Plasma Metabolite Signatures in Male Carriers of Genetic Variants Associated with Non-Alcoholic Fatty Liver Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Clinical and Laboratory Measurements
2.3. Metabolomics Analysis
2.4. Calculations
2.5. Genetic Analysis
2.6. Statistical Analysis
3. Results
3.1. Clinical and Laboratory Measurements
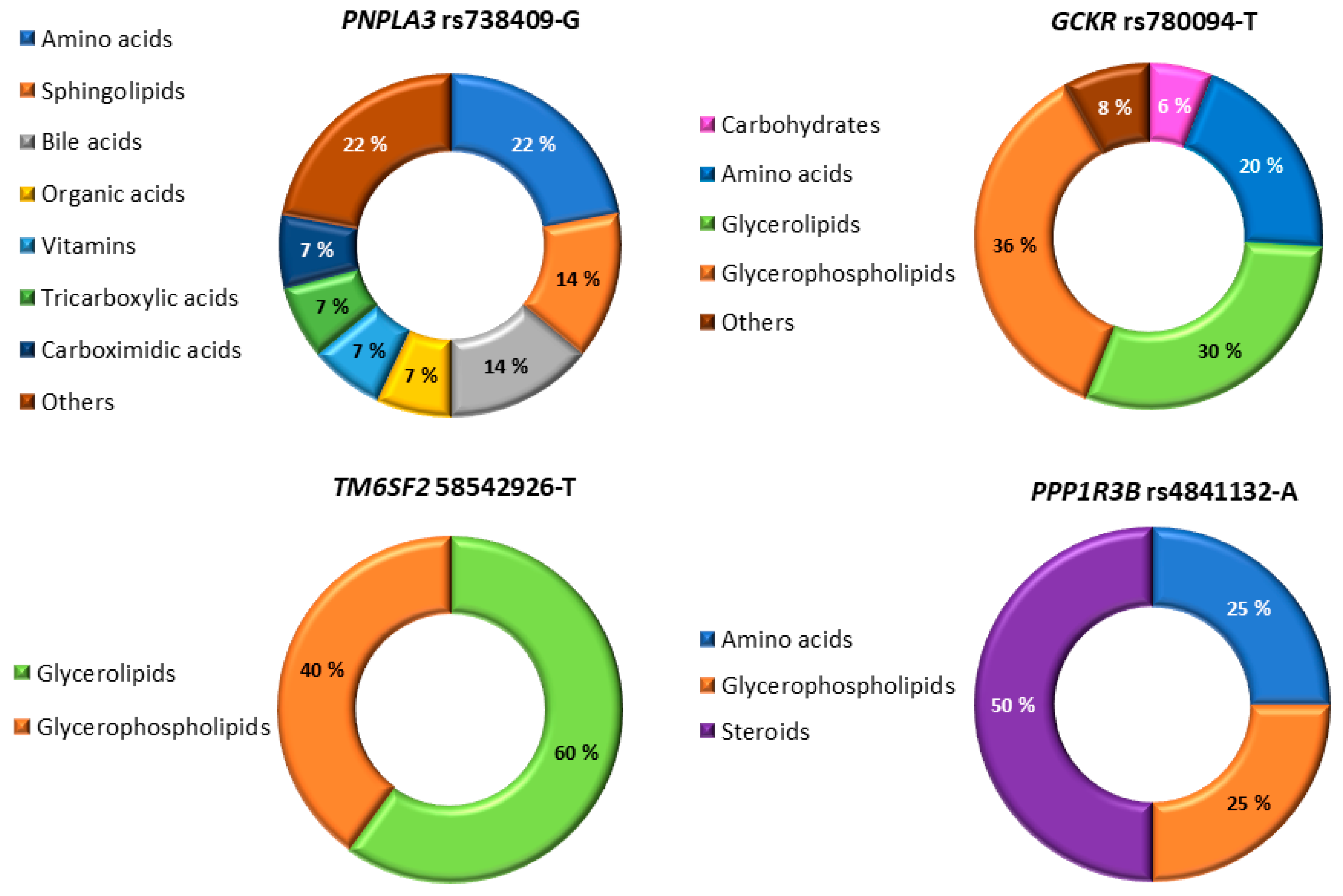
3.2. Association of the Six NAFDL Genetic Risk Variants with Metabolites
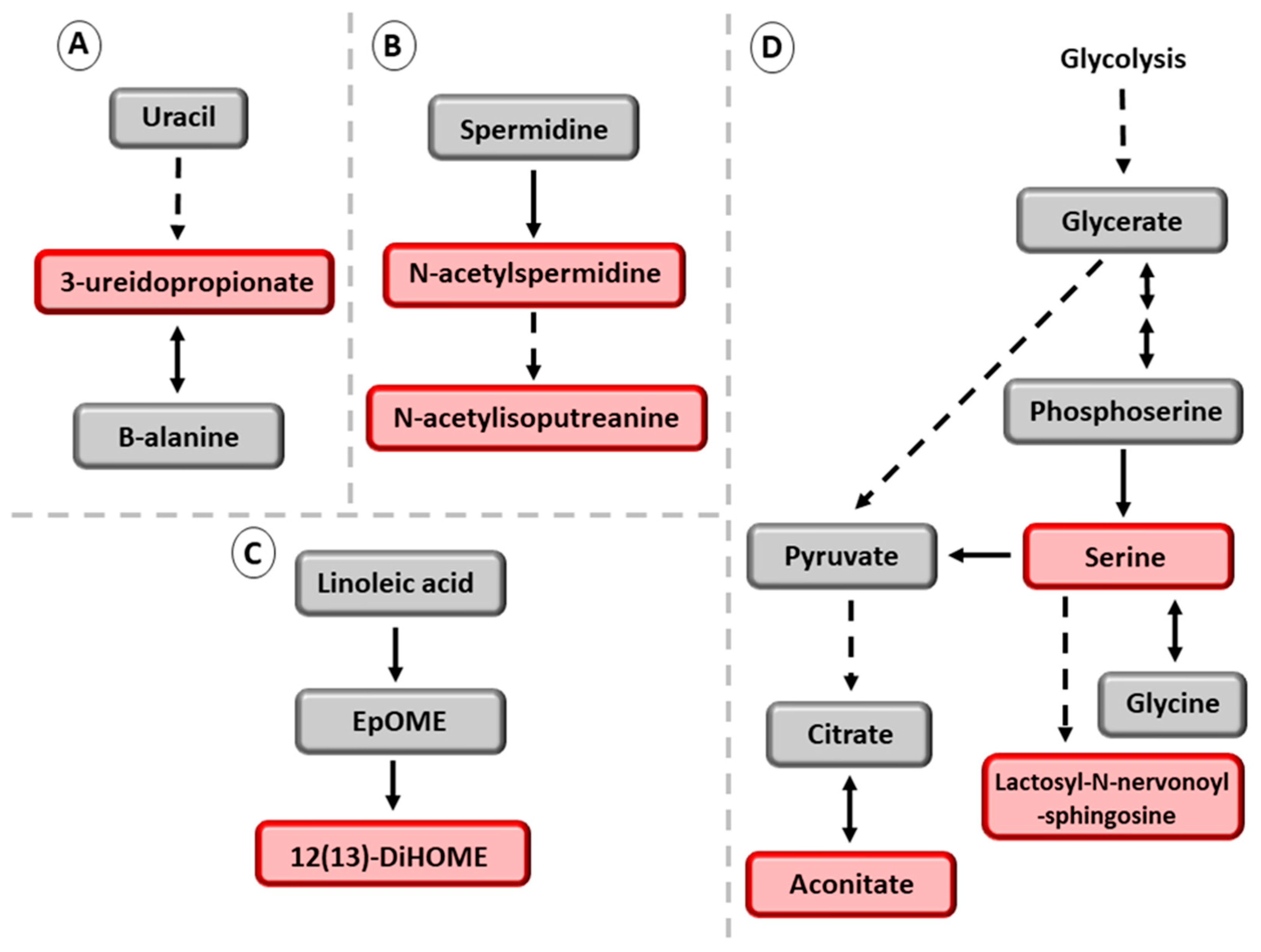
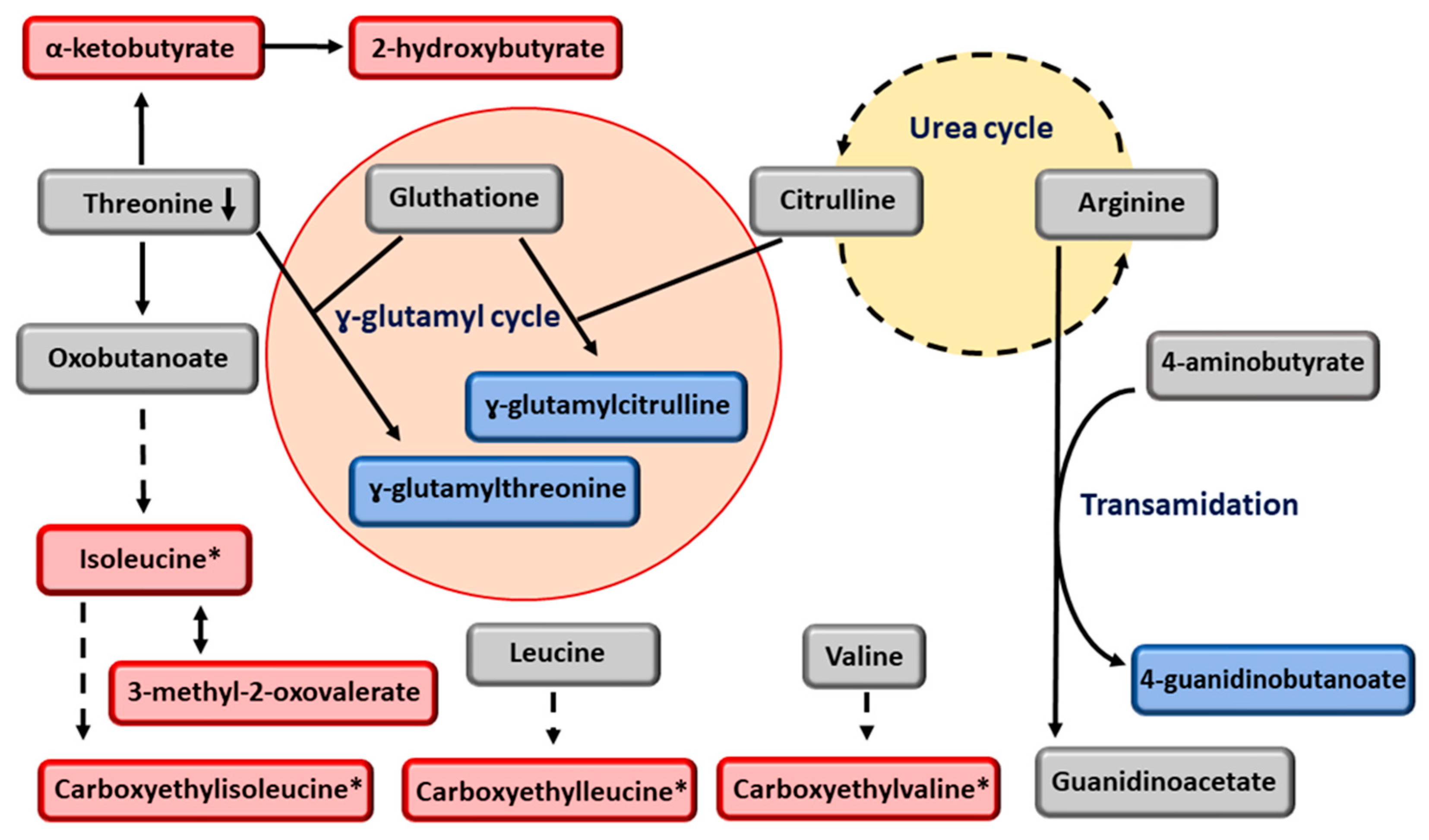
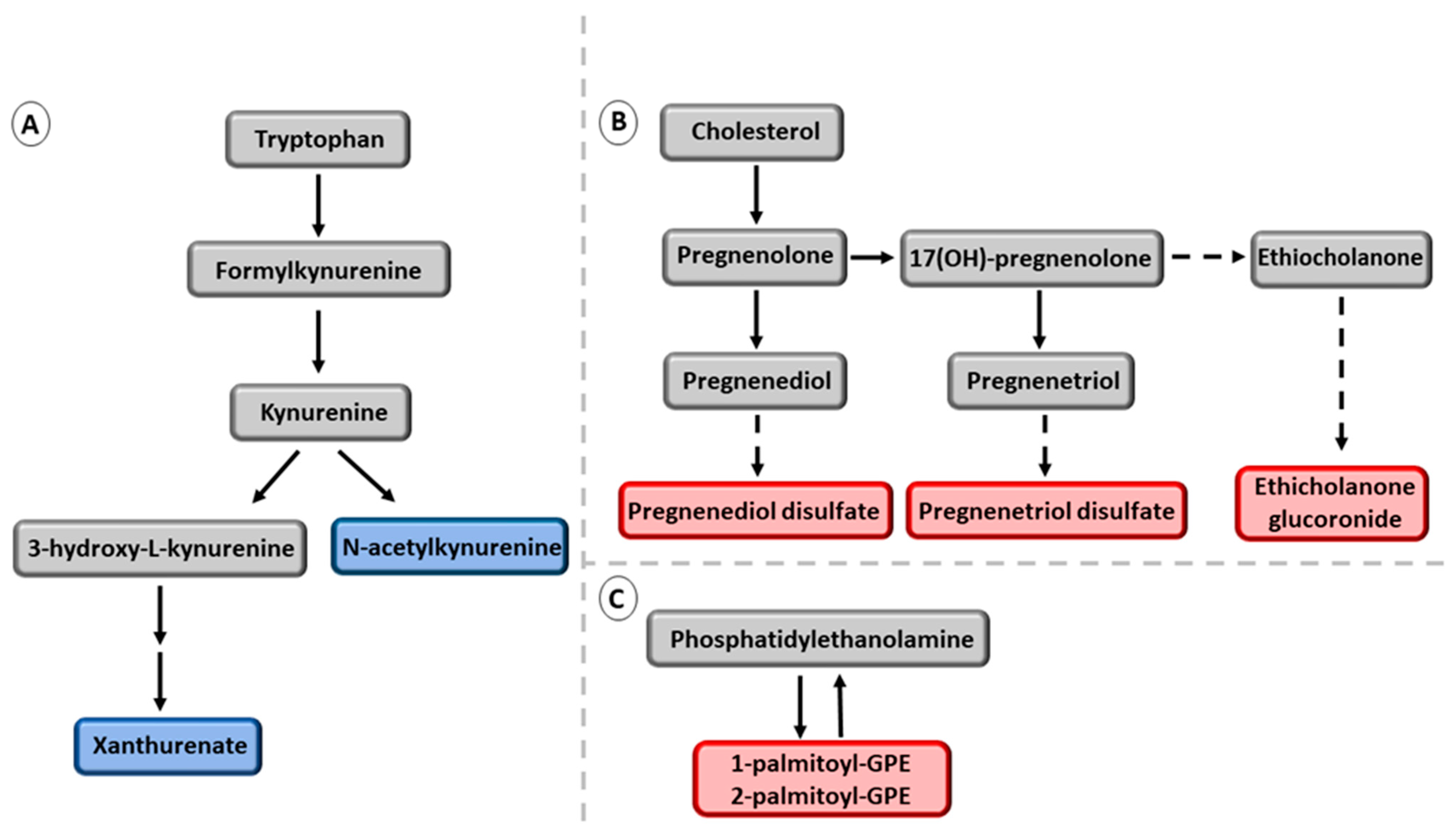
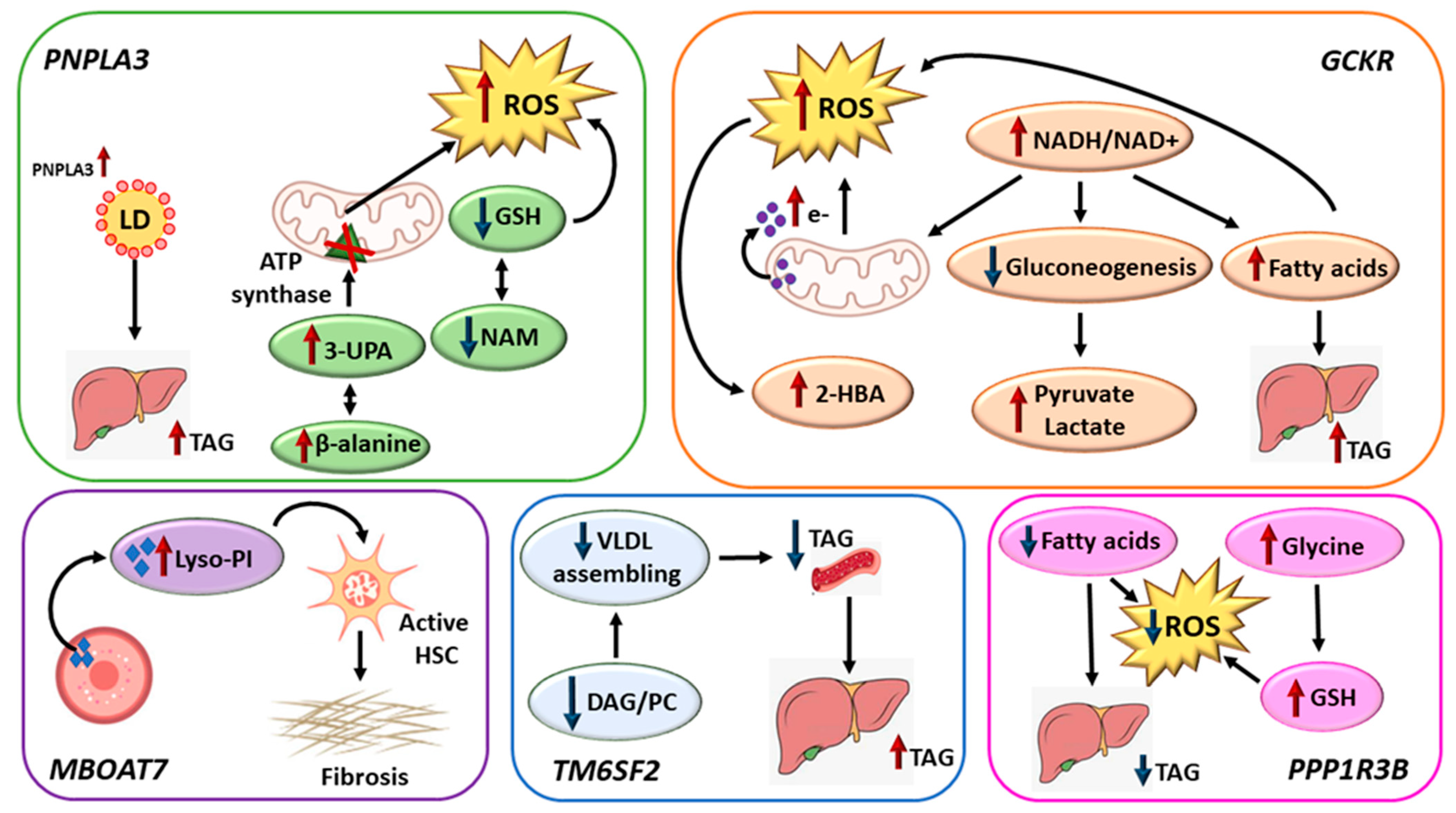
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Adams, L.A.; Canbay, A.; Syn, W.-K. Extrahepatic complications of nonalcoholic fatty liver disease. Hepatology 2014, 59, 1174–1197. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Tabibian, J.H.; Ekstedt, M.; Kechagias, S.; Hamaguchi, M.; Hultcrantz, R.; Hagström, H.; Yoon, S.K.; Charatcharoenwitthaya, P.; et al. Association of non-alcoholic fatty liver disease with chronic kidney disease: A systematic review and meta-analysis. PLoS Med. 2014, 11, e1001680. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Tilg, H.; Byrne, C.D. Non-alcoholic fatty liver disease: A multisystem disease requiring a multidisciplinary and holistic approach. Lancet Gastroenterol. Hepatol. 2021, 6, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Rotman, Y.; Koh, C.; Zmuda, J.M.; Kleiner, D.E.; Liang, T.J.; NASH CRN. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 2010, 52, 894–903. [Google Scholar] [CrossRef]
- Speliotes, E.K.; Yerges-Armstrong, L.M.; Wu, J.; Hernaez, R.; Kim, L.J.; Palmer, C.D.; Gudnason, V.; Eiriksdottir, G.; Garcia, M.E.; Launer, L.J.; et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011, 7, e1001324. [Google Scholar] [CrossRef]
- Smagris, E.; BasuRay, S.; Li, J.; Huang, Y.; Lai, K.M.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Pnpla3I148M knock-in mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015, 61, 108–118. [Google Scholar] [CrossRef]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjærg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef]
- Buch, S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- Mancina, R.M.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Borén, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of european descent. Gastroenterology 2016, 150, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wen, J.; Tao, X.; Lu, B.; Du, Y.; Wang, M.; Wang, X.; Zhang, W.; Gong, W.; Ling, C.; et al. Genetic variation in the GCKR gene is associated with non-alcoholic fatty liver disease in Chinese people. Mol. Biol. Rep. 2011, 38, 1145–1150. [Google Scholar] [CrossRef]
- Hernaez, R.; McLean, J.; Lazo, M.; Brancati, F.L.; Hirschhorn, J.N.; Borecki, I.B.; Harris, T.B.; Genetics of obesity-related liver dsease (GOLD) Consortium; Nguyen, T.; Kamel, I.R.; et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third national health and nutrition examination survey. Clin. Gastroenterol. Hepatol. 2013, 11, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Wang, Y.; Jia, X.; Wu, W.; Li, L.; Tian, X.; Li, S.; Wang, C.; Xu, H.; Cao, J.; et al. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11437–11442. [Google Scholar] [CrossRef] [PubMed]
