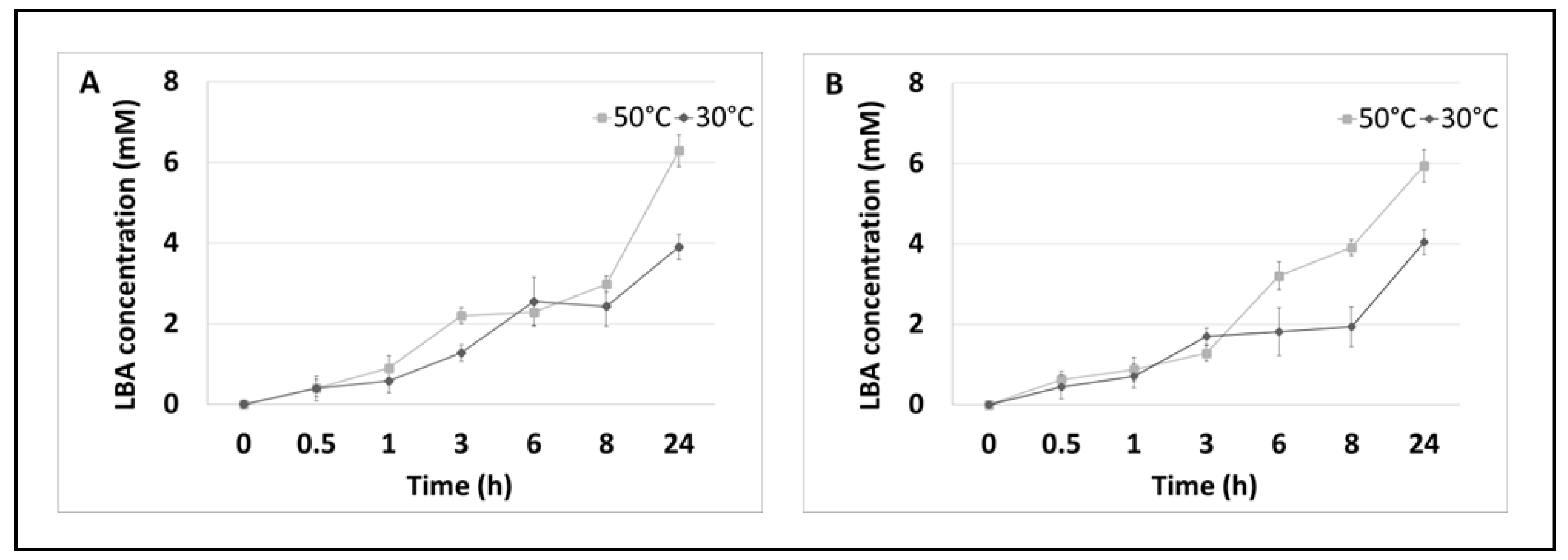

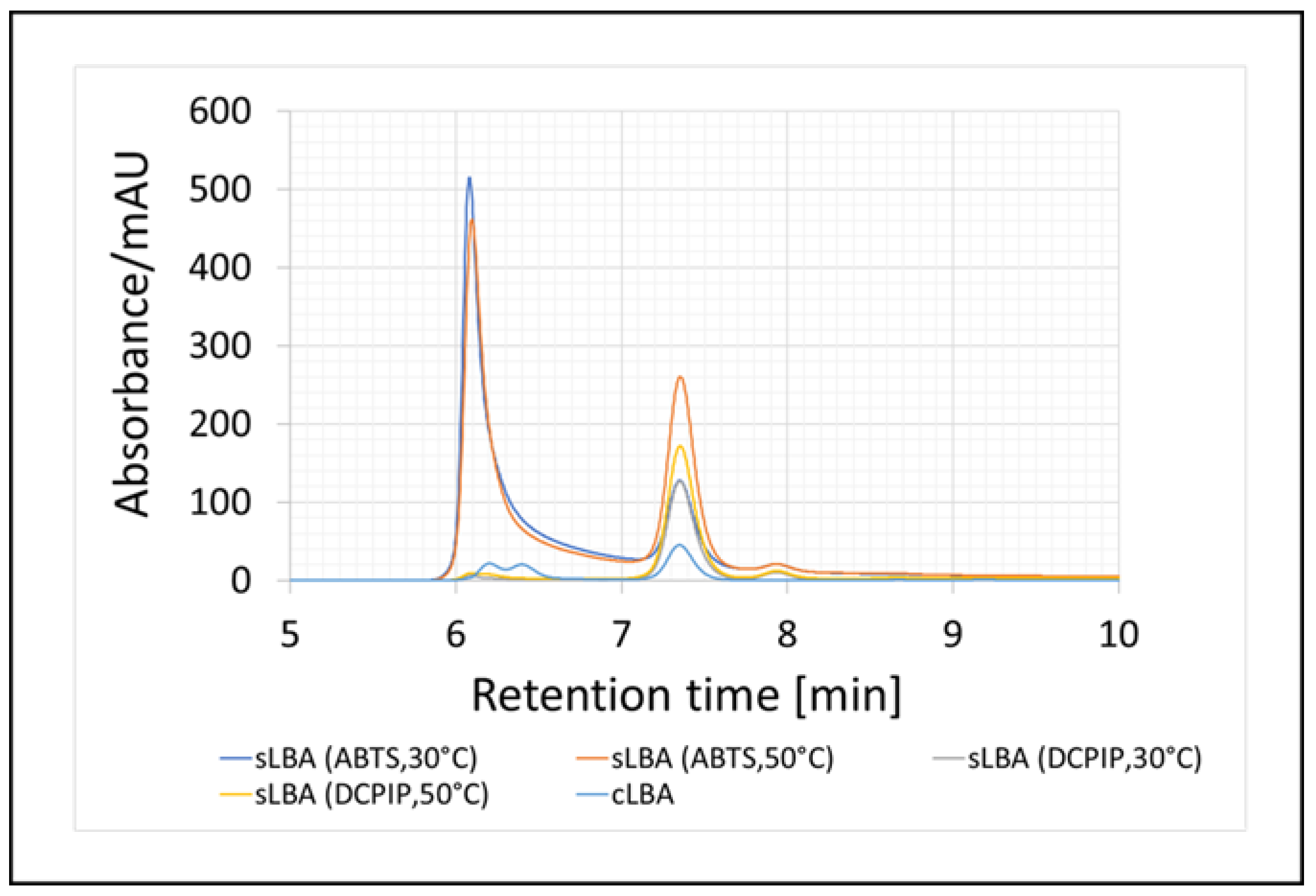
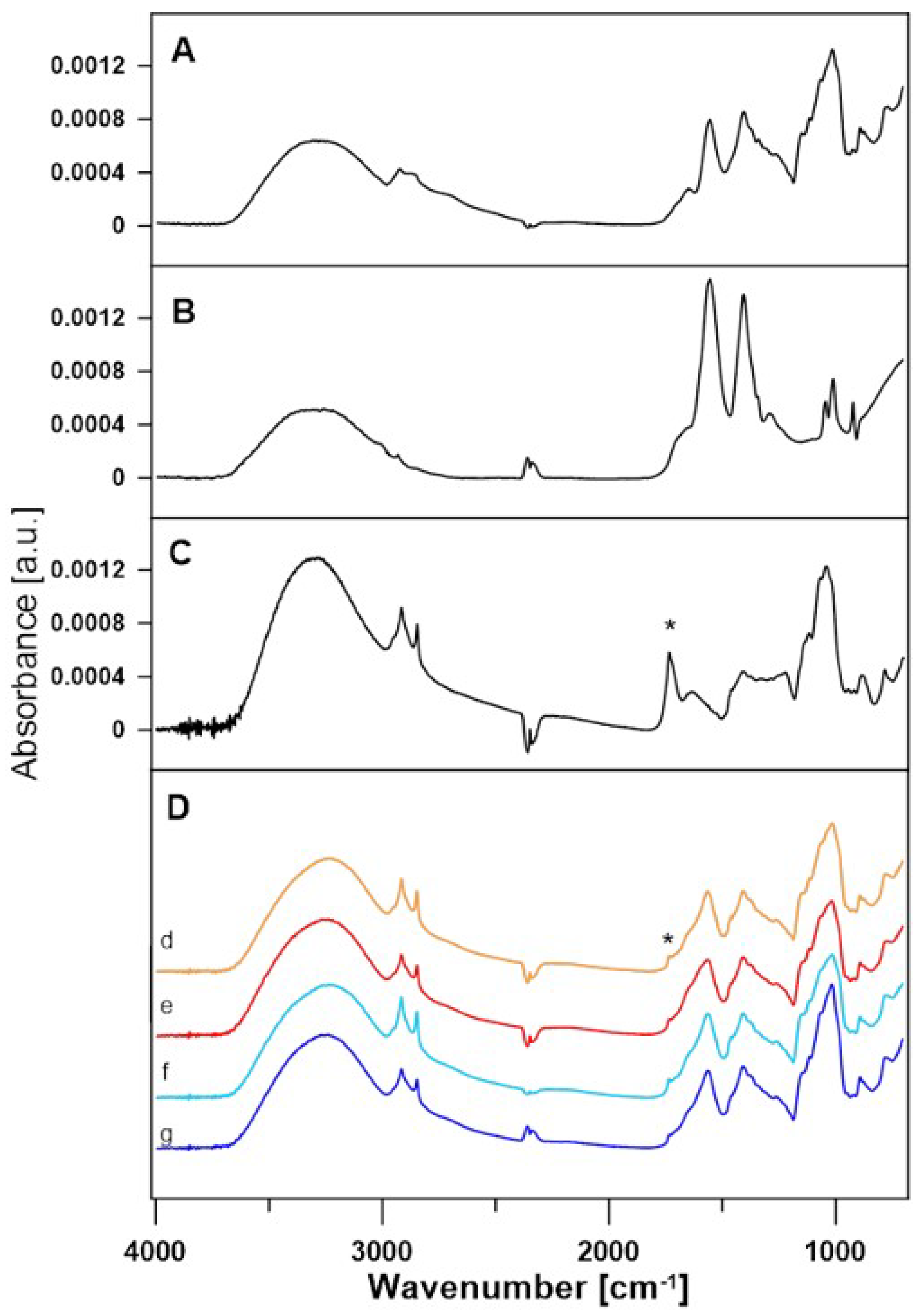
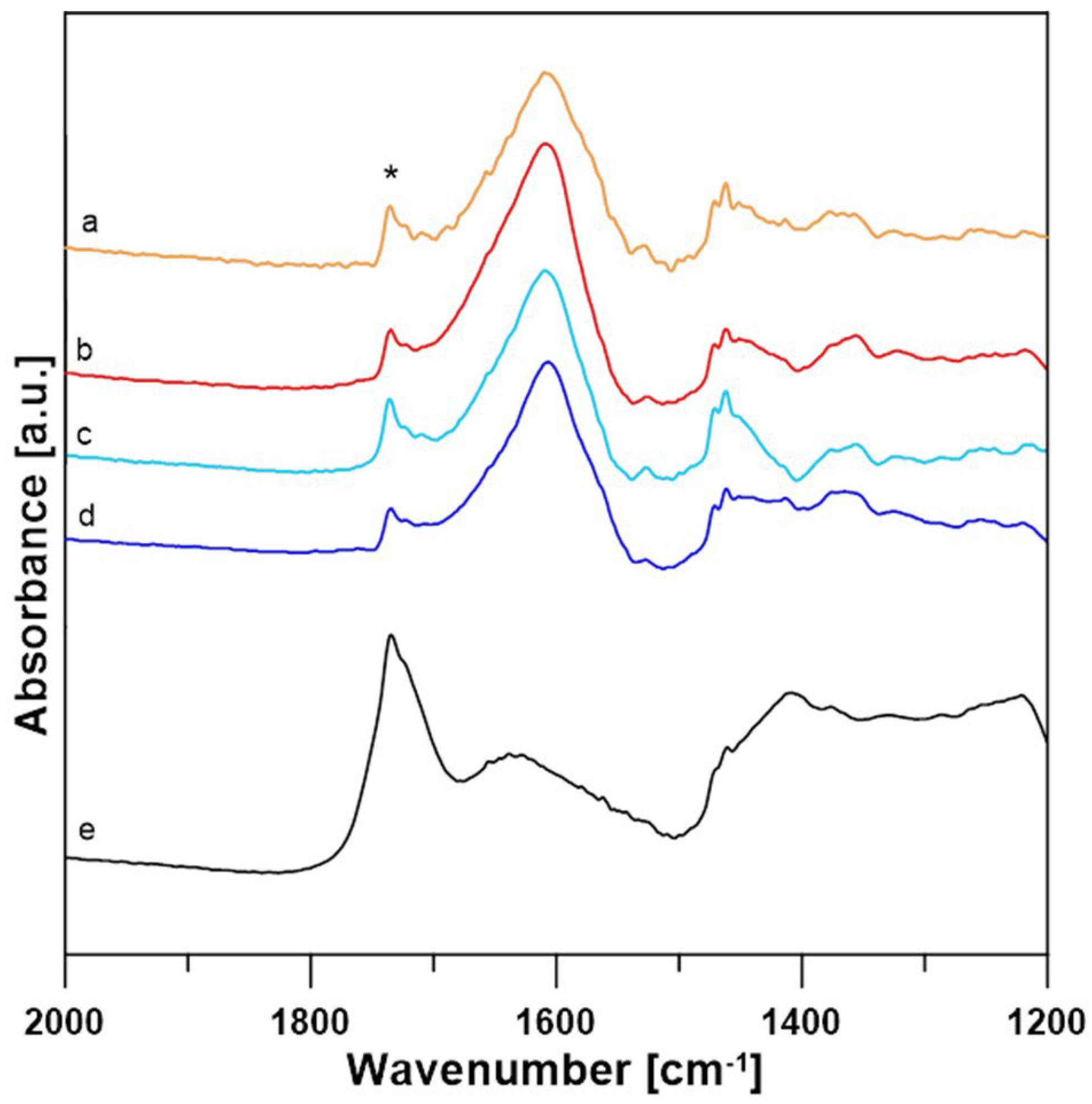
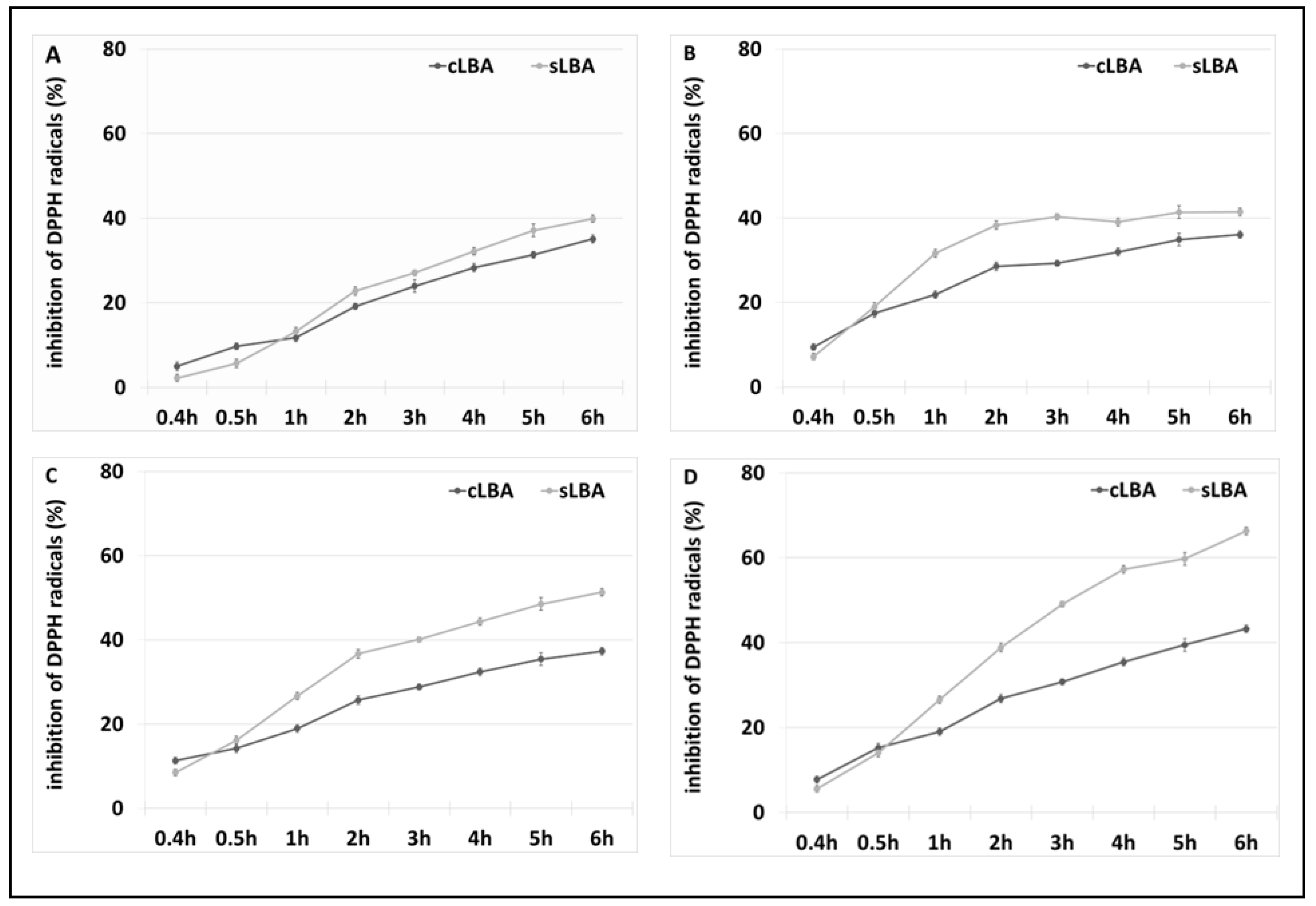
| Enzymatic System | Redox Mediator | Temperature | Lactobionic Acid Concentration [mM] | Conversion Efficiencies [%] |
|---|---|---|---|---|
| PlCDH CuLAC Lactose | ABTS | 30 °C | 12.00 | 24.00 |
| 50 °C | 21.28 | 42.56 | ||
| PlCDH CuLAC Lactose | DCPIP | 30 °C | 9.69 | 19.38 |
| 50 °C | 12.97 | 25.94 |
| Enzymatic System/ Bioactive Substances | % Bacterial Growth Inhibition | |||
|---|---|---|---|---|
| S. aureus ATCC 25923 MIC (cLBA) = 1 mg/mL | S. epidermidis ATCC 14990 MIC (cLBA) = 2 mg/mL | E. coli ATCC 25922 MIC (cLBA) = 4 mg/mL | P. aeruginosa ATCC 27853 MIC (cLBA) = 9 mg/mL | |
| PlCDH, CuLAC, Lactose ABTS, T = 30 °C | 45.3 ± 0.8 | 1.3 ± 0.4 | 89.2 ± 0.4 | 100.0 ± 0.2 |
| PlCDH, CuLAC, Lactose ABTS, T = 50 °C | 13.8 ± 0.5 | 0.0 ± 0.0 | 72.9 ± 0.8 | 81.1 ± 0.7 |
| PlCDH, CuLAC, Lactose DCPIP, T = 30 °C | 41.5 ± 0.3 | 67.4 ± 0.5 | 92.6 ± 0.7 | 100.0 ± 0.3 |
| PlCDH, CuLAC, Lactose DCPIP, T = 50 °C | 42.4 ± 0.5 | 44.6 ± 0.4 | 92.0 ± 0.8 | 100.0 ± 0.3 |
| Enzymatic System/ Bioactive Substances | % of Metabolically Active Cells | |||
|---|---|---|---|---|
| S. aureus ATCC 25923 | S. epidermidis ATCC 14990 | E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | |
| PlCDH, CuLAC, Lactose ABTS, T = 30 °C | 27.1 ± 0.6 | 53.9 ± 0.5 | 12.6 ± 0.3 | 14.2 ± 0.5 |
| PlCDH, CuLAC, Lactose ABTS, T = 50 °C | 45.4 ± 0.3 | 52.5 ± 0.3 | 25.8 ± 0.5 | 24.7 ± 0.6 |
| PlCDH, CuLAC, Lactose DCPIP, T = 30 °C | 25.6 ± 0.7 | 41.4 ± 0.6 | 12.5 ± 0.4 | 12.9 ± 0.8 |
| PlCDH, CuLAC, Lactose DCPIP, T = 50 °C | 26.0 ± 0.8 | 38.0 ± 0.4 | 12.6 ± 0.3 | 12.6 ± 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piątek-Gołda, W.; Sulej, J.; Grąz, M.; Waśko, P.; Janik-Zabrotowicz, E.; Osińska-Jaroszuk, M. Multi-Enzymatic Synthesis of Lactobionic Acid Using Wood-Degrading Enzymes Produced by White Rot Fungi. Metabolites 2023, 13, 469. https://doi.org/10.3390/metabo13040469
Piątek-Gołda W, Sulej J, Grąz M, Waśko P, Janik-Zabrotowicz E, Osińska-Jaroszuk M. Multi-Enzymatic Synthesis of Lactobionic Acid Using Wood-Degrading Enzymes Produced by White Rot Fungi. Metabolites. 2023; 13(4):469. https://doi.org/10.3390/metabo13040469
Chicago/Turabian StylePiątek-Gołda, Wiktoria, Justyna Sulej, Marcin Grąz, Piotr Waśko, Ewa Janik-Zabrotowicz, and Monika Osińska-Jaroszuk. 2023. "Multi-Enzymatic Synthesis of Lactobionic Acid Using Wood-Degrading Enzymes Produced by White Rot Fungi" Metabolites 13, no. 4: 469. https://doi.org/10.3390/metabo13040469
APA StylePiątek-Gołda, W., Sulej, J., Grąz, M., Waśko, P., Janik-Zabrotowicz, E., & Osińska-Jaroszuk, M. (2023). Multi-Enzymatic Synthesis of Lactobionic Acid Using Wood-Degrading Enzymes Produced by White Rot Fungi. Metabolites, 13(4), 469. https://doi.org/10.3390/metabo13040469






