Genetic Markers of Insulin Resistance and Atherosclerosis in Type 2 Diabetes Mellitus Patients with Coronary Artery Disease
Abstract
1. Background
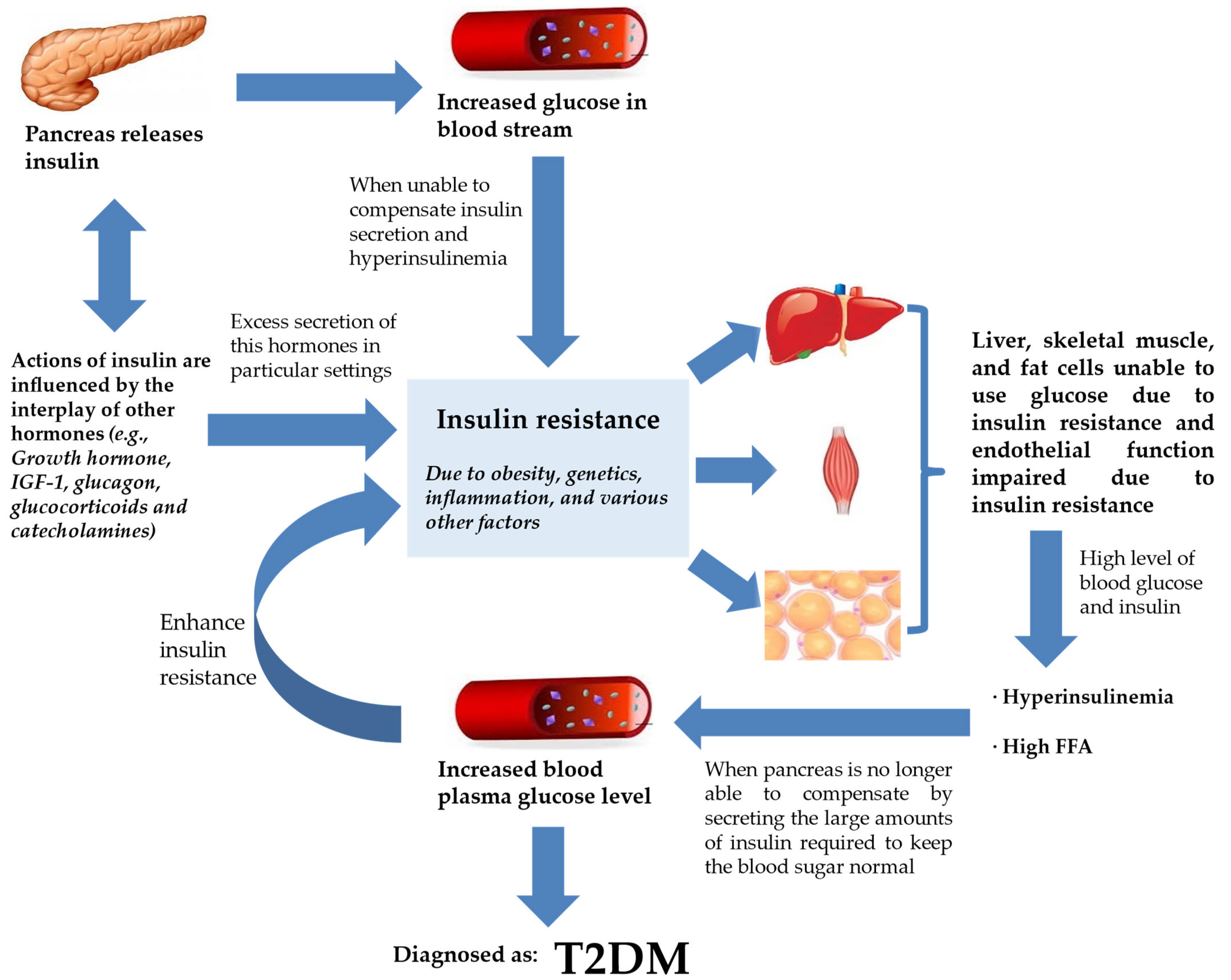
2. How Does IR Result in T2DM?
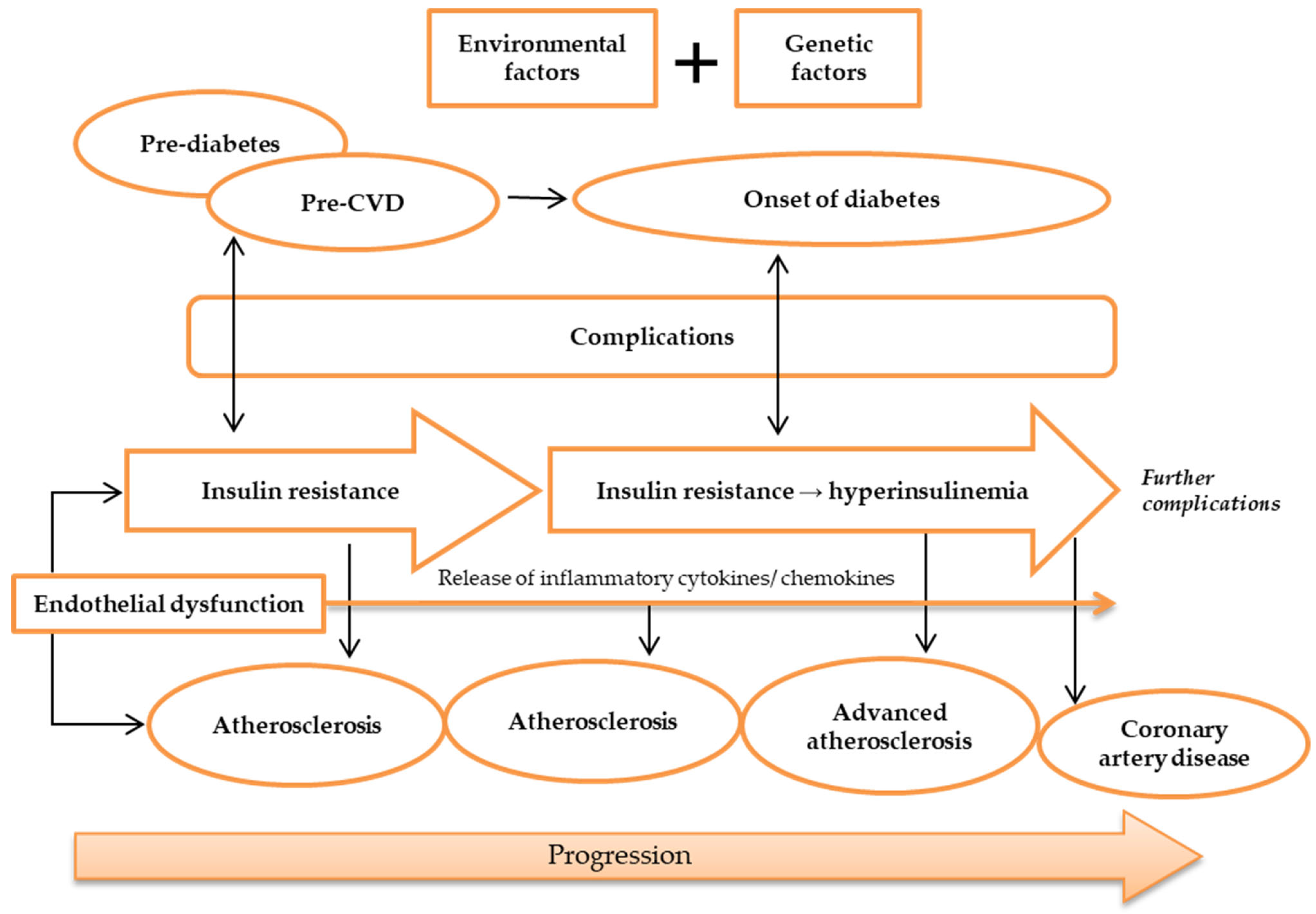
3. Atherosclerosis and Its Relationship with IR, T2DM, and CAD
4. How Do IR and Atherosclerosis Link Genetically?
5. Genes and SNPs That May Be Associated with IR and Atherosclerosis in T2DM Patients with CAD
5.1. CHI3L1 Gene
5.2. CD36 Genes
5.3. LEPR Gene
5.4. RETN Genes
5.5. IL-18 Genes
5.6. RBP-4 Genes
5.7. RARRES2 Gene
6. Executive Summary
6.1. Genetic Markers and Disease
6.2. Mechanisms Involved in the Identification of Genetic Markers
6.3. Association of Potential Genetic Markers with IR and Atherosclerosis
7. Conclusions
8. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- IDF. Type 2 diabetes. In IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021; p. 14. [Google Scholar]
- Grant, P.J.; Cosentino, F.; Marx, N. Diabetes and coronary artery disease: Not just a risk factor. Heart 2020, 106, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Boersma, G.J.; Johansson, E.; Pereira, M.J.; Heurling, K.; Skrtic, S.; Lau, J.; Katsogiannos, P.; Panagiotou, G.; Lubberink, M.; Kullberg, J.; et al. Altered Glucose Uptake in Muscle, Visceral Adipose Tissue, and Brain Predict Whole-Body Insulin Resistance and may Contribute to the Development of Type 2 Diabetes: A Combined PET/MR Study. Horm. Metab. Res. 2018, 50, 627–639. [Google Scholar] [PubMed]
- Honka, M.J.; Latva-Rasku, A.; Bucci, M.; Virtanen, K.A.; Hannukainen, J.C.; Kalliokoski, K.K.; Nuutila, P. Insulin-stimulated glucose uptake in skeletal muscle, adipose tissue and liver: A positron emission tomography study. Eur. J. Endocrinol. 2018, 178, 523–531. [Google Scholar] [CrossRef]
- Takeda, Y.; Matoba, K.; Sekiguchi, K.; Nagai, Y.; Yokota, T.; Utsunomiya, K.; Nishimura, R. Endothelial Dysfunction in Diabetes. Biomedicines 2020, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Malakar, A.K.; Choudhury, D.; Halder, B.; Paul, P.; Uddin, A.; Chakraborty, S. A review on coronary artery disease, its risk factors, and therapeutics. J. Cell. Physiol. 2019, 234, 16812–16823. [Google Scholar] [CrossRef]
- Singh, K.B.; Nnadozie, M.C.; Abdal, M.; Shrestha, N.; Abe, R.A.M.; Masroor, A.; Khorochkov, A.; Prieto, J.; Mohammed, L. Type 2 Diabetes and Causes of Sudden Cardiac Death: A Systematic Review. Cureus 2021, 13, e18145. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ilyas, I.; Little, P.J.; Li, H.; Kamato, D.; Zheng, X.; Luo, S.; Li, Z.; Liu, P.; Han, J.; et al. Endothelial Dysfunction in Atherosclerotic Cardiovascular Diseases and Beyond: From Mechanism to Pharmacotherapies. Pharmacol. Rev. 2021, 73, 924–967. [Google Scholar] [CrossRef]
- Jebari-Benslaiman, S.; Galicia-García, U.; Larrea-Sebal, A.; Olaetxea, J.R.; Alloza, I.; Vandenbroeck, K.; Benito-Vicente, A.; Martín, C. Pathophysiology of Atherosclerosis. Int. J. Mol. Sci. 2022, 23, 3346. [Google Scholar] [CrossRef]
- Poredos, P.; Poredos, A.V.; Gregoric, I. Endothelial Dysfunction and Its Clinical Implications. Angiology 2021, 72, 604–615. [Google Scholar] [CrossRef]
- Syed, R.; Biyabani, M.U.; Prasad, S.; Deeba, F.; Jamil, K. Correlation and Identification of Variable number of Tandem repeats of eNOS Gene in Coronary artery disease (CAD). Saudi J. Biol. Sci. 2010, 17, 209–213. [Google Scholar] [CrossRef]
- Rai, H.; Parveen, F.; Kumar, S.; Kapoor, A.; Sinha, N. Association of Endothelial Nitric Oxide Synthase Gene Polymorphisms with Coronary Artery Disease: An Updated Meta-Analysis and Systematic Review. PLoS ONE 2014, 9, e113363. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Ping, Y.; Wang, Y.; Zhang, Y. The roles of endothelial nitric oxide synthase gene polymorphisms in diabetes mellitus and its associated vascular complications: A systematic review and meta-analysis. Endocrine 2018, 62, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Król, M.; Kepinska, M. Human Nitric Oxide Synthase—Its Functions, Polymorphisms, and Inhibitors in the Context of Inflammation, Diabetes and Cardiovascular Diseases. Int. J. Mol. Sci. 2021, 22, 56. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S. Obesity as a Risk Factor for Diabetes Mellitus in the Local Population of Pakistan. Univers. J. Clin. Med. 2014, 2, 58–64. [Google Scholar] [CrossRef]
- Fu, Z.; Gilbert, E.R.; Liu, D. Regulation of insulin synthesis and secretion and pancreatic Beta-cell dysfunction in diabetes. Curr. Diabetes Rev. 2013, 9, 25–53. [Google Scholar] [CrossRef] [PubMed]
- Rogowicz-Frontczak, A.; Majchrzak, A.; Zozulińska-Ziółkiewicz, D. Insulin resistance in endocrine disorders—Treatment options. Endokrynol. Polska 2017, 68, 334–351. [Google Scholar] [CrossRef] [PubMed]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 2014, 6, a009191. [Google Scholar] [CrossRef]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 2014, 105, 141–150. [Google Scholar] [CrossRef]
- Xin, Y.; Wang, Y.; Chi, J.; Zhu, X.; Zhao, H.; Zhao, S.; Wang, Y. Elevated free fatty acid level is associated with insulin-resistant state in nondiabetic Chinese people. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 139–147. [Google Scholar] [CrossRef]
- Da Silva Rosa, S.C.; Nayak, N.; Caymo, A.M.; Gordon, J.W. Mechanisms of muscle insulin resistance and the cross-talk with liver and adipose tissue. Physiol. Rep. 2020, 8, e14607. [Google Scholar] [CrossRef]
- Roberts, C.K.; Hevener, A.L.; Barnard, R.J. Metabolic syndrome and insulin resistance: Underlying causes and modification by exercise training. Compr. Physiol. 2013, 3, 1–58. [Google Scholar]
- Wondmkun, Y.T. Obesity, Insulin Resistance, and Type 2 Diabetes: Associations and Therapeutic Implications. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 3611–3616. [Google Scholar] [CrossRef] [PubMed]
- Ebbert, J.O.; Jensen, M.D. Fat depots, free fatty acids, and dyslipidemia. Nutrients 2013, 5, 498–508. [Google Scholar] [CrossRef]
- Khound, R.; Taher, J.; Baker, C.; Adeli, K.; Su, Q. GLP-1 Elicits an Intrinsic Gut–Liver Metabolic Signal to Ameliorate Diet-Induced VLDL Overproduction and Insulin Resistance. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 2252–2259. [Google Scholar] [CrossRef]
- Petersen, K.S.; Bowen, K.J.; Tindall, A.M.; Sullivan, V.K.; Johnston, E.A.; Fleming, J.A.; Kris-Etherton, P.M. The Effect of Inflammation and Insulin Resistance on Lipid and Lipoprotein Responsiveness to Dietary Intervention. Curr. Dev. Nutr. 2020, 4, nzaa160. [Google Scholar] [CrossRef]
- Tohidi, M.; Baghbani-Oskouei, A.; Ahanchi, N.S.; Azizi, F.; Hadaegh, F. Fasting plasma glucose is a stronger predictor of diabetes than triglyceride–glucose index, triglycerides/high-density lipoprotein cholesterol, and homeostasis model assessment of insulin resistance: Tehran Lipid and Glucose Study. Acta. Diabetol. 2018, 55, 1067–1074. [Google Scholar] [CrossRef]
- Akash, M.S.; Rehman, K.; Chen, S. Role of inflammatory mechanisms in pathogenesis of type 2 diabetes mellitus. J. Cell. Biochem. 2013, 114, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Chadt, A.; Al-Hasani, H. Glucose transporters in adipose tissue, liver, and skeletal muscle in metabolic health and disease. Pflügers Arch.-Eur. J. Physiol. 2020, 472, 1273–1298. [Google Scholar] [CrossRef]
- Zhou, M.S.; Wang, A.; Yu, H. Link between insulin resistance and hypertension: What is the evidence from evolutionary biology? Diabetol. Metab. Syndr. 2014, 6, 12. [Google Scholar] [CrossRef]
- Roden, M.; Petersen, K.; Shulman, G. Insulin Resistance in Type 2 Diabetes. In Textbook of Diabetes; John Wiley & Sons Ltd.: West Sussex, UK, 2017; pp. 174–186. [Google Scholar]
- Tabák, A.G.; Jokela, M.; Akbaraly, T.N.; Brunner, E.J.; Kivimäki, M.; Witte, D.R. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: An analysis from the Whitehall II study. Lancet 2009, 373, 2215–2221. [Google Scholar] [CrossRef]
- Di Pino, A.; DeFronzo, R.A. Insulin Resistance and Atherosclerosis: Implications for Insulin-Sensitizing Agents. Endocr. Rev. 2019, 40, 1447–1467. [Google Scholar] [CrossRef] [PubMed]
- Aboonabi, A.; Meyer, R.R.; Singh, I. The association between metabolic syndrome components and the development of atherosclerosis. J. Hum. Hypertens. 2019, 33, 844–855. [Google Scholar] [CrossRef]
- Mariana, M.; Lijun, M.; Barry, I.F. Genetic and Environmental Factors Associated With Type 2 Diabetes and Diabetic Vascular Complications. Rev. Diabet. Stud. 2012, 9, 6–22. [Google Scholar]
- Nigro, J.; Osman, N.; Dart, A.M.; Little, P.J. Insulin Resistance and Atherosclerosis. Endocr. Rev. 2006, 27, 242–259. [Google Scholar] [CrossRef]
- Sniderman, A.D.; Thanassoulis, G.; Glavinovic, T.; Navar, A.M.; Pencina, M.; Catapano, A.; Ference, B.A. Apolipoprotein B Particles and Cardiovascular Disease: A Narrative Review. JAMA Cardiol. 2019, 4, 1287–1295. [Google Scholar] [CrossRef]
- Bornfeldt, K.E.; Tabas, I. Insulin Resistance, Hyperglycemia, and Atherosclerosis. Cell Metab. 2011, 14, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Poznyak, A.; Grechko, A.V.; Poggio, P.; Myasoedova, V.A.; Alfieri, V.; Orekhov, A.N. The Diabetes Mellitus–Atherosclerosis Connection: The Role of Lipid and Glucose Metabolism and Chronic Inflammation. Int. J. Mol. Sci. 2020, 21, 1835. [Google Scholar] [CrossRef]
- Raut, P.; Dhawale, S.; Kulkarni, D.; Pekamwar, S.; Shelke, S.; Panzade, P.; Paliwal, A. Pharmacodynamic findings for the usefulness of Luffa cylindrica (L.) leaves in atherosclerosis therapy with supporting antioxidant potential. Future J. Pharm. Sci. 2021, 7, 38. [Google Scholar] [CrossRef]
- Chopra, S.; Peter, S. Screening for coronary artery disease in patients with type 2 diabetes mellitus: An evidence-based review. Indian J. Endocrinol. Metab. 2012, 16, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K. Emerging Risk Biomarkers in Cardiovascular Diseases and Disorders. J. Lipids 2015, 2015, 971453. [Google Scholar] [CrossRef] [PubMed]
- Sakakura, K.; Nakano, M.; Otsuka, F.; Ladich, E.; Kolodgie, F.D.; Virmani, R. Pathophysiology of Atherosclerosis Plaque Progression. Heart Lung Circ. 2013, 22, 399–411. [Google Scholar] [CrossRef]
- Mota, R.; Homeister, J.W.; Willis, M.S.; Bahnson, E.M. Atherosclerosis: Pathogenesis, Genetics and Experimental Models. In eLS.; John Wiley & Sons Ltd.: Chichester, UK, 2017; pp. 1–10. [Google Scholar]
- Poznyak, A.V.; Grechko, A.V.; Wetzker, R.; Orekhov, A.N. In Search for Genes Related to Atherosclerosis and Dyslipidemia Using Animal Models. Int. J. Mol. Sci. 2020, 21, 2097. [Google Scholar] [CrossRef]
- Kappala, S.S. Risk Factors and Blood-Borne Biochemical Markers in Type 2 Diabetes Mellitus. Ph.D. Thesis, University of Central Lancashire, Preston, UK, 2012. [Google Scholar]
- Tran, N.; Garcia, T.; Aniqa, M.; Ali, S.; Ally, A.; Nauli, S.M. Endothelial Nitric Oxide Synthase (eNOS) and the Cardiovascular System: In Physiology and in Disease States. Am. J. Biomed. Sci. Res. 2022, 15, 153–177. [Google Scholar] [PubMed]
- Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T. Advanced glycation end products and oxidative stress in type 2 diabetes mellitus. Biomolecules 2015, 5, 194–222. [Google Scholar] [CrossRef] [PubMed]
- Indranil, B.; Gausal, A.K. Endothelial Dysfunction in Cardiovascular Diseases. In Basic and Clinical Understanding of Microcirculation; Kaneez Fatima, S., Seyed Soheil Saeedi, S., Nazar Luqman, B., Eds.; IntechOpen: Rijeka, Croatia, 2019; Chapter 5. [Google Scholar]
- Jung, U.J.; Choi, M.-S. Obesity and Its Metabolic Complications: The Role of Adipokines and the Relationship between Obesity, Inflammation, Insulin Resistance, Dyslipidemia and Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef]
- Li, H.; Meng, Y.; He, S.; Tan, X.; Zhang, Y.; Zhang, X.; Wang, L.; Zheng, W. Macrophages, Chronic Inflammation, and Insulin Resistance. Cells 2022, 11, 3001. [Google Scholar] [CrossRef]
- Nielsen, K.R.; Steffensen, R.; Boegsted, M.; Baech, J.; Lundbye-Christensen, S.; Hetland, M.L.; Krintel, S.B.; Johnsen, H.E.; Nyegaard, M.; Johansen, J.S. Promoter polymorphisms in the chitinase 3-like 1 gene influence the serum concentration of YKL-40 in Danish patients with rheumatoid arthritis and in healthy subjects. Arthritis. Res. Ther. 2011, 13, R109. [Google Scholar] [CrossRef]
- Bokor, S.; Legry, V.; Meirhaeghe, A.; Ruiz, J.R.; Mauro, B.; Widhalm, K.; Manios, Y.; Amouyel, P.; Moreno, L.A.; Molnàr, D.; et al. Single-nucleotide Polymorphism of CD36 Locus and Obesity in European Adolescents. Obesity 2010, 18, 1398–1403. [Google Scholar] [CrossRef]
- Matteini, A.M.; Li, J.; Lange, E.M.; Tanaka, T.; Lange, L.A.; Tracy, R.P.; Wang, Y.; Biggs, M.L.; Arking, D.E.; Fallin, M.D.; et al. Novel gene variants predict serum levels of the cytokines IL-18 and IL-1ra in older adults. Cytokine 2014, 65, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Müssig, K.; Staiger, H.; Machicao, F.; Thamer, C.; Machann, J.; Schick, F.; Claussen, C.D.; Stefan, N.; Fritsche, A.; Häring, H.-U. RARRES2, encoding the novel adipokine chemerin, is a genetic determinant of disproportionate regional body fat distribution: A comparative magnetic resonance imaging study. Metab. Clin. Exp. 2009, 58, 519–524. [Google Scholar] [CrossRef]
