LC-QToF-Based Metabolomics Identifies Aberrant Tissue Metabolites Associated with a Higher-Fat Diet and Their ‘Reversion to Healthy’ with Dietary Probiotic Supplementation
Abstract
1. Introduction
2. Materials and Methods
2.1. Maintenance of Animals and Experimentation
2.2. Sample Preparation for Metabolite Profiling
2.3. LC-QToF Analysis
2.4. Data Processing, Chemometrics, and Statistical Analysis
3. Results and Discussion
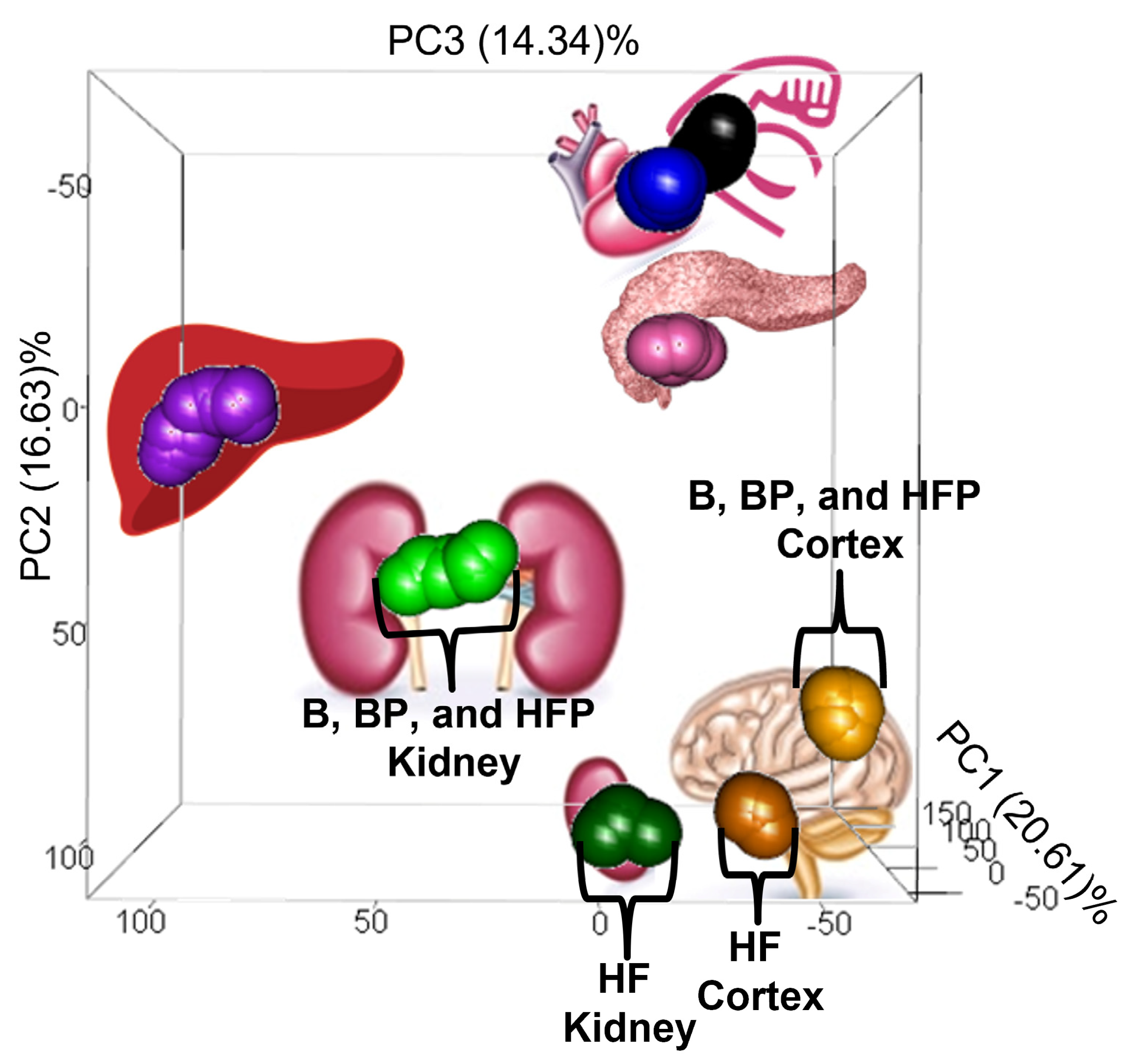
3.1. Dietary Effects on the Pig Tissue Metabolomes
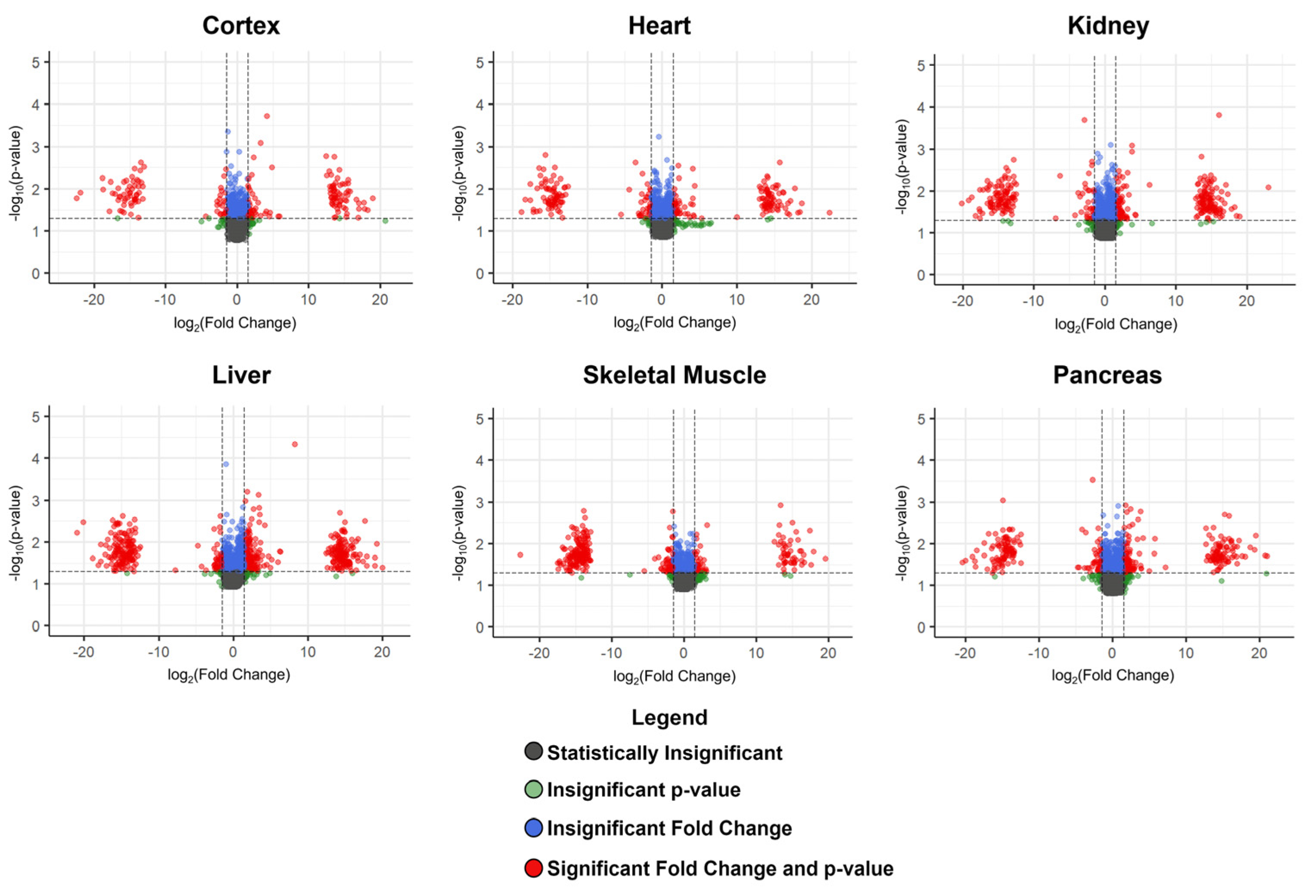
3.2. Metabolites Altered Due to a Higher-Fat Diet
3.2.1. Diet-Associated Alterations in the Brain Cortex
3.2.2. Diet-Associated Alterations in the Heart and Pancreas
3.2.3. Diet-Associated Alterations in the Kidney
3.2.4. A Reduction in the Membrane PC/PE Ratio
3.2.5. Diet-Associated Alterations in the Liver
3.2.6. Diet-Associated Alterations in the Skeletal Muscle
3.3. Metabolites Altered Due to Probiotic Supplementation
3.3.1. Phosphatidylcholines
3.3.2. Uridine Diphosphate-N-Acetylglucosamine
3.3.3. Saccharopine
3.4. Probiotic-Induced Metabolic Reversions
3.4.1. Probiotic-Induced Reversion in the Brain Cortex
3.4.2. Probiotic-Induced Reversion in the Heart
3.4.3. Probiotic-Induced Reversion in the Kidney
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NIH. Oral Probiotics: An Introduction. 2012. Available online: http://nccam.nih.gov/health/probiotics/introduction.htm (accessed on 27 April 2014).
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Mu, J.; Li, X.; Zhao, X. Relationship between Probiotics and Obesity: A Review of Recent Research. Food Sci. Technol. 2022, 42. [Google Scholar] [CrossRef]
- Christensen, H.R.; Frøkiaer, H.; Pestka, J.J. Lactobacilli Differentially Modulate Expression of Cytokines and Maturation Surface Markers in Murine Dendritic Cells. J. Immunol. 2002, 168, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Roberfroid, M.B. Dietary Modulation of the Human Colonic Microbiota: Introducing the Concept of Prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef]
- Varcoe, J.J.; Krejcarek, G.; Busta, F.; Brady, L. Prophylactic Feeding of Lactobacillus acidophilus NCFM to Mice Attenuates Overt Colonic Hyperplasia. J. Food Prot. 2003, 66, 457–465. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An Overview of Beneficial Effects. Antonie Van Leeuwenhoek 2002, 82, 279–289. [Google Scholar] [CrossRef]
- Corthésy, B.; Gaskins, H.R.; Mercenier, A. Cross-Talk between Probiotic Bacteria and the Host Immune System. J. Nutr. 2007, 137 (Suppl. 2), 781S–790S. [Google Scholar] [CrossRef]
- Rousseaux, C.; Thuru, X.; Gelot, A.; Barnich, N.; Neut, C.; Dubuquoy, L.; Dubuquoy, C.; Merour, E.; Geboes, K.; Chamaillard, M.; et al. Lactobacillus acidophilus Modulates Intestinal Pain and Induces Opioid and Cannabinoid Receptors. Nat. Med. 2007, 13, 35–37. [Google Scholar] [CrossRef]
- Mohamadzadeh, M.; Olson, S.; Kalina, W.V.; Ruthel, G.; Demmin, G.L.; Warfield, K.L.; Bavari, S.; Klaenhammer, T.R. Lactobacilli Activate Human Dendritic Cells That Skew T Cells toward T Helper 1 Polarization. Proc. Natl. Acad. Sci. USA 2005, 102, 2880–2885. [Google Scholar] [CrossRef]
- Hong, Y.-S.; Hong, K.S.; Park, M.-H.; Ahn, Y.-T.; Lee, J.-H.; Huh, C.-S.; Lee, J.; Kim, I.-K.; Hwang, G.-S.; Kim, J.S. Metabonomic Understanding of Probiotic Effects in Humans With Irritable Bowel Syndrome. J. Clin. Gastroenterol. 2011, 45, 415–425. [Google Scholar] [CrossRef]
- Kumar, M.; Nagpal, R.; Verma, V.; Kumar, A.; Kaur, N.; Hemalatha, R.; Gautam, S.K.; Singh, B. Probiotic Metabolites as Epigenetic Targets in the Prevention of Colon Cancer. Nutr. Rev. 2013, 71, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Madsen, K. Using Metabolomics to Decipher Probiotic Effects in Patients With Irritable Bowel Syndrome. J. Clin. Gastroenterol. 2011, 45, 389–390. [Google Scholar] [CrossRef]
- Ma, Y.-Y.; Li, L.; Yu, C.-H.; Shen, Z.; Chen, L.-H.; Li, Y.-M. Effects of Probiotics on Nonalcoholic Fatty Liver Disease: A Meta-Analysis. World J. Gastroenterol. WJG 2013, 19, 6911–6918. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective Increases of Bifidobacteria in Gut Microflora Improve High-Fat-Diet-Induced Diabetes in Mice through a Mechanism Associated with Endotoxaemia. Diabetologia 2007, 50, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Park, J.-H.; Seok, S.-H.; Baek, M.-W.; Kim, D.-J.; Lee, K.-E.; Paek, K.-S.; Lee, Y.; Park, J.-H. Human Originated Bacteria, Lactobacillus rhamnosus PL60, Produce Conjugated Linoleic Acid and Show Anti-Obesity Effects in Diet-Induced Obese Mice. Biochim. Biophys. Acta 2006, 1761, 736–744. [Google Scholar] [CrossRef]
- Hanhineva, K.; Barri, T.; Kolehmainen, M.; Pekkinen, J.; Pihlajamäki, J.; Vesterbacka, A.; Solano-Aguilar, G.; Mykkänen, H.; Dragsted, L.O.; Urban, J.F.; et al. Comparative Nontargeted Profiling of Metabolic Changes in Tissues and Biofluids in High-Fat Diet-Fed Ossabaw Pig. J. Proteome Res. 2013, 12, 3980–3992. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Monagas, M.; Jang, S.; Molokin, A.; Harnly, J.M.; Urban, J.F.; Solano-Aguilar, G.; Chen, P. A High Fat, High Cholesterol Diet Leads to Changes in Metabolite Patterns in Pigs—A Metabolomic Study. Food Chem. 2015, 173, 171–178. [Google Scholar] [CrossRef]
- Trasino, S.E.; Dawson, H.D.; Urban, J.F.; Wang, T.T.Y.; Solano-Aguilar, G. Feeding Probiotic Lactobacillus paracasei to Ossabaw Pigs on a High-Fat Diet Prevents Cholesteryl-Ester Accumulation and LPS Modulation of the Liver X Receptor and Inflammatory Axis in Alveolar Macrophages. J. Nutr. Biochem. 2013, 24, 1931–1939. [Google Scholar] [CrossRef]
- Lee, L.; Alloosh, M.; Saxena, R.; Van Alstine, W.; Watkins, B.A.; Klaunig, J.E.; Sturek, M.; Chalasani, N. Nutritional Model of Steatohepatitis and Metabolic Syndrome in the Ossabaw Miniature Swine. Hepatology 2009, 50, 56–67. [Google Scholar] [CrossRef]
- Bjerg, A.T.; Kristensen, M.; Ritz, C.; Holst, J.J.; Rasmussen, C.; Leser, T.D.; Wellejus, A.; Astrup, A. Lactobacillus paracasei Subsp Paracasei L. Casei W8 Suppresses Energy Intake Acutely. Appetite 2014, 82, 111–118. [Google Scholar] [CrossRef]
- Bjerg, A.T.; Kristensen, M.; Ritz, C.; Stark, K.; Holst, J.; Leser, T.; Wellejus, A.; Astrup, A. Four Weeks Supplementation with Lactobacillus paracasei Subsp. Paracasei L. Casei W8® Shows Modest Effect on Triacylglycerol in Young Healthy Adults. Benef. Microbes 2015, 6, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Kårlund, A.; Hanhineva, K.; Lehtonen, M.; Karjalainen, R.O.; Sandell, M. Nontargeted Metabolite Profiles and Sensory Properties of Strawberry Cultivars Grown Both Organically and Conventionally. J. Agric. Food Chem. 2015, 63, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Pekkinen, J.; Rosa-Sibakov, N.; Micard, V.; Keski-Rahkonen, P.; Lehtonen, M.; Poutanen, K.; Mykkänen, H.; Hanhineva, K. Amino Acid-Derived Betaines Dominate as Urinary Markers for Rye Bran Intake in Mice Fed High-Fat Diet—A Nontargeted Metabolomics Study. Mol. Nutr. Food Res. 2015, 59, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Couch, R.D.; Dailey, A.; Zaidi, F.; Navarro, K.; Forsyth, C.B.; Mutlu, E.; Engen, P.A.; Keshavarzian, A. Alcohol Induced Alterations to the Human Fecal VOC Metabolome. PLoS ONE 2015, 10, e0119362. [Google Scholar] [CrossRef]
- Pizzorno, J. Glutathione! Integr. Med. 2014, 13, 8–12. [Google Scholar]
- Dringen, R.; Hirrlinger, J. Glutathione Pathways in the Brain. Biol. Chem. 2003, 384, 505–516. [Google Scholar] [CrossRef]
- Labban, R.S.M.; Alfawaz, H.; Almnaizel, A.T.; Hassan, W.M.; Bhat, R.S.; Moubayed, N.M.; Bjørklund, G.; El-Ansary, A. High-Fat Diet-Induced Obesity and Impairment of Brain Neurotransmitter Pool. Transl. Neurosci. 2020, 11, 147–160. [Google Scholar] [CrossRef]
- Cavaliere, G.; Trinchese, G.; Penna, E.; Cimmino, F.; Pirozzi, C.; Lama, A.; Annunziata, C.; Catapano, A.; Mattace Raso, G.; Meli, R.; et al. High-Fat Diet Induces Neuroinflammation and Mitochondrial Impairment in Mice Cerebral Cortex and Synaptic Fraction. Front. Cell. Neurosci. 2019, 13, 509. [Google Scholar] [CrossRef]
- Andrich, D.E.; Melbouci, L.; Ou, Y.; Auclair, N.; Mercier, J.; Grenier, J.-C.; Lira, F.S.; Barreiro, L.B.; Danialou, G.; Comtois, A.-S.; et al. A Short-Term High-Fat Diet Alters Glutathione Levels and IL-6 Gene Expression in Oxidative Skeletal Muscles of Young Rats. Front. Physiol. 2019, 10, 372. [Google Scholar] [CrossRef]
- Norris, K.M.; Okie, W.; Kim, W.K.; Adhikari, R.; Yoo, S.; King, S.; Pazdro, R. A High-Fat Diet Differentially Regulates Glutathione Phenotypes in the Obesity-Prone Mouse Strains DBA/2J, C57BL/6J, and AKR/J. Nutr. Res. 2016, 36, 1316–1324. [Google Scholar] [CrossRef]
- Hao, X.; Huang, Y.; Qiu, M.; Yin, C.; Ren, H.; Gan, H.; Li, H.; Zhou, Y.; Xia, J.; Li, W.; et al. Immunoassay of S-Adenosylmethionine and S-Adenosylhomocysteine: The Methylation Index as a Biomarker for Disease and Health Status. BMC Res. Notes 2016, 9, 498. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Herrmann, W. Homocysteine and Lipids: S-Adenosyl Methionine as a Key Intermediate. FEBS Lett. 2009, 583, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- KEGG PATHWAY: Cysteine and Methionine Metabolism—Reference Pathway. Available online: http://www.genome.jp/kegg-bin/show_pathway?map00270+C00021 (accessed on 2 July 2017).
- Barroso, M.; Florindo, C.; Kalwa, H.; Silva, Z.; Turanov, A.A.; Carlson, B.A.; de Almeida, I.T.; Blom, H.J.; Gladyshev, V.N.; Hatfield, D.L.; et al. Inhibition of Cellular Methyltransferases Promotes Endothelial Cell Activation by Suppressing Glutathione Peroxidase 1 Protein Expression. J. Biol. Chem. 2014, 289, 15350–15362. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.B.; Bottiglieri, T.; Arning, E.; Ziegler, G.M.; Hansen, A.L.; Masliah, E. Elevated S-Adenosylhomocysteine in Alzheimer Brain: Influence on Methyltransferases and Cognitive Function. J. Neural Transm. 2004, 111, 547–567. [Google Scholar] [CrossRef] [PubMed]
- Selley, M.L. A Metabolic Link between S-Adenosylhomocysteine and Polyunsaturated Fatty Acid Metabolism in Alzheimer’s Disease. Neurobiol. Aging 2007, 28, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in Inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Zhou, R.; Chen, X.-L.; Zhou, Z.; Zhang, Y.; Lan, Q.; Liao, G.; Chen, Y.; Zhu, H. Higher Dietary Intakes of Choline and Betaine Are Associated with a Lower Risk of Primary Liver Cancer: A Case-Control Study. Sci. Rep. 2017, 7, 679. [Google Scholar] [CrossRef]
- Olthof, M.R.; van Vliet, T.; Boelsma, E.; Verhoef, P. Low Dose Betaine Supplementation Leads to Immediate and Long Term Lowering of Plasma Homocysteine in Healthy Men and Women. J. Nutr. 2003, 133, 4135–4138. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Zhao, M.; Zheng, L.; Fan, D. Changes in the Concentrations of Trimethylamine N-Oxide (TMAO) and Its Precursors in Patients with Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 15198. [Google Scholar] [CrossRef]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef]
- Yang, S.; Li, X.; Yang, F.; Zhao, R.; Pan, X.; Liang, J.; Tian, L.; Li, X.; Liu, L.; Xing, Y.; et al. Gut Microbiota-Dependent Marker TMAO in Promoting Cardiovascular Disease: Inflammation Mechanism, Clinical Prognostic, and Potential as a Therapeutic Target. Front. Pharmacol. 2019, 10, 1360. [Google Scholar] [CrossRef] [PubMed]
- Krueger, E.S.; Beales, J.L.; Elison, W.S.; Tessem1, J.S. Gut Metabolite Trimethylamine N-Oxide Protects β Cell Insulin Secretion by Reducing Oxidative Stress and Maintaining Insulin Granule Formation. Curr. Dev. Nutr. 2021, 5 (Suppl. 2), 57. [Google Scholar] [CrossRef]
- Vinnere Pettersson, O.; Leong, S.L. Fungal Xerophiles (Osmophiles). In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011. [Google Scholar] [CrossRef]
- d’Enfert, C.; Kaune, A.-K.; Alaban, L.-R.; Chakraborty, S.; Cole, N.; Delavy, M.; Kosmala, D.; Marsaux, B.; Fróis-Martins, R.; Morelli, M.; et al. The Impact of the Fungus-Host-Microbiota Interplay upon Candida albicans Infections: Current Knowledge and New Perspectives. FEMS Microbiol. Rev. 2021, 45, fuaa060. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Jiang, B.; Zhang, T.; Chen, J. Combined Mutagenesis and Metabolic Regulation to Enhance D-Arabitol Production from Candida Parapsilosis. J. Ind. Microbiol. Biotechnol. 2020, 47, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Seiboth, B.; Metz, B. Fungal Arabinan and L-Arabinose Metabolism. Appl. Microbiol. Biotechnol. 2011, 89, 1665–1673. [Google Scholar] [CrossRef]
- Heisel, T.; Montassier, E.; Johnson, A.; Al-Ghalith, G.; Lin, Y.W.; Wei, L.N. High-Fat Diet Changes Fungal Microbiomes and Interkingdom Relationships in the Murine Gut. mSphere 2017, 2, e00351-17. [Google Scholar] [CrossRef]
- Pérez, J.C. Fungi of the Human Gut Microbiota: Roles and Significance. Int. J. Med. Microbiol. 2021, 311, 151490. [Google Scholar] [CrossRef]
- Tran, D.H.; Wang, Z.V. Glucose Metabolism in Cardiac Hypertrophy and Heart Failure. J. Am. Heart Assoc. 2019, 8, e012673. Available online: https://www.ahajournals.org/doi/10.1161/JAHA.119.012673 (accessed on 11 October 2022). [CrossRef]
- KEGG PATHWAY: Tryptophan Metabolism—Homo Sapiens (Human). Available online: http://www.genome.jp/kegg-bin/show_pathway?hsa00380+1644 (accessed on 7 May 2017).
- Leong, S.C.; Sirich, T.L. Indoxyl Sulfate-Review of Toxicity and Therapeutic Strategies. Toxins 2016, 8, 358. [Google Scholar] [CrossRef]
- Kim, J.; Hong, H.; Heo, A.; Park, W. Indole Toxicity Involves the Inhibition of Adenosine Triphosphate Production and Protein Folding in Pseudomonas putida. FEMS Microbiol. Lett. 2013, 343, 89–99. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta (BBA) Biomembr. 2017, 1859 Pt B, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Perona, J.S. Membrane Lipid Alterations in the Metabolic Syndrome and the Role of Dietary Oils. Biochim. Biophys. Acta (BBA) Biomembr. 2017, 1859 Pt B, 1690–1703. [Google Scholar] [CrossRef] [PubMed]
- Maev, I.V.; Samsonov, A.A.; Palgova, L.K.; Pavlov, C.S.; Shirokova, E.N.; Vovk, E.I.; Starostin, K.M. Effectiveness of Phosphatidylcholine as Adjunctive Therapy in Improving Liver Function Tests in Patients with Non-Alcoholic Fatty Liver Disease and Metabolic Comorbidities: Real-Life Observational Study from Russia. BMJ Open Gastroenterol. 2020, 7, e000368. [Google Scholar] [CrossRef] [PubMed]
- Afshinnia, F.; Rajendiran, T.M.; Karnovsky, A.; Soni, T.; Wang, X.; Xie, D.; Yang, W.; Shafi, T.; Weir, M.P.; He, J.; et al. Lipidomic Signature of Progression of Chronic Kidney Disease in the Chronic Renal Insufficiency Cohort. Kidney Int. Rep. 2016, 1, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ge, X.; Li, X.; He, J.; Wei, X.; Du, J.; Sun, J.; Li, X.; Xun, Z.; Liu, W.; et al. High-Fat Diet Promotes Renal Injury by Inducing Oxidative Stress and Mitochondrial Dysfunction. Cell Death Dis. 2020, 11, 914. [Google Scholar] [CrossRef]
- Amaral, J.D.; Viana, R.J.S.; Ramalho, R.M.; Steer, C.J.; Rodrigues, C.M.P. Bile Acids: Regulation of Apoptosis by Ursodeoxycholic Acid. J. Lipid Res. 2009, 50, 1721–1734. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile Acid Metabolism and Signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Ferdinandusse, S.; Denis, S.; Faust, P.L.; Wanders, R.J.A. Bile Acids: The Role of Peroxisomes. J. Lipid Res. 2009, 50, 2139–2147. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Zhou, H.; Pandak, W.M.; Ren, S.; Gil, G.; Dent, P. Bile Acids as Regulatory Molecules. J. Lipid Res. 2009, 50, 1509–1520. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Shimizu, H.; Hagio, M.; Fukiya, S.; Watanabe, M.; Tanaka, Y.; Joe, G.-H.; Iwaya, H.; Yoshitsugu, R.; Kikuchi, K.; et al. 12α-Hydroxylated Bile Acid Induces Hepatic Steatosis with Dysbiosis in Rats. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2020, 1865, 158811. [Google Scholar] [CrossRef]
- Abrigo, J.; Gonzalez, F.; Aguirre, F.; Tacchi, F.; Gonzalez, A.; Meza, M.P.; Simon, F.; Cabrera, D.; Arrese, M.; Karpen, S.; et al. Cholic Acid and Deoxycholic Acid Induce Skeletal Muscle Atrophy through a Mechanism Dependent on TGR5 Receptor. J. Cell. Physiol. 2021, 236, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Azer, S.A.; Hasanato, R. Use of Bile Acids as Potential Markers of Liver Dysfunction in Humans: A Systematic Review. Medicine 2021, 100, e27464. [Google Scholar] [CrossRef] [PubMed]
- Staels, B.; Fonseca, V.A. Bile Acids and Metabolic Regulation. Diabetes Care 2009, 32 (Suppl. 2), S237–S245. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Floris, F.; Andersson, U.; Pikuleva, I.; Lövgren-Sandblom, A.; Bjerke, M.; Paucar, M.; Wallin, A.; Svenningsson, P.; Björkhem, I. 7α-Hydroxy-3-Oxo-4-Cholestenoic Acid in Cerebrospinal Fluid Reflects the Integrity of the Blood-Brain Barrier. J. Lipid Res. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Borg, A.J.E.; Dennig, A.; Weber, H.; Nidetzky, B. Mechanistic Characterization of UDP-Glucuronic Acid 4-Epimerase. FEBS J. 2021, 288, 1163–1178. [Google Scholar] [CrossRef]
- Roy-Chowdhury, J.; Roy-Chowdhury, N. 58—Bilirubin Metabolism and Its Disorders. In Zakim and Boyer’s Hepatology, 7th ed.; Sanyal, A.J., Boyer, T.D., Lindor, K.D., Terrault, N.A., Eds.; Elsevier: Philadelphia, PA, USA, 2018; pp. 898–925.e8. [Google Scholar] [CrossRef]
- Xu, J.; Kulkarni, S.R.; Li, L.; Slitt, A.L. UDP-Glucuronosyltransferase Expression in Mouse Liver Is Increased in Obesity- and Fasting-Induced Steatosis. Drug Metab. Dispos. 2012, 40, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Ghose, R.; Omoluabi, O.; Gandhi, A.; Shah, P.; Strohacker, K.; Carpenter, K.C.; McFarlin, B.; Guo, T. Role of High-Fat Diet in Regulation of Gene Expression of Drug Metabolizing Enzymes and Transporters. Life Sci. 2011, 89, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Li, S.; Yan, C.; Sun, R.; He, J.; Xie, Y.; Peng, Y.; Wang, G.; Aa, J. Inhibitory Effects of Endogenous Linoleic Acid and Glutaric Acid on the Renal Glucuronidation of Berberrubine in Mice and on Recombinant Human UGT1A7, 1A8, and 1A9. Mol. Pharmacol. 2018, 93, 216–227. [Google Scholar] [CrossRef]
- Shibuya, A.; Itoh, T.; Tukey, R.H.; Fujiwara, R. Impact of Fatty Acids on Human UDP-Glucuronosyltransferase 1A1 Activity and Its Expression in Neonatal Hyperbilirubinemia. Sci. Rep. 2013, 3, 2903. [Google Scholar] [CrossRef]
- Miller, S.G.; Hafen, P.S.; Brault, J.J. Increased Adenine Nucleotide Degradation in Skeletal Muscle Atrophy. Int. J. Mol. Sci. 2020, 21, 88. [Google Scholar] [CrossRef]
- Pan, Z.; Mao, B.; Zhang, Q.; Tang, X.; Yang, B.; Zhao, J.; Cui, S.; Zhang, H. Postbiotics Prepared Using Lactobacillus paracasei CCFM1224 Prevent Nonalcoholic Fatty Liver Disease by Modulating the Gut Microbiota and Liver Metabolism. Int. J. Mol. Sci. 2022, 23, 13522. [Google Scholar] [CrossRef] [PubMed]
- Buse, M.G. Hexosamines, Insulin Resistance and the Complications of Diabetes: Current Status. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E1–E8. [Google Scholar] [CrossRef] [PubMed]
- Fantus, I.G.; Goldberg, H.J.; Whiteside, C.I.; Topic, D. The Hexosamine Biosynthesis Pathway. In The Diabetic Kidney; Contemporary Diabetes; Humana Press: Totowa, NJ, USA, 2006; pp. 117–133. [Google Scholar] [CrossRef]
- Bond, M.R.; Hanover, J.A. O-GlcNAc Cycling: A Link between Metabolism and Chronic Disease. Annu. Rev. Nutr. 2013, 33, 205–229. [Google Scholar] [CrossRef] [PubMed]
- Maszczak-Seneczko, D.; Sosicka, P.; Olczak, T.; Jakimowicz, P.; Majkowski, M.; Olczak, M. UDP-N-Acetylglucosamine Transporter (SLC35A3) Regulates Biosynthesis of Highly Branched N-Glycans and Keratan Sulfate. J. Biol. Chem. 2013, 288, 21850–21860. [Google Scholar] [CrossRef] [PubMed]
- Toma, L.; Pinhal, M.A.; Dietrich, C.P.; Nader, H.B.; Hirschberg, C.B. Transport of UDP-Galactose into the Golgi Lumen Regulates the Biosynthesis of Proteoglycans. J. Biol. Chem. 1996, 271, 3897–3901. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.B.; Kim, J.; Cartee, G.D. Prolonged Incubation in PUGNAc Results in Increased Protein O-Linked Glycosylation and Insulin Resistance in Rat Skeletal Muscle. Diabetes 2004, 53, 921–930. [Google Scholar] [CrossRef]
- Silva-Aguiar, R.P.; Peruchetti, D.B.; Pinheiro, A.A.S.; Caruso-Neves, C.; Dias, W.B. O-GlcNAcylation in Renal (Patho)Physiology. Int. J. Mol. Sci. 2022, 23, 11260. [Google Scholar] [CrossRef]
- Pena, I.A.; Marques, L.A.; Laranjeira, Â.B.A.; Yunes, J.A.; Eberlin, M.N.; MacKenzie, A.; Arruda, P. Mouse Lysine Catabolism to Aminoadipate Occurs Primarily through the Saccharopine Pathway; Implications for Pyridoxine Dependent Epilepsy (PDE). Biochim. Biophys. Acta 2017, 1863, 121–128. [Google Scholar] [CrossRef]
- Papes, F.; Kemper, E.L.; Cord-Neto, G.; Langone, F.; Arruda, P. Lysine Degradation through the Saccharopine Pathway in Mammals: Involvement of Both Bifunctional and Monofunctional Lysine-Degrading Enzymes in Mouse. Biochem. J. 1999, 344, 555–563. [Google Scholar] [CrossRef]
- Gatrell, S.K.; Berg, L.E.; Barnard, J.T.; Grimmett, J.G.; Barnes, K.M.; Blemings, K.P. Tissue Distribution of Indices of Lysine Catabolism in Growing Swine. J. Anim. Sci. 2013, 91, 238–247. [Google Scholar] [CrossRef]
- KEGG PATHWAY: Lysine Degradation—Reference Pathway. Available online: http://www.genome.jp/kegg-bin/show_pathway?map00310+C00449 (accessed on 2 July 2017).
- Galili, G.; Tang, G.; Zhu, X.; Gakiere, B. Lysine Catabolism: A Stress and Development Super-Regulated Metabolic Pathway. Curr. Opin. Plant Biol. 2001, 4, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Makide, K.; Kitamura, H.; Sato, Y.; Okutani, M.; Aoki, J. Emerging Lysophospholipid Mediators, Lysophosphatidylserine, Lysophosphatidylthreonine, Lysophosphatidylethanolamine and Lysophosphatidylglycerol. Prostaglandins Other Lipid Mediat. 2009, 89, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.T.; Ramesh, T.; Toh, X.R.; Nguyen, L.N. Emerging Roles of Lysophospholipids in Health and Disease. Prog. Lipid Res. 2020, 80, 101068. [Google Scholar] [CrossRef]
- Tan, S.H.; Koh, H.W.L.; Chua, J.Y.; Burla, B.; Ong, C.C.; Teo, L.S.L.; Yang, X.; Benke, P.I.; Choi, H.; Torta, F.; et al. Variability of the Plasma Lipidome and Subclinical Coronary Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 100–112. [Google Scholar] [CrossRef]
- Wang, Z.; Peters, B.A.; Usyk, M.; Xing, J.; Hanna, D.B.; Wang, T.; Post, W.S.; Landay, A.L.; Hodis, H.N.; Weber, K.; et al. Gut Microbiota, Plasma Metabolomic Profiles, and Carotid Artery Atherosclerosis in HIV Infection. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Schober, A.; Siess, W. Lysophosphatidic Acid in Atherosclerotic Diseases. Br. J. Pharmacol. 2012, 167, 465–482. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Sakurai, T.; Chen, Z.; Inoue, N.; Chiba, H.; Hui, S.-P. Lysophosphatidylethanolamine Affects Lipid Accumulation and Metabolism in a Human Liver-Derived Cell Line. Nutrients 2022, 14, 579. [Google Scholar] [CrossRef]
- Chorell, E.; Olsson, T.; Jansson, J.-H.; Wennberg, P. Lysophospholipids as Predictive Markers of ST-Elevation Myocardial Infarction (STEMI) and Non-ST-Elevation Myocardial Infarction (NSTEMI). Metabolites 2020, 11, 25. [Google Scholar] [CrossRef]
- Gilbert, M.S.; Ijssennagger, N.; Kies, A.K.; van Mil, S.W.C. Protein Fermentation in the Gut; Implications for Intestinal Dysfunction in Humans, Pigs, and Poultry. Am. J. Physiol.-Gastrointest. Liver Physiol. 2018, 315, G159–G170. [Google Scholar] [CrossRef]

| Tissue | Metabolite Name | log2(Fold Change) | −log10(p-Value) | Frequency: Basal (n = 5) | Frequency: Higher-Fat (n = 5) |
|---|---|---|---|---|---|
| Brain Cortex | S-adenosylhomocysteine | 18.99 | 1.77 | 0 | 3 |
| Brain Cortex | Glutathione | −21.95 | 1.90 | 4 | 0 |
| Heart | Arabitol | −17.62 | 2.12 | 3 | 0 |
| Heart | Betaine | −17.19 | 1.45 | 4 | 0 |
| Kidney | 1-(Octadecenoyl)-sn-glycero-3-phosphoethanolamine | 15.31 | 2.27 | 0 | 3 |
| Kidney | Indoxyl sulfate | −17.57 | 1.35 | 3 | 0 |
| Liver | 1-(Octadecenoyl)-sn-glycero-3-phosphoethanolamine | 14.88 | 2.06 | 0 | 4 |
| Liver | Glycocholic acid | 4.73 | 1.66 | 1 | 5 |
| Liver | 1-O-Hexadecyl-sn-glycero-3-phosphocholine | −17.12 | 1.92 | 4 | 0 |
| Skeletal Muscle | Inosine | 19.55 | 1.64 | 0 | 4 |
| Skeletal Muscle | Uridine diphosphate glucuronic acid | 15.00 | 1.65 | 0 | 4 |
| Skeletal Muscle | 7α-Hydroxy-3-oxo-4-cholestenoic acid | 1.85 | 1.81 | 4 | 5 |
| Skeletal Muscle | 1-(Octadecenoyl)-sn-glycero-3-phosphoethanolamine | 1.83 | 1.46 | 1 | 5 |
| Pancreas | 1-(Octadecenoyl)-sn-glycero-3-phosphoethanolamine | 2.38 | 1.42 | 1 | 5 |
| Pancreas | Trimethylamine N-oxide | −2.64 | 1.54 | 5 | 4 |
| Pancreas | Betaine | −2.45 | 1.85 | 5 | 3 |
| Dietary Comparisions | ||||||
|---|---|---|---|---|---|---|
| Basal versus Basal+probiotic | Tissue | Metabolite Name | log2(Fold Change) | −log10(p-Value) | Frequency: Basal (n = 5) | Frequency: Basal+Probiotic (n = 5) |
| Brain Cortex | Phosphatidylethanolamine lyso alkenyl 18:1 | 15.79 | 3.13 | 3 | 0 | |
| Brain Cortex | 1-Octadecanoyl-sn-glycero-3-phosphocholine | 3.28 | 1.39 | 3 | 3 | |
| Brain Cortex | Taurine | −18.63 | 7.75 | 0 | 3 | |
| Heart | 6-amino-2-[[3-methyl-2-[[pyrrolidine-2-carbonyl]amino]butanoyl]amino]hexanoic acid | 14.90 | 2.02 | 4 | 0 | |
| Heart | (4-amino-4,6-dimethyl-5-sulfooxy-tetrahydropyran-2-yl) [hydroxy-[[3-hydroxy-5-(5-methyl-2,4-dioxo-pyrimidin-1-yl)tetrahydrofuran-2-yl]methoxy]phosphoryl] hydrogen phosphate | 1.69 | 1.32 | 5 | 3 | |
| Heart | tert-butyl N-[1-[[1-cyclohexyl-2-hydroxy-2-[6-(phenylcarbamoyl)-3,3a,4,5,6,6a-hexahydro-2H-pyrrolo [3,2-b]pyrrol-1-yl]ethyl]amino]-1-oxopropan-2-yl]-N-methylcarbamate | −15.54 | 3.10 | 0 | 4 | |
| Kidney | Uridine diphosphate-N-acetylglucosamine * | 22.1 | 4.62 | 4 | 0 | |
| Kidney | Urothion | 15.62 | 4.40 | 4 | 0 | |
| Kidney | Uridine 5′-diphosphogalactose | −14.86 | 6.61 | 0 | 3 | |
| Liver | Saccharopine * | 16.94 | 4.51 | 4 | 0 | |
| Liver | LysoPC(18:0) | 15.81 | 4.14 | 3 | 0 | |
| Liver | [5-(2,6-dihydroxy-2,3-dihydropurin-9-yl)-3,4-dihydroxy-tetrahydrofuran-2-yl]methyl phosphono hydrogen phosphate | −18.17 | 3.34 | 0 | 3 | |
| Skeletal Muscle | 1-O-Hexadecyl-2-O-acetyl-sn-glyceryl-3-phosphorylcholine | 20.67 | 2.86 | 3 | 0 | |
| Skeletal Muscle | 4-Hydroxy-3-methoxymandelic acid | 15.14 | 3.35 | 3 | 0 | |
| Skeletal Muscle | Uridine 5′-diphosphogalactose | −15.27 | 1.75 | 0 | 3 | |
| Pancreas | N-(2-aminoacetyl)-1-(4-phenyldiazenylphenyl)pyrrolidine-2-carboxamide | 18.39 | 4.39 | 3 | 0 | |
| Pancreas | 4-hydroxypyrrolidine-2-carbaldehyde | −6.04 | 2.69 | 5 | 5 | |
| Pancreas | Guanosine diphosphate mannose | −15.39 | 3.52 | 0 | 3 | |
| Higher-fat versus Higher-fat+probiotic | Tissue | Metabolite Name | log2(Fold Change) | −log10(p-value) | Frequency: Higher-fat (n = 5) | Frequency: Higher-fat+probiotic (n = 5) |
| Brain Cortex | S-adenosylhomocysteine * | 18.99 | 3.74 | 3 | 0 | |
| Brain Cortex | 1-Stearoyl-2-hydroxy-sn-glycero-3-phosphocholine | 18.39 | 1.40 | 3 | 0 | |
| Brain Cortex | 5,6,7,8-Tetrahydromethanopterin | −18.71 | 5.05 | 0 | 3 | |
| Heart | 2′-α-mannosyl-L-tryptophan | 15.31 | 6.63 | 3 | 0 | |
| Heart | N’-{[8-({5-[2-(2-aminopyridin-4-yl)ethyl]-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl}oxy)-1-hydroxy-5-(hydroxymethyl)-6-methoxy-9,10-dioxo-3-(2-phenylethyl)-9,10-dihydroanthracen-2-yl]methyl}-N-methylguanidine | 15.56 | 7.09 | 3 | 0 | |
| Heart | 3,4,5-Trinitrofuran-2-thiol | −16.58 | 3.45 | 0 | 3 | |
| Kidney | Uridine diphosphate-N-acetylglucosamine * | 22.45 | 8.07 | 3 | 0 | |
| Kidney | 3-sulfolactic acid | −16.66 | 3.92 | 0 | 3 | |
| Kidney | Indoxyl sulfate * | −19.18 | 2.79 | 0 | 3 | |
| Liver | Adenosine monophosphate | 20.96 | 2.21 | 3 | 0 | |
| Liver | Phosphodimethylethanolamine | −19.45 | 3.73 | 0 | 4 | |
| Liver | 6-(7-hydroxy-4,6-dimethylhepta-2,4-dien-2-yl)-4-methoxy-5-methyl-2H-pyran-2-one | −22.02 | 5.30 | 0 | 4 | |
| Skeletal Muscle | Inosine * | 19.55 | 2.79 | 4 | 0 | |
| Skeletal Muscle | Xylitol | 16.88 | 4.37 | 4 | 0 | |
| Skeletal Muscle | 2-[[5-acetamido-6-(2-amino-2-carboxy-1-methyl-ethoxy)-3,4-dihydroxy-tetrahydropyran-2-yl]methoxy]-4-hydroxy-5-[[2-(2-methoxyethoxycarbonylamino)acetyl]amino]-6-(1,2,3-trihydroxypropyl)tetrahydropyran-2-carboxylic acid | −14.75 | 4.02 | 0 | 3 | |
| Pancreas | LysoPE(20:1) | 20.76 | 2.94 | 4 | 0 | |
| Pancreas | Guanosine monophosphate | 17.74 | 5.19 | 3 | 0 | |
| Pancreas | N-({6-[(5-tert-butyl-1,2-oxazol-3-yl)methyl]-3-hydroxyoxan-2-yl}methyl)propanamide | −17.82 | 4.60 | 0 | 3 | |
| Tissue | Metabolite Name | log2(Fold Change) Higher-Fat vs. Basal | log2(Fold-Change) Higher-Fat+Probiotic vs. Basal | Frequency: Higher-Fat (n = 5) | Frequency: Basal (n = 5) | Frequency: Higher-Fat+Probiotic (n = 5) |
|---|---|---|---|---|---|---|
| Brain Cortex | S-Adenosylhomocysteine * | 19.00 | 0.00 | 3 | 0 | 0 |
| Brain Cortex | Tryptophan | 4.58 | −0.24 | 3 | 3 | 4 |
| Brain Cortex | 4-(Methylsulfanyl)-2-oxobutanoic acid | 3.91 | −0.46 | 3 | 3 | 3 |
| Brain Cortex | LysoPC(22:4) | −14.06 | 0.04 | 0 | 4 | 3 |
| Brain Cortex | 2-[[2-(hexadecanoylamino)acetyl]amino]-3-(1H-imidazol-5-yl)propanoic acid | −14.67 | 0.01 | 0 | 3 | 3 |
| Brain Cortex | LysoPC(22:6) | −21.26 | −0.21 | 0 | 3 | 3 |
| Brain Cortex | Glutathione * | −21.95 | 0.16 | 0 | 4 | 2 |
| Heart | LysoPE(18:2) | 14.36 | 0.00 | 4 | 0 | 0 |
| Kidney | LysoPC(16:1) | 2.04 | −0.19 | 3 | 2 | 3 |
| Kidney | Indoxyl sulfate * | −17.57 | 1.61 | 0 | 3 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dailey, A.; Solano-Aguilar, G.; Urban, J.F., Jr.; Couch, R.D. LC-QToF-Based Metabolomics Identifies Aberrant Tissue Metabolites Associated with a Higher-Fat Diet and Their ‘Reversion to Healthy’ with Dietary Probiotic Supplementation. Metabolites 2023, 13, 358. https://doi.org/10.3390/metabo13030358
Dailey A, Solano-Aguilar G, Urban JF Jr., Couch RD. LC-QToF-Based Metabolomics Identifies Aberrant Tissue Metabolites Associated with a Higher-Fat Diet and Their ‘Reversion to Healthy’ with Dietary Probiotic Supplementation. Metabolites. 2023; 13(3):358. https://doi.org/10.3390/metabo13030358
Chicago/Turabian StyleDailey, Allyson, Gloria Solano-Aguilar, Joseph F. Urban, Jr., and Robin D. Couch. 2023. "LC-QToF-Based Metabolomics Identifies Aberrant Tissue Metabolites Associated with a Higher-Fat Diet and Their ‘Reversion to Healthy’ with Dietary Probiotic Supplementation" Metabolites 13, no. 3: 358. https://doi.org/10.3390/metabo13030358
APA StyleDailey, A., Solano-Aguilar, G., Urban, J. F., Jr., & Couch, R. D. (2023). LC-QToF-Based Metabolomics Identifies Aberrant Tissue Metabolites Associated with a Higher-Fat Diet and Their ‘Reversion to Healthy’ with Dietary Probiotic Supplementation. Metabolites, 13(3), 358. https://doi.org/10.3390/metabo13030358






