Cross-Platform Comparison of Amino Acid Metabolic Profiling in Three Model Organisms Used in Environmental Metabolomics
Abstract
1. Introduction
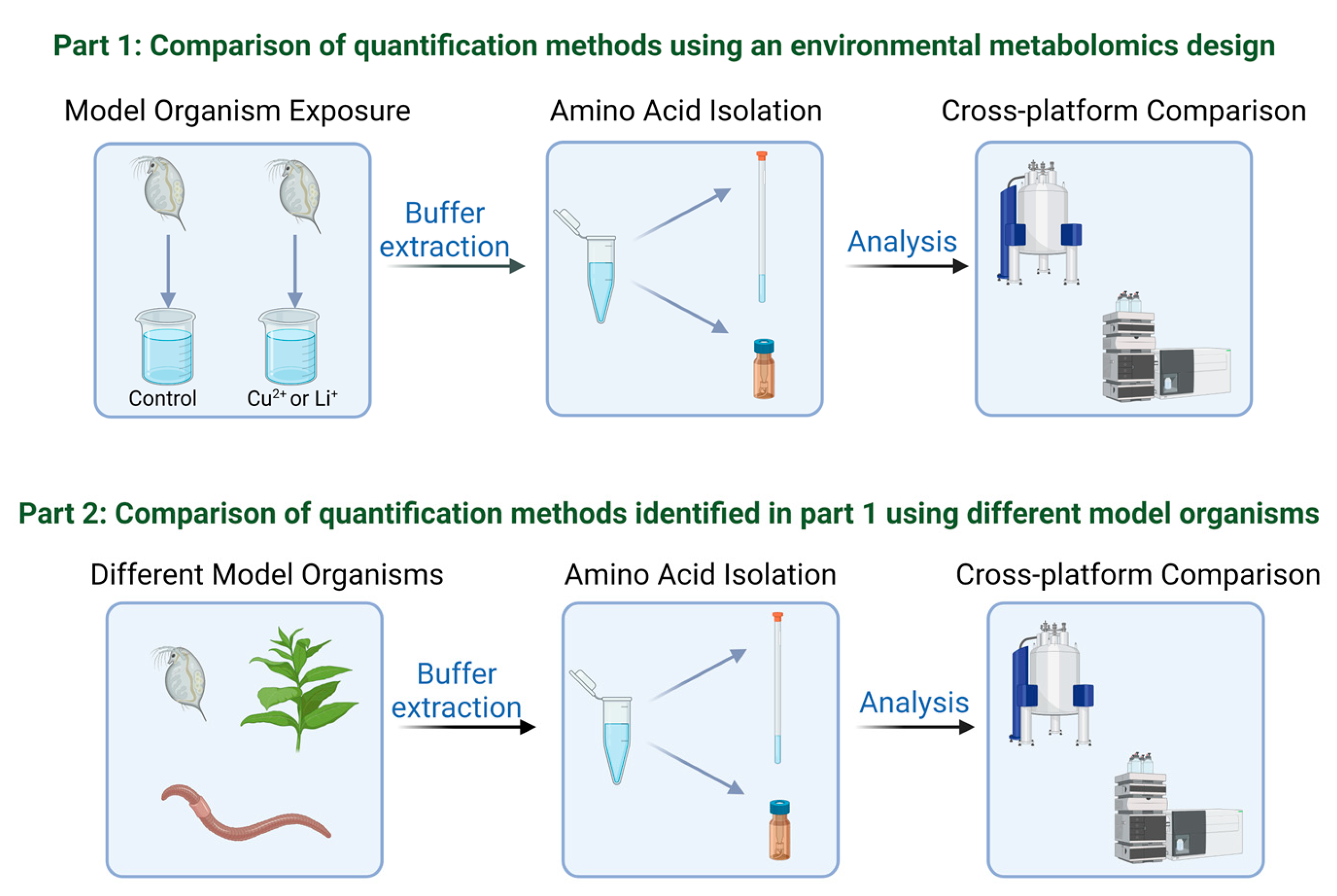
2. Materials and Methods
2.1. Daphnia magna Amino Acid Extraction
2.2. Eisenia fetida and Nicotiana tabacum Amino Acid Extraction
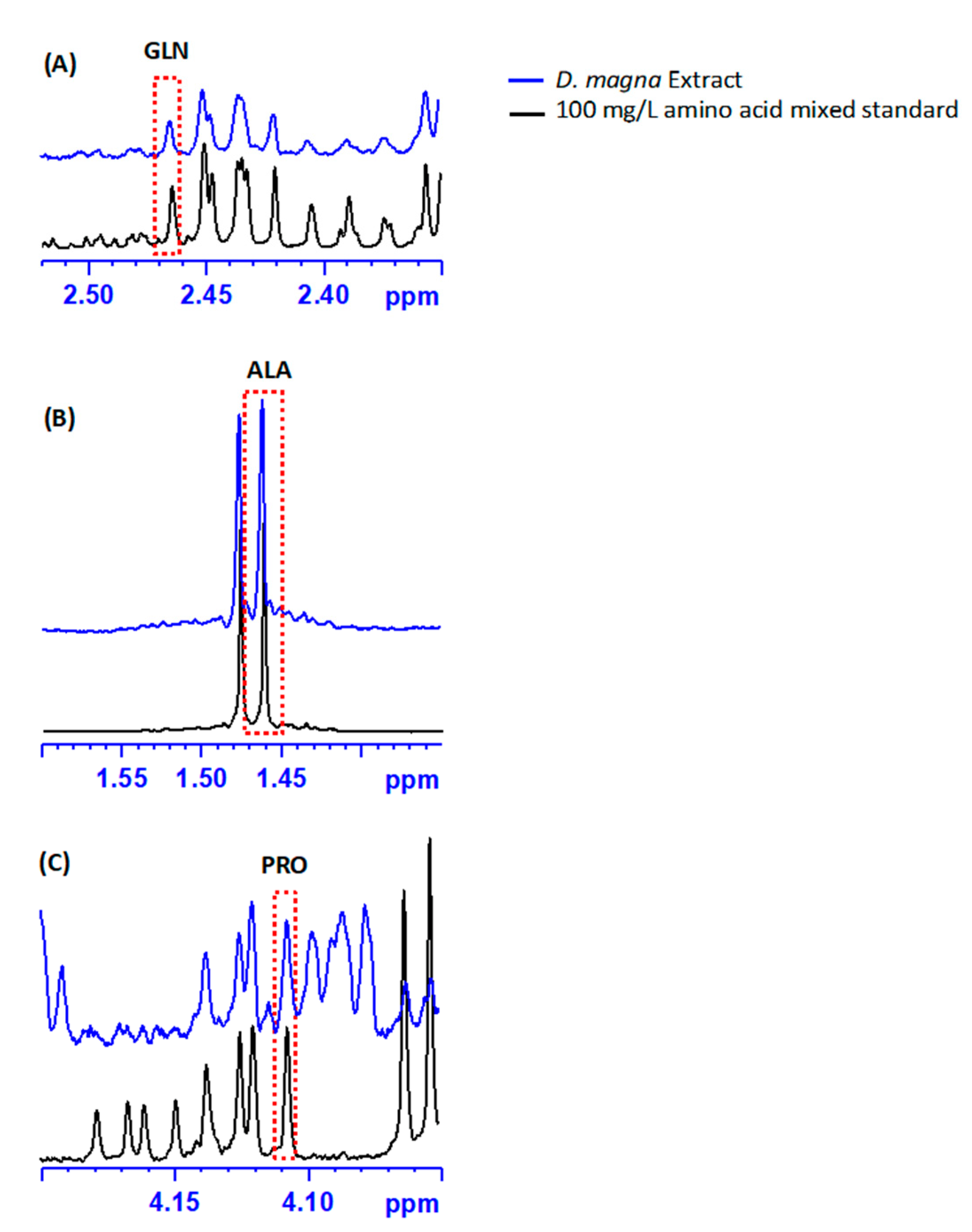
2.3. Amino Acid Profiling via 1H NMR
2.4. Amino Acid Profiling via LC-MS/MS
2.5. Statistical Analysis and Comparison of Method Agreement
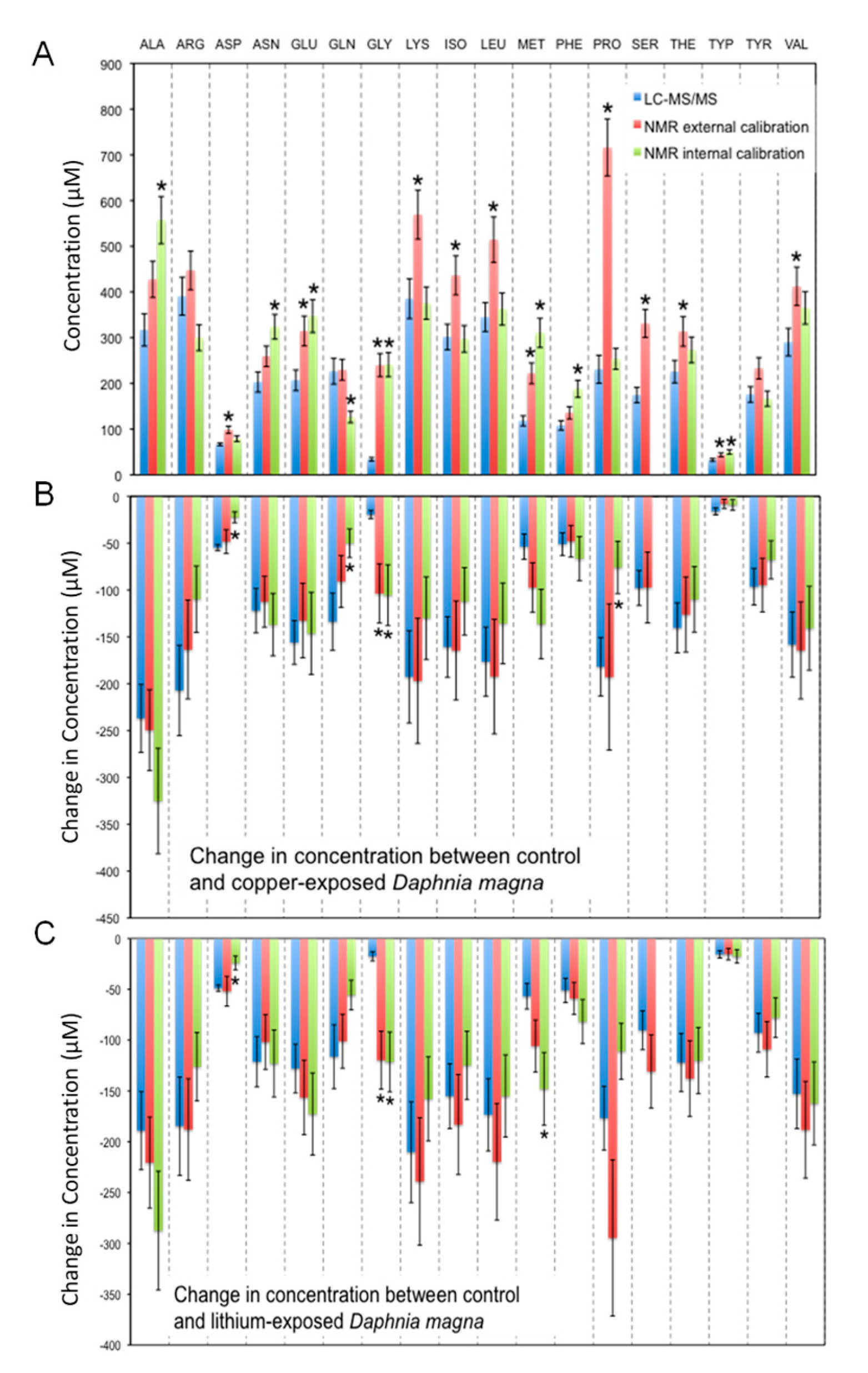
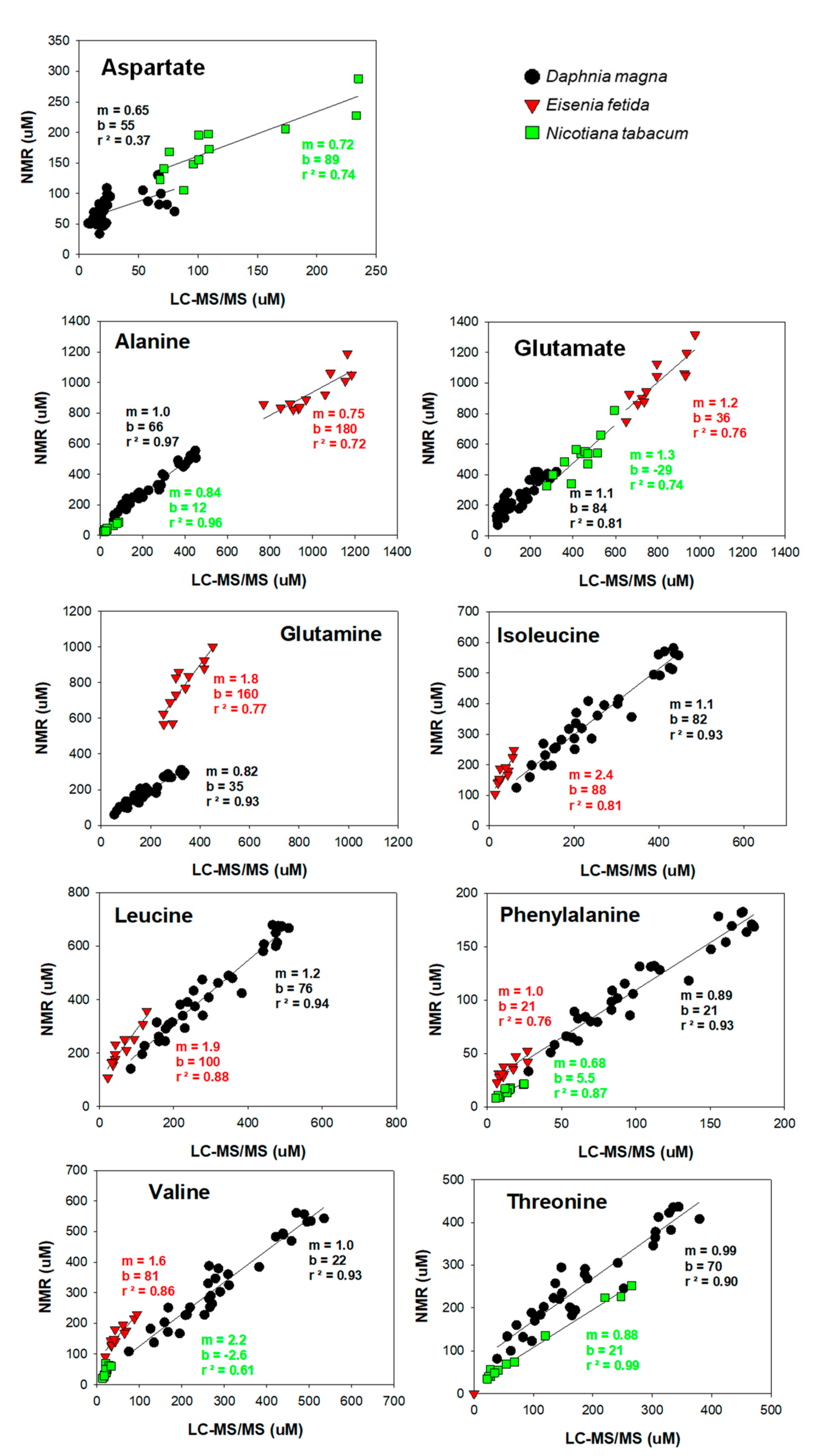
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Viant, M.R. Applications of metabolomics to the environmental sciences. Metabolomics 2009, 5, 1–2. [Google Scholar] [CrossRef]
- Kim, H.M.; Kang, J.S. Metabolomic Studies for the Evaluation of Toxicity Induced by Environmental Toxicants on Model Organisms. Metabolites 2021, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-J.; Qian, L.; Ding, L.-Y.; Wang, L.; Wong, M.H.; Tao, H.-C. Ecological and toxicological assessments of anthropogenic contaminants based on environmental metabolomics. Environ. Sci. Ecotechnology 2021, 5, 100081. [Google Scholar] [CrossRef] [PubMed]
- Lankadurai, B.P.; Nagato, E.G.; Simpson, M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressors. Environ. Rev. 2013, 21, 180–205. [Google Scholar] [CrossRef]
- Viant, M.R.; Sommer, U. Mass spectrometry based environmental metabolomics: A primer and review. Metabolomics 2012, 9, 144–158. [Google Scholar] [CrossRef]
- Wishart, D.S. Quantitative metabolomics using NMR. TrAC 2008, 27, 228–237. [Google Scholar] [CrossRef]
- Simpson, M.J.; Bearden, D.W. Environmental Metabolomics: NMR Techniques. eMagRes 2013, 2, 549–560. [Google Scholar] [CrossRef]
- Viant, M.R.; Bearden, D.W.; Bundy, J.G.; Burton, I.W.; Collette, T.W.; Ekman, D.R.; Ezernieks, V.; Karakach, T.K.; Lin, C.-Y.; Rochfort, S.; et al. International NMR-Based Environmental Metabolomics Intercomparison Exercise. Environ. Sci. Technol. 2009, 43, 219–225. [Google Scholar] [CrossRef]
- Cui, Q.; Lewis, I.A.; Hegeman, A.D.; Anderson, M.E.; Li, J.; Schulte, C.F.; Westler, W.M.; Eghbalnia, H.R.; Sussman, M.R.; Markley, J.L. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008, 26, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Larive, C.K.; Barding, G.A.; Dinges, M.M. NMR Spectroscopy for Metabolomics and Metabolic Profiling. Anal. Chem. 2015, 87, 133–146. [Google Scholar] [CrossRef]
- Brown, S.A.; Simpson, A.J.; Simpson, M.J. Evaluation of sample preparation methods for nuclear magnetic resonance metabolic profiling studies with eisenia fetida. Environ. Toxicol. Chem. 2008, 27, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Nagato, E.G.; Lankadurai, B.P.; Soong, R.; Simpson, A.J.; Simpson, M.J. Development of an NMR microprobe procedure for high-throughput environmental metabolomics of Daphnia magna. Magn. Reson. Chem. 2015, 53, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef]
- Burton, I.W.; Quilliam, M.A.; Walter, J.A. Quantitative 1H NMR with External Standards: Use in Preparation of Calibration Solutions for Algal Toxins and Other Natural Products. Anal. Chem. 2005, 77, 3123–3131. [Google Scholar] [CrossRef]
- Caligiani, A.; Acquotti, D.; Palla, G.; Bocchi, V. Identification and quantification of the main organic components of vinegars by high resolution 1H NMR spectroscopy. Anal. Chim. Acta 2007, 585, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, V.; Pinciroli, V. Quantitative NMR in synthetic and combinatorial chemistry. J. Pharm. Biomed. Anal. 2005, 38, 851–857. [Google Scholar] [CrossRef]
- Majumdar, R.D.; Akhter, M.; Fortier-McGill, B.; Soong, R.; Liaghati-Mobarhan, Y.; Simpson, A.J.; Spraul, M.; Schmidt, S.; Heumann, H. In Vivo Solution-State NMR-Based Environmental Metabolomics. eMagRes 2017, 6, 133–148. [Google Scholar] [CrossRef]
- Ekman, D.R.; Skelton, D.M.; Davis, J.M.; Villeneuve, D.L.; Cavallin, J.E.; Schroeder, A.; Jensen, K.M.; Ankley, G.T.; Collette, T.W. Metabolite Profiling of Fish Skin Mucus: A Novel Approach for Minimally-Invasive Environmental Exposure Monitoring and Surveillance. Environ. Sci. Technol. 2015, 49, 3091–3100. [Google Scholar] [CrossRef]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16. [Google Scholar] [CrossRef]
- Gowda, G.A.N.; Raftery, D. Recent Advances in NMR-Based Metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Lankadurai, B.P.; Wolfe, D.M.; Whitfield Åslund, M.L.; Simpson, A.J.; Simpson, M.J. 1H NMR-based metabolomic analysis of polar and non-polar earthworm metabolites after sub-lethal exposure to phenanthrene. Metabolomics 2012, 9, 44–56. [Google Scholar] [CrossRef]
- Lankadurai, B.P.; Simpson, A.J.; Simpson, M.J. 1H NMR metabolomics of Eisenia fetida responses after sub-lethal exposure to perfluorooctanoic acid and perfluorooctane sulfonate. Environ. Chem. 2012, 9, 502–511. [Google Scholar] [CrossRef]
- McKelvie, J.R.; Yuk, J.; Xu, Y.; Simpson, A.J.; Simpson, M. 1H NMR and GC/MS metabolomics of earthworm responses to sub-lethal DDT and endosulfan exposure. Metabolomics 2008, 5, 84–94. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, L.; Zhang, X.; Shi, C.; Ma, L.; Zhang, X.; Wang, G. Similarities and differences among the responses to three chlorinated organophosphate esters in earthworm: Evidences from biomarkers, transcriptomics and metabolomics. Sci. Total. Environ. 2022, 815, 152853. [Google Scholar] [CrossRef]
- Nagato, E.G.; Simpson, A.J.; Simpson, M.J. Metabolomics reveals energetic impairments in Daphnia magna exposed to diazinon, malathion and bisphenol-A. Aquat. Toxicol. 2016, 170, 175–186. [Google Scholar] [CrossRef]
- Poynton, H.C.; Lazorchak, J.M.; Impellitteri, C.A.; Blalock, B.J.; Rogers, K.; Allen, H.J.; Loguinov, A.; Heckman, J.L.; Govindasmawy, S. Toxicogenomic Responses of Nanotoxicity in Daphnia magna Exposed to Silver Nitrate and Coated Silver Nanoparticles. Environ. Sci. Technol. 2012, 46, 6288–6296. [Google Scholar] [CrossRef]
- Taylor, N.; Weber, R.J.; Southam, A.; Payne, T.; Hrydziuszko, O.; Arvanitis, T.; Viant, M. A new approach to toxicity testing in Daphnia magna: Application of high throughput FT-ICR mass spectrometry metabolomics. Metabolomics 2009, 5, 44–58. [Google Scholar] [CrossRef]
- Wang, P.; Li, Q.-Q.; Hui, J.; Xiang, Q.-Q.; Yan, H.; Chen, L.-Q. Metabolomics reveals the mechanism of polyethylene microplastic toxicity to Daphnia magna. Chemosphere 2022, 307, 135887. [Google Scholar] [CrossRef]
- Ekman, D.R.; Teng, Q.; Villeneuve, D.L.; Kahl, M.D.; Jensen, K.M.; Durhan, E.J.; Ankley, G.T.; Collette, T.W. Investigating Compensation and Recovery of Fathead Minnow (Pimephales promelas) Exposed to 17α-Ethynylestradiol with Metabolite Profiling. Environ. Sci. Technol. 2008, 42, 4188–4194. [Google Scholar] [CrossRef]
- Ekman, D.R.; Teng, Q.; Villeneuve, D.L.; Kahl, M.D.; Jensen, K.M.; Durhan, E.J.; Ankley, G.T.; Collette, T.W. Profiling lipid metabolites yields unique information on sex- and time-dependent responses of fathead minnows (Pimephales promelas) exposed to 17α-ethynylestradiol. Metabolomics 2008, 5, 22–32. [Google Scholar] [CrossRef]
- Teng, Q.; Ekman, D.R.; Huang, W.; Collette, T.W. Impacts of 17α-ethynylestradiol exposure on metabolite profiles of zebrafish (Danio rerio) liver cells. Aquat. Toxicol. 2013, 130, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Rosenblum, E.S.; Tjeerdema, R.S. NMR-Based Metabolomics: A Powerful Approach for Characterizing the Effects of Environmental Stressors on Organism Health. Environ. Sci. Technol. 2003, 37, 4982–4989. [Google Scholar] [CrossRef] [PubMed]
- Collette, T.W.; Teng, Q.; Jensen, K.M.; Kahl, M.D.; Makynen, E.A.; Durhan, E.J.; Villeneuve, D.L.; Martinović-Weigelt, D.; Ankley, G.T.; Ekman, D.R. Impacts of an Anti-Androgen and an Androgen/Anti-Androgen Mixture on the Metabolite Profile of Male Fathead Minnow Urine. Environ. Sci. Technol. 2010, 44, 6881–6886. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, W.; Wang, D.; Teng, M.; Yan, J.; Miao, J.; Zhou, Z. 1H NMR-based metabolomics analysis of adult zebrafish (Danio rerio) after exposure to diniconazole as well as its bioaccumulation behavior. Chemosphere 2017, 168, 1571–1577. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Canela, C.; Prats, E.; Piña, B.; Tauler, R. Assessment of chlorpyrifos toxic effects in zebrafish (Danio rerio) metabolism. Environ. Pollut. 2017, 220, 1231–1243. [Google Scholar] [CrossRef]
- Barding, G.A.; Orr, D.J.; Larive, C.K. Plant Metabolomics. eMagRes 2011, 1, 85–99. [Google Scholar] [CrossRef]
- White, R.A.; Borkum, M.I.; Rivas-Ubach, A.; Bilbao, A.; Wendler, J.P.; Colby, S.M.; Köberl, M.; Jansson, C. From data to knowledge: The future of multi-omics data analysis for the rhizosphere. Rhizosphere 2017, 3, 222–229. [Google Scholar] [CrossRef]
- Tomita, S.; Ikeda, S.; Tsuda, S.; Someya, N.; Asano, K.; Kikuchi, J.; Chikayama, E.; Ono, H.; Sekiyama, Y. A survey of metabolic changes in potato leaves by NMR-based metabolic profiling in relation to resistance to late blight disease under field conditions. Magn. Reson. Chem. 2017, 55, 120–127. [Google Scholar] [CrossRef]
- Bijttebier, S.; Van der Auwera, A.; Foubert, K.; Voorspoels, S.; Pieters, L.; Apers, S. Bridging the gap between comprehensive extraction protocols in plant metabolomics studies and method validation. Anal. Chim. Acta 2016, 935, 136–150. [Google Scholar] [CrossRef]
- Fabres, P.J.; Collins, C.; Cavagnaro, T.R.; Rodríguez López, C.M. A Concise Review on Multi-Omics Data Integration for Terroir Analysis in Vitis vinifera. Front. Plant Sci. 2017, 8, 1065–1073. [Google Scholar] [CrossRef]
- Jorge, T.F.; Rodrigues, J.A.; Caldana, C.; Schmidt, R.; van Dongen, J.T.; Thomas-Oates, J.; António, C. Mass spectrometry-based plant metabolomics: Metabolite responses to abiotic stress. Mass Spectrom. Rev. 2015, 35, 620–649. [Google Scholar] [CrossRef]
- Yuk, J.; Simpson, M.J.; Simpson, A.J. 1-D and 2-D NMR-based metabolomics of earthworms exposed to endosulfan and endosulfan sulfate in soil. Environ. Pollut. 2013, 175, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Moradi, P.; Ford-Lloyd, B.; Pritchard, J. Metabolomic approach reveals the biochemical mechanisms underlying drought stress tolerance in thyme. Anal. Biochem. 2017, 527, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 29265–29276. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bai, J.; Wen, L.; Wang, W.; Zhang, L.; Liu, Z.; Liu, H. Phytotoxicity of microplastics to the floating plant Spirodela polyrhiza (L.): Plant functional traits and metabolomics. Environ. Pollut. 2023, 322, 121199. [Google Scholar] [CrossRef]
- Wagner, N.D.; Lankadurai, B.P.; Simpson, M.J.; Simpson, A.J.; Frost, P.C. Metabolomic Differentiation of Nutritional Stress in an Aquatic Invertebrate. Physiol. Biochem. Zool. 2015, 88, 43–52. [Google Scholar] [CrossRef]
- Edison, A.S.; Hall, R.D.; Junot, C.; Karp, P.D.; Kurland, I.J.; Mistrik, R.; Reed, L.K.; Saito, K.; Salek, R.M.; Steinbeck, C.; et al. The Time Is Right to Focus on Model Organism Metabolomes. Metabolites 2016, 6, 8–15. [Google Scholar] [CrossRef]
- Sun, H.; Liu, X.; Li, F.; Li, W.; Zhang, J.; Xiao, Z.; Shen, L.; Li, Y.; Wang, F.; Yang, J. First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum. Sci. Rep. 2017, 7, 3013–3027. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of Altered Metabolomic States in Nicotiana tabacum Cells Induced by Priming Agents. Front. Plant Sci. 2016, 7, 1527–1543. [Google Scholar] [CrossRef]
- Walter, A.; Schurr, U. The modular character of growth in Nicotiana tabacum plants under steady-state nutrition. J. Exp. Bot. 1999, 50, 1169–1177. [Google Scholar] [CrossRef]
- Wang, J.; Cheung, M.; Rasooli, L.; Amirsadeghi, S.; Vanlerberghe, G.C. Plant respiration in a high CO2 world: How will alternative oxidase respond to future atmospheric and climatic conditions? Can. J. Plant Sci. 2014, 94, 1091–1101. [Google Scholar] [CrossRef]
- Alber, N.A.; Sivanesan, H.; Vanlerberghe, G.C. The occurrence and control of nitric oxide generation by the plant mitochondrial electron transport chain. Plant Cell Environ. 2016, 40, 1074–1085. [Google Scholar] [CrossRef]
- Nagato, E.G.; D’Eon, J.C.; Lankadurai, B.P.; Poirier, D.G.; Reiner, E.J.; Simpson, A.J.; Simpson, M.J. 1H NMR-based metabolomics investigation of Daphnia magna responses to sub-lethal exposure to arsenic, copper and lithium. Chemosphere 2013, 93, 331–337. [Google Scholar] [CrossRef]
- Åslund, M.W.; Celejewski, M.; Lankadurai, B.P.; Simpson, A.J.; Simpson, M.J. Natural variability and correlations in the metabolic profile of healthy Eisenia fetida earthworms observed using 1H NMR metabolomics. Chemosphere 2011, 83, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Brown, S.A. Purge NMR: Effective and easy solvent suppression. J. Magn. Reson. 2005, 175, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Gu, L.; Jones, A.D.; Last, R.L. LC−MS/MS Assay for Protein Amino Acids and Metabolically Related Compounds for Large-Scale Screening of Metabolic Phenotypes. Anal. Chem. 2007, 79, 8067–8075. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 327, 307–310. [Google Scholar] [CrossRef]
- Malz, F.; Jancke, H. Validation of quantitative NMR. J. Pharm. Biomed. Anal. 2005, 38, 813–823. [Google Scholar] [CrossRef]
- Taylor, P.J. Matrix effects: The Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry. Clin. Biochem. 2005, 38, 328–334. [Google Scholar] [CrossRef]
- Matuszewski, B.K.; Constanzer, M.L.; Chavez-Eng, C.M. Strategies for the Assessment of Matrix Effect in Quantitative Bioanalytical Methods Based on HPLC−MS/MS. Anal. Chem. 2003, 75, 3019–3030. [Google Scholar] [CrossRef] [PubMed]
- Horton, H.R.; Moran, L.A.; Scrimgeour, K.G.; Perry, M.D.; Rawn, J.D. Principles of Biochemistry, 4th ed.; Pearson Prentice Hall: Hoboken, NJ, USA, 2006. [Google Scholar]
- Nowick, J.S.; Khakshoor, O.; Hashemzadeh, M.; Brower, J.O. DSA: A New Internal Standard for NMR Studies in Aqueous Solution. Org. Lett. 2003, 5, 3511–3513. [Google Scholar] [CrossRef] [PubMed]
- Alum, M.F.; Shaw, P.A.; Sweatman, B.C.; Ubhi, B.K.; Haselden, J.N.; Connor, S.C. 4,4-Dimethyl-4-silapentane-1-ammonium trifluoroacetate (DSA), a promising universal internal standard for NMR-based metabolic profiling studies of biofluids, including blood plasma and serum. Metabolomics 2008, 4, 122–127. [Google Scholar] [CrossRef]
- Akoka, S.; Barantin, L.; Trierweiler, M. Concentration Measurement by Proton NMR Using the ERETIC Method. Anal. Chem. 1999, 71, 2554–2557. [Google Scholar] [CrossRef] [PubMed]
- Cullen, C.H.; Ray, G.J.; Szabo, C.M. A comparison of quantitative nuclear magnetic resonance methods: Internal, external, and electronic referencing. Magn. Reson. Chem. 2013, 51, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Li, L. Matrix effect on chemical isotope labeling and its implication in metabolomic sample preparation for quantitative metabolomics. Metabolomics 2015, 11, 1733–1742. [Google Scholar] [CrossRef]
- Wu, Y.; Li, L. Sample normalization methods in quantitative metabolomics. J. Chromatogr. A 2016, 1430, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Sänger van de Griend, C. Enantiomeric Separation of Glycyl Dipeptides by Capillary Electrophoresis with Cyclodextrins as Chiral Selectors. Electrophoresis 1999, 20, 3417–3423. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’eon, J.C.; Lankadurai, B.P.; Simpson, A.J.; Reiner, E.J.; Poirier, D.G.; Vanlerberghe, G.C.; Simpson, M.J. Cross-Platform Comparison of Amino Acid Metabolic Profiling in Three Model Organisms Used in Environmental Metabolomics. Metabolites 2023, 13, 402. https://doi.org/10.3390/metabo13030402
D’eon JC, Lankadurai BP, Simpson AJ, Reiner EJ, Poirier DG, Vanlerberghe GC, Simpson MJ. Cross-Platform Comparison of Amino Acid Metabolic Profiling in Three Model Organisms Used in Environmental Metabolomics. Metabolites. 2023; 13(3):402. https://doi.org/10.3390/metabo13030402
Chicago/Turabian StyleD’eon, Jessica C., Brian P. Lankadurai, André J. Simpson, Eric J. Reiner, David G. Poirier, Greg C. Vanlerberghe, and Myrna J. Simpson. 2023. "Cross-Platform Comparison of Amino Acid Metabolic Profiling in Three Model Organisms Used in Environmental Metabolomics" Metabolites 13, no. 3: 402. https://doi.org/10.3390/metabo13030402
APA StyleD’eon, J. C., Lankadurai, B. P., Simpson, A. J., Reiner, E. J., Poirier, D. G., Vanlerberghe, G. C., & Simpson, M. J. (2023). Cross-Platform Comparison of Amino Acid Metabolic Profiling in Three Model Organisms Used in Environmental Metabolomics. Metabolites, 13(3), 402. https://doi.org/10.3390/metabo13030402