- Tönjes, A.; Scholz, M.; Breitfeld, J.; Marzi, C.; Grallert, H.; Gross, A.; Ladenvall, C.; Schleinitz, D.; Krause, K.; Kirsten, H.; et al. Genome Wide Meta-analysis Highlights the Role of Genetic Variation in RARRES2 in the Regulation of Circulating Serum Chemerin. PLoS Genet. 2014, 10, e1004854. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xing, S.; Zheng, F.; Xing, Q. Increased expression of chitinase 3-like 1 in aorta of patients with atherosclerosis and suppression of atherosclerosis in apolipoprotein E-knockout mice by chitinase 3-like 1 gene silencing. Mediat. Inflamm. 2014, 2014, 905463. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, J.B. Genetic insights into cardiometabolic risk factors. Clin. Biochem. Rev. 2014, 35, 15–36. [Google Scholar] [PubMed]
- Johnston, H.R.; Keats, B.J.B.; Sherman, S.L. 12-Population Genetics. In Emery and Rimoin’s Principles and Practice of Medical Genetics and Genomics, 7th ed.; Pyeritz, R.E., Korf, B.R., Grody, W.W., Eds.; Academic Press: Massachusetts, USA, 2019; pp. 359–373. [Google Scholar]
- Habib, P.T.; Alsamman, A.M.; Shereif, G.A.; Hamwieh, A. SNPector: SNP inspection tool for diagnosing gene pathogenicity and drug response in a naked sequence. F1000Research 2020, 8, 2133. [Google Scholar] [CrossRef]
- Rong, Y.; Dong, S.-S.; Hu, W.-X.; Guo, Y.; Chen, Y.-X.; Chen, J.-B.; Zhu, D.-L.; Chen, H.; Yang, T.-L. DDRS: Detection of drug response SNPs specifically in patients receiving drug treatment. Comput. Struct. Biotechnol. J. 2021, 19, 3650–3657. [Google Scholar] [CrossRef]
- Ridker, P.M.; Chasman, D.I.; Rose, L.; Loscalzo, J.; Elias, J.A. Plasma levels of the proinflammatory chitin-binding glycoprotein YKL-40, variation in the chitinase 3-like 1 gene (CHI3L1), and incident cardiovascular events. J. Am. Heart Assoc. 2014, 3, e000897. [Google Scholar] [CrossRef]
- Thomsen, S.B.; Rathcke, C.N.; Skaaby, T.; Linneberg, A.; Vestergaard, H. The Association between genetic variations of CHI3L1, levels of the encoded glycoprotein YKL-40 and the lipid profile in a Danish population. PLoS ONE 2012, 7, e47094. [Google Scholar] [CrossRef]
- Lee, H.; Kang, R.; Jee, S.H.; Yoon, Y. A promoter polymorphism −2122C>T of CHI3L1 is associated with serum low density lipoprotein cholesterol level in Korean subjects. Clin. Biochem. 2010, 43, 1195–1200. [Google Scholar] [CrossRef]
- Banerjee, M.; Gautam, S.; Saxena, M.; Bid, H.K.; Agrawal, C.G. Association of CD36 gene variants rs1761667 (G>A) and rs1527483 (C>T) with Type 2 diabetes in North Indian population. Int. J. Diabetes Mellit. 2010, 2, 179–183. [Google Scholar] [CrossRef]
- Boghdady, A.; Arafa, U.A.; Sabet, E.A.; Salama, E.; El Sharawy, A.; Elbadry, M.I. Association between rs1761667 polymorphism of CD36 gene and risk of coronary atherosclerosis in Egyptian population. Cardiovasc. Diagn. Ther. 2016, 6, 120–130. [Google Scholar] [CrossRef]
- Saukko, M.; Kesäniemi, Y.A.; Ukkola, O. Leptin receptor Lys109Arg and Gln223Arg polymorphisms are associated with early atherosclerosis. Metab. Syndr. Relat. Disord. 2010, 8, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Osawa, H.; Onuma, H.; Ochi, M.; Murakami, A.; Yamauchi, J.; Takasuka, T.; Tanabe, F.; Shimizu, I.; Kato, K.; Nishida, W.; et al. Resistin SNP-420 determines its monocyte mRNA and serum levels inducing type 2 diabetes. Biochem. Biophys. Res. Commun. 2005, 335, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S. Resistin and Tumor Necrosis Factor—Alpha Gene Polymorphism and the Risk for Cardiovascular Disease in a Pakistani Population. Ph.D. Thesis, Quaid-i-Azam University, Islamabad, Pakistan, 2013. [Google Scholar]
- Suriyaprom, K.; Phonrat, B.; Namjuntra, P.; Chanchay, S.; Tungtrongchitr, R. The +299(G>A) resistin gene polymorphism and susceptibility to type 2 diabetes in Thais. J. Clin. Biochem. Nutr. 2009, 44, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Koch, W.; Wolferstetter, H.; Schatke, A.; Schömig, A.; Kastrati, A. Interleukin 18 gene variation and risk of acute myocardial infarction. Cytokine 2011, 56, 786–791. [Google Scholar] [CrossRef]
- Fischer, C.P.; Perstrup, L.B.; Berntsen, A.; Eskildsen, P.; Pedersen, B.K. Elevated plasma interleukin-18 is a marker of insulin-resistance in type 2 diabetic and non-diabetic humans. J. Clin. Immunol. 2005, 117, 152–160. [Google Scholar] [CrossRef]
- Wan, K.; Zhao, J.; Deng, Y.; Chen, X.; Zhang, Q.; Zeng, Z.; Zhang, L.; Chen, Y. A genetic polymorphism in RBP4 is associated with coronary artery disease. Int. J. Mol. Sci. 2014, 15, 22309–22319. [Google Scholar] [CrossRef]
- Munkhtulga, L.; Nakayama, K.; Utsumi, N.; Yanagisawa, Y.; Gotoh, T.; Omi, T.; Kumada, M.; Erdenebulgan, B.; Zolzaya, K.; Lkhagvasuren, T.; et al. Identification of a regulatory SNP in the retinol binding protein 4 gene associated with type 2 diabetes in Mongolia. Hum. Genet. 2007, 120, 879–888. [Google Scholar] [CrossRef]
- Zhao, T.; Su, Z.; Li, Y.; Zhang, X.; You, Q. Chitinase-3 like-protein-1 function and its role in diseases. Signal Transduct. Target. Ther. 2020, 5, 201. [Google Scholar] [CrossRef]
- Huan, W.; Yandong, L.; Chao, W.; Sili, Z.; Jun, B.; Mingfang, L.; Yu, C.; Lefeng, Q. YKL-40 Aggravates Early-Stage Atherosclerosis by Inhibiting Macrophage Apoptosis in an Aven-dependent Way. Front. Cell Dev. Biol. 2021, 9, 752773. [Google Scholar] [CrossRef]
- Wu, S.; Hsu, L.A.; Cheng, S.T.; Teng, M.S.; Yeh, C.H.; Sun, Y.C.; Huang, H.L.; Ko, Y.L. Circulating YKL-40 level, but not CHI3L1 gene variants, is associated with atherosclerosis-related quantitative traits and the risk of peripheral artery disease. Int. J. Mol. Sci. 2014, 15, 22421–22437. [Google Scholar] [CrossRef]
- Di Rosa, M.; Szychlinska, M.A.; Tibullo, D.; Malaguarnera, L.; Musumeci, G. Expression of CHI3L1 and CHIT1 in osteoarthritic rat cartilage model. A morphological study. Eur. J. Histochem. 2014, 58, 2423. [Google Scholar] [CrossRef]
- Perumalsamy, S.; Huri, H.Z.; Ahmad, W.A.W. Association of YKL-40 Encoding Gene CHI3L1 rs946263 with Insulin Resistance and Severity of Coronary Artery Disease in Type 2 Diabetes Mellitus Patients. Metab. Clin. Exp. 2021, 116, 154520. [Google Scholar] [CrossRef]
- Syed Ikmal, S.I.; Zaman Huri, H.; Vethakkan, S.R.; Wan Ahmad, W.A. Potential biomarkers of insulin resistance and atherosclerosis in type 2 diabetes mellitus patients with coronary artery disease. Int. J. Endocrinol. 2013, 2013, 698567. [Google Scholar] [CrossRef]
- Armesilla, A.L.; Vega, M.A. Structural organization of the gene for human CD36 glycoprotein. J. Biol. Chem. 1994, 269, 18985–18991. [Google Scholar] [CrossRef] [PubMed]
- Rać, M.E.; Safranow, K.; Poncyljusz, W. Molecular basis of human CD36 gene mutations. Mol. Med. 2007, 13, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Collot-Teixeira, S.; Martin, J.; McDermott-Roe, C.; Poston, R.; McGregor, J.L. CD36 and macrophages in atherosclerosis. Cardiovasc. Res. 2007, 75, 468–477. [Google Scholar] [CrossRef]
- Gautam, S.; Agrawal, C.G.; Bid, H.K.; Banerjee, M. Preliminary studies on CD36 gene in type 2 diabetic patients from north India. Indian J. Med. Res. 2011, 134, 107–112. [Google Scholar]
- Tian, K.; Xu, Y.; Sahebkar, A.; Xu, S. CD36 in Atherosclerosis: Pathophysiological Mechanisms and Therapeutic Implications. Curr. Atheroscler. Rep. 2020, 22, 59. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Peng, Y.; Hang, W.; Nie, J.; Zhou, N.; Wang, D.W. The role of CD36 in cardiovascular disease. Cardiovasc. Res. 2020, 118, 115–129. [Google Scholar] [CrossRef]
- Akter, K.; Lanza, E.A.; Martin, S.A.; Myronyuk, N.; Rua, M.; Raffa, R.B. Diabetes mellitus and Alzheimer’s disease: Shared pathology and treatment? Br. J. Clin. Pharmacol. 2011, 71, 365–376. [Google Scholar] [CrossRef]
- Hsieh, F.-L.; Turner, L.; Bolla, J.R.; Robinson, C.V.; Lavstsen, T.; Higgins, M.K. The structural basis for CD36 binding by the malaria parasite. Nat. Commun. 2016, 7, 12837. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, H.; Waggiallah, H.; Alagib, A. Oxidative Low Density Lipoprotien Prohibited Plasmodium Falciparum Clearance in type 2 Diabetes Mellitus Via Cluster Differentiation 36. N. Am. J. Med. Sci. 2013, 5, 703–706. [Google Scholar]
- Love-Gregory, L.; Sherva, R.; Schappe, T.; Qi, J.S.; McCrea, J.; Klein, S.; Connelly, M.A.; Abumrad, N.A. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum. Mol. Genet. 2011, 20, 193–201. [Google Scholar] [CrossRef]
- Yang, M.M.; Wang, J.; Fan, J.J.; Ng, T.K.; Sun, D.J.; Guo, X.; Teng, Y.; Li, Y.-B. Variations in the Obesity Gene “LEPR” Contribute to Risk of Type 2 Diabetes Mellitus: Evidence from a Meta-Analysis. J. Diabetes Res. 2016, 2016, 5412084. [Google Scholar] [CrossRef]
- Fischer, A.W.; Cannon, B.; Nedergaard, J. Leptin: Is It Thermogenic? Endocr. Rev. 2020, 41, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Unger, R.H.; Roth, M.G. A new biology of diabetes revealed by leptin. Cell Metab. 2015, 21, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Coppari, R.; Bjørbæk, C. Leptin revisited: Its mechanism of action and potential for treating diabetes. Nat. Rev. Drug Discov. 2012, 11, 692–708. [Google Scholar] [CrossRef]
- Adiga, U.; Banawalikar, N.; Mayur, S.; Bansal, R.; Ameera, N.; Rao, S. Association of insulin resistance and leptin receptor gene polymorphism in type 2 diabetes mellitus. J. Chin. Med. Assoc. 2021, 84, 383–388. [Google Scholar] [CrossRef]
- Wu, L.; Sun, D. Leptin Receptor Gene Polymorphism and the Risk of Cardiovascular Disease: A Systemic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2017, 14, 375. [Google Scholar] [CrossRef]
- Shin, M.-K.; Eraso, C.C.; Mu, Y.-P.; Gu, C.; Yeung, B.H.Y.; Kim, L.J.; Liu, X.-R.; Wu, Z.-J.; Paudel, O.; Pichard, L.E.; et al. Leptin Induces Hypertension Acting on Transient Receptor Potential Melastatin 7 Channel in the Carotid Body. Circ. Res. 2019, 125, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Äijälä, M. Studies about Contribution of Leptin Receptor in Cardiovascular Risk; University of Oulu: Oulu, Finland, 2013. [Google Scholar]
- Sun, Q.; Cornelis, M.C.; Kraft, P.; Qi, L.; van Dam, R.M.; Girman, C.J.; Laurie, C.C.; Mirel, D.B.; Gong, H.; Sheu, C.C.; et al. Genome-wide association study identifies polymorphisms in LEPR as determinants of plasma soluble leptin receptor levels. Hum. Mol. Genet. 2010, 19, 1846–1855. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.; Cerda, A.; Genvigir, F.D.; Sampaio, M.F.; Armaganijan, D.; Bernik, M.M.; Dorea, E.L.; Hirata, M.H.; Hinuy, H.M.; Hirata, R.D. Leptin receptor gene polymorphisms are associated with adiposity and metabolic alterations in Brazilian individuals. Arq. Bras. Endocrinol. Metabol. 2013, 57, 677–684. [Google Scholar] [CrossRef]
- Cao, X.; Huo, P.; Li, W.; Li, P.; He, L.; Meng, H. Interactions among moderate/severe periodontitis, ADIPOQ-rs1501299, and LEPR-rs1137100 polymorphisms on the risk of type 2 diabetes in a Chinese population. Arch. Oral Biol. 2019, 103, 26–32. [Google Scholar] [CrossRef]
- Li, X.L.; Sui, J.Q.; Lu, L.L.; Zhang, N.N.; Xu, X.; Dong, Q.Y.; Xin, Y.N.; Xuan, S.Y. Gene polymorphisms associated with non-alcoholic fatty liver disease and coronary artery disease: A concise review. Lipids Health Dis. 2016, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, Q.; Cai, D.; Guo, H.; Fang, J.; Cui, H.; Gou, L.; Deng, J.; Wang, Z.; Zuo, Z. Resistin, a Novel Host Defense Peptide of Innate Immunity. Front. Immunol. 2021, 12, 699807. [Google Scholar] [CrossRef]
- Al-Hilali, H.A.; Abduljaleel, A.K. The role of TNF and Resistin Gene +299 (GA) Polymorphism in the Development of Insulin Resistance in non obese Type 2 Diabetes Mellitus Iraqi Patients. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 475–486. [Google Scholar]
- Su, K.-Z.; Li, Y.-R.; Zhang, D.; Yuan, J.-H.; Zhang, C.-S.; Liu, Y.; Song, L.-M.; Lin, Q.; Li, M.-W.; Dong, J. Relation of Circulating Resistin to Insulin Resistance in Type 2 Diabetes and Obesity: A Systematic Review and Meta-Analysis. Front. Physiol. 2019, 10, 1399. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Yu, L.; Zhou, L. Association between serum resistin concentration and hypertension: A systematic review and meta-analysis. Oncotarget 2017, 8, 41529–41537. [Google Scholar] [CrossRef]
- Niaz, S.; Latif, J.; Hussain, S. Serum resistin: A possible link between inflammation, hypertension and coronary artery disease. Pak. J. Med. Sci. 2019, 35, 641–646. [Google Scholar] [CrossRef]
- Rashad, N.M.; Allam, R.M.; Said, D.; Ali, A.E.; Mohy, N.M.; Abomandour, H.G. Influence of +299G>A and +62G˃A resistin gene promoter variants on cardiovascular risk in Egyptian women with systemic lupus erythematosus. Egypt. Rheumatol. 2019, 41, 215–220. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, J.; Bala, K.; Singh, J. Association of resistin (rs3745367) and urotensin II (rs228648 and rs2890565) gene polymorphisms with risk of type 2 diabetes mellitus in Indian population. Mol. Biol. Rep. 2020, 47, 9489–9497. [Google Scholar] [CrossRef] [PubMed]
- Suriyaprom, K.; Tungtrongchitr, R.; Namjuntra, P. Associations of Resistin Levels with Resistin Gene Polymorphism and Metabolic Syndrome in Thais. J. Med. Biochem. 2015, 34, 170–178. [Google Scholar] [CrossRef]
- Chung, C.-M.; Lin, T.-H.; Chen, J.-W.; Leu, H.-B.; Yin, W.-H.; Ho, H.-Y.; Sheu, S.-H.; Tsai, W.-C.; Chen, J.-H.; Lin, S.-J.; et al. Common quantitative trait locus downstream of RETN gene identified by genome-wide association study is associated with risk of type 2 diabetes mellitus in Han Chinese: A Mendelian randomization effect. Diabetes Metab. Res. Rev. 2014, 30, 232–240. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Izaola, O.; Primo, D.; de la Fuente, B.; Mulero, I.; Aller, R. The rs1862513 Variant in Resistin Gene-Modified Insulin Resistance and Insulin Levels after Weight Loss Secondary to Hypocaloric Diet. Ann. Nutr. Metab. 2016, 69, 256–262. [Google Scholar] [CrossRef]
- Qiao, X.; Xu, D.; Sun, D.; Sun, S.; Huang, Z.; Cui, W. Association analysis of interleukin-18 gene promoter region polymorphisms and susceptibility to sporadic breast cancer in Chinese Han women. J. Clin. Lab. Anal. 2018, 32, e22591. [Google Scholar] [CrossRef] [PubMed]
- Hassuna, N.A.; El Feky, M.; Hussein, A.A.R.M.; Mahmoud, M.A.; Idriss, N.K.; Abdelwahab, S.F.; Ibrahim, M.A. Interleukin-18 and interferon-γ single nucleotide polymorphisms in Egyptian patients with tuberculosis. PLoS ONE 2021, 16, e0244949. [Google Scholar] [CrossRef] [PubMed]
- Motavaf, M.; Safari, S.; Alavian, S.M. Interleukin 18 gene promoter polymorphisms and susceptibility to chronic hepatitis B infection: A review study. Hepat. Mon. 2014, 14, e19879. [Google Scholar] [CrossRef]
- Cheng, D.; Hao, Y.; Zhou, W.; Ma, Y. The relationship between interleukin-18 polymorphisms and allergic disease: A meta-analysis. BioMed Res. Int. 2014, 2014, 290687. [Google Scholar] [CrossRef]
- Jefferis, B.J.; Papacosta, O.; Owen, C.G.; Wannamethee, S.G.; Humphries, S.E.; Woodward, M.; Lennon, L.T.; Thomson, A.; Welsh, P.; Rumley, A.; et al. Interleukin 18 and coronary heart disease: Prospective study and systematic review. Atherosclerosis 2011, 217, 227–233. [Google Scholar] [CrossRef]
- Farias, T.D.; Canto, L.M.; Medeiros, M.D.; Sereia, A.F.; Back, L.K.; Mello, F.M.; Zimmermann, A.F.; Pereira, I.A.; Muniz, Y.C.; Marrero, A.R.; et al. Lack of association between interleukin-18 polymorphisms and rheumatoid arthritis. Rev. Bras. Reumatol. 2013, 53, 199–205. [Google Scholar] [PubMed]
- He, M.; Cornelis, M.C.; Kraft, P.; van Dam, R.M.; Sun, Q.; Laurie, C.C.; Mirel, D.B.; Chasman, D.I.; Ridker, P.M.; Hunter, D.J.; et al. Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 885–890. [Google Scholar] [CrossRef]
- Opstad, T.B.; Pettersen, A.Å.; Arnesen, H.; Seljeflot, I. Circulating levels of IL-18 are significantly influenced by the IL-18 +183 A/G polymorphism in coronary artery disease patients with diabetes type 2 and the metabolic syndrome: An Observational Study. Cardiovasc. Diabetol. 2011, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Rychter, A.M.; Skrzypczak-Zielińska, M.; Zielińska, A.; Eder, P.; Souto, E.B.; Zawada, A.; Ratajczak, A.E.; Dobrowolska, A.; Krela-Kaźmierczak, I. Is the Retinol-Binding Protein 4 a Possible Risk Factor for Cardiovascular Diseases in Obesity? Int. J. Mol. Sci. 2020, 21, 5229. [Google Scholar] [CrossRef] [PubMed]
- González-Alvarez, R.; Garza-Rodríguez, M.d.L.; Delgado-Enciso, I.; Treviño-Alvarado, V.M.; Canales-Del-Castillo, R.; Martínez-De-Villarreal, L.E.; Lugo-Trampe, Á.; Tejero, M.E.; Schlabritz-Loutsevitch, N.E.; Rocha-Pizaña, M.D.R.; et al. Molecular evolution and expression profile of the chemerine encoding gene RARRES2 in baboon and chimpanzee. Biol. Res. 2015, 48, 31. [Google Scholar] [CrossRef]
- Fatima, S.S.; Bozaoglu, K.; Rehman, R.; Alam, F.; Memon, A.S. Elevated Chemerin Levels in Pakistani Men: An Interrelation with Metabolic Syndrome Phenotypes. PLoS ONE 2013, 8, e57113. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, S.; Brandão Proença, J.; Santos-Silva, A.; Neuparth, M.J. Adiponectin, Leptin, and Chemerin in Elderly Patients with Type 2 Diabetes Mellitus: A Close Linkage with Obesity and Length of the Disease. BioMed Res. Int. 2014, 2014, 701915. [Google Scholar] [CrossRef]
- Sun, J.-X.; Zhang, C.; Cheng, Z.-B.; Tang, M.-Y.; Liu, Y.-Z.; Jiang, J.-F.; Xiao, X.; Huang, L. Chemerin in atherosclerosis. Clin. Chim. Acta 2021, 520, 8–15. [Google Scholar] [CrossRef]
- Gao, X.; Mi, S.; Zhang, F.; Gong, F.; Lai, Y.; Gao, F.; Zhang, X.; Wang, L.; Tao, H. Association of chemerin mRNA expression in human epicardial adipose tissue with coronary atherosclerosis. Cardiovasc. Diabetol. 2011, 10, 87. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, J.; Wang, C.; Sun, J.; Lai, X.; Xu, Y.; Lan, X.; Lei, C.; Zhang, C.; Yang, D.; et al. Exploring polymorphisms of the bovine RARRES2 gene and their associations with growth traits. Mol. Biol. Rep. 2012, 39, 2305–2311. [Google Scholar] [CrossRef]
- Perumalsamy, S.; Wan Ahmad, W.A.; Zaman Huri, H. Single Nucleotide Polymorphism rs17173608 in the Chemerin Encoding Gene: Is It a Predictor of Insulin Resistance and Severity of Coronary Artery Disease in Non-Obese Type 2 Diabetes? Healthcare 2021, 9, 623. [Google Scholar] [CrossRef] [PubMed]

| Gene | SNP ID | Position | Parameter Association | Population | N | p-Value | Ref |
|---|---|---|---|---|---|---|---|
| CHI3L1 | rs946263 | −9639 C > G | G-allele was nominally found to be associated with T2DM | Danish | 9438 | 0.027 | [63] |
| Was associated with serum Ykl-40 levels | Danish | 6784 | <0.0001 | [64] | |||
| −2122 C > T | Was associated with LDL level (cause of atherosclerosis) | Korean | 290 | 0.005 | [65] | ||
| Was associated with a significantly increased CHI3L1 mRNA level in peripheral blood cells and elevated nuclear factor binding | 0.008 | ||||||
| CD36 | rs1761667 | −31118 G > A | Was associated with T2DM | North Indian | 400 | <0.001 | [66] |
| Was associated with lipid profile | <0.001 | ||||||
| Was associated with LDL | <0.05 | ||||||
| Was associated with VLDL | 0.029 | ||||||
| A > G | CAD patients with an AG genotype had higher plasma levels of LDL | Egyptian | 100 | 0.046 | [67] | ||
| LEPR | rs1137100 | 109 T > A | Was independently associated with early atherosclerosis | Finnish | 526 | 0.042 | [68] |
| Was associated with high total cholesterol | 0.005 | ||||||
| Was associated with insulin levels | |||||||
| RETN | rs1862513 | −420 C > G | Was associated with resistin levels in DM patients | Japanese | 198 | 2.9 × 10−7 | [69] |
| Was associated with HbA1c levels | |||||||
| Was associated with total cholesterol levels (CAD marker) | Pakistani | 350 | <0.0001 | [70] | |||
| Was associated with LDL levels (CAD marker) | 0.0067 | ||||||
| Was associated with resistin levels | 0.0009 | ||||||
| Was associated with hs-CRP levels (CAD marker) | <0.0001 | ||||||
| rs3745367 | +299 G > A | Significantly associated with T2DM | Thai | 95 | 0.004 | [71] | |
| Associated with total cholesterol levels | Pakistani | <0.0001 | [70] | ||||
| Associated with LDL levels | 0.0153 | ||||||
| Associated with resistin levels | <0.0001 | ||||||
| Associated with hs-CRP levels | <0.0001 | ||||||
| IL-18 | rs1834481 | G > C | Associated with CAD/ MI risk | European | 3202 | 0.0021 | [72] |
| Associated with IL-18 levels | European | 200 | <0.005 | [73] | |||
| Associated with glucose levels | <0.005 | ||||||
| RBP-4 | rs7094671 | G > A | Associated with CAD | Chinese | 392 | <0.0001 | [74] |
| +5169 C > T | Associated with T2DM | Mongolian | 281 | <0.005 | [75] | ||
| RARRES2 | rs17173608 | T > G | Associated with risk of metabolic syndrome (IR, high LDL) | Iranian | 300 | 0.012 | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perumalsamy, S.; Huri, H.Z.; Abdullah, B.M.; Mazlan, O.; Wan Ahmad, W.A.; Vethakkan, S.R.D.B. Genetic Markers of Insulin Resistance and Atherosclerosis in Type 2 Diabetes Mellitus Patients with Coronary Artery Disease. Metabolites 2023, 13, 427. https://doi.org/10.3390/metabo13030427
Perumalsamy S, Huri HZ, Abdullah BM, Mazlan O, Wan Ahmad WA, Vethakkan SRDB. Genetic Markers of Insulin Resistance and Atherosclerosis in Type 2 Diabetes Mellitus Patients with Coronary Artery Disease. Metabolites. 2023; 13(3):427. https://doi.org/10.3390/metabo13030427
Chicago/Turabian StylePerumalsamy, Sangeetha, Hasniza Zaman Huri, Bashar Mudhaffar Abdullah, Othman Mazlan, Wan Azman Wan Ahmad, and Shireene Ratna D. B. Vethakkan. 2023. "Genetic Markers of Insulin Resistance and Atherosclerosis in Type 2 Diabetes Mellitus Patients with Coronary Artery Disease" Metabolites 13, no. 3: 427. https://doi.org/10.3390/metabo13030427
APA StylePerumalsamy, S., Huri, H. Z., Abdullah, B. M., Mazlan, O., Wan Ahmad, W. A., & Vethakkan, S. R. D. B. (2023). Genetic Markers of Insulin Resistance and Atherosclerosis in Type 2 Diabetes Mellitus Patients with Coronary Artery Disease. Metabolites, 13(3), 427. https://doi.org/10.3390/metabo13030427