- Agius, L. Role of glycogen phosphorylase in liver glycogen metabolism. Mol. Asp. Med. 2015, 46, 34–45. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef] [PubMed]
- Sonsuz, A.; Basaranoglu, M.; Ozbay, G. Relationship between aminotransferase levels and histopathological findings in patients with nonalcoholic steatohepatitis. Am. J. Gastroenterol. 2000, 95, 1370–1371. [Google Scholar] [CrossRef]
- Barata, L.; Feitosa, M.F.; Bielak, L.F.; Halligan, B.; Baldridge, A.S.; Guo, X.; Yerges-Armstrong, L.M.; Smith, A.V.; Yao, J.; Palmer, N.D.; et al. Insulin resistance exacerbates genetic predisposition to nonalcoholic fatty liver disease in individuals without diabetes. Hepatol. Commun. 2019, 3, 894–907. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Qadri, S.; Ahlholm, N.; Porthan, K.; Männistö, V.; Sammalkorpi, H.; Penttilä, A.K.; Hakkarainen, A.; Lehtimäki, T.E.; Gaggini, M.; et al. Distinct contributions of metabolic dysfunction and genetic risk factors in the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2022, 76, 526–535. [Google Scholar] [CrossRef]
- Bedogni, G.; Kahn, H.S.; Bellentani, S.; Tiribelli, C. A simple index of lipid overaccumulation is a good marker of liver steatosis. BMC Gastroenterol. 2010, 10, 98. [Google Scholar] [CrossRef]
- Laakso, M.; Kuusisto, J.; Stančáková, A.; Kuulasmaa, T.; Pajukanta, P.; Lusis, A.J.; Collins, F.S.; Mohlke, K.L.; Boehnke, M. The Metabolic Syndrome in Men study: A resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 2017, 58, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Stancáková, A.; Javorský, M.; Kuulasmaa, T.; Haffner, S.M.; Kuusisto, J.; Laakso, M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes 2009, 58, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2013, 36, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, M.; DeFronzo, R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing, Comparison with the euglycemic insulin clamp. Diabetes Care 1999, 22, 1462–1470. [Google Scholar] [CrossRef]
- Stancáková, A.; Civelek, M.; Saleem, N.K.; Soininen, P.; Kangas, A.J.; Cederberg, H.; Paananen, J.; Pihlajamäki, J.; Bonnycastle, L.L.; Morken, M.A.; et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9369 Finnish men. Diabetes 2012, 61, 1895–1902. [Google Scholar] [CrossRef]
- Altman, D.G.; Gardner, M.J. Statistics in medicine, calculating confidence intervals for regression and correlation. Br. Med. J. 1988, 296, 1238–1242. [Google Scholar] [CrossRef]
- BasuRay, S.; Wang, Y.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. Accumulation of PNPLA3 on lipid droplets is the basis of as-sociated hepatic steatosis. Proc. Natl. Acad. Sci. USA 2019, 116, 9521–9526. [Google Scholar] [CrossRef]
- Mann, J.P.; Pietzner, M.; Wittemans, L.B.; Rolfe, E.L.; Kerrison, N.D.; Imamura, F.; Forouhi, N.G.; Fauman, E.; Allison, M.E.; Griffin, J.L.; et al. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum. Mol. Genet. 2020, 29, 3451–3463. [Google Scholar] [CrossRef]
- Le, T.T.; Ziemba, A.; Urasaki, Y.; Hayes, E.; Brotman, S.; Pizzorno, G. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. J. Lipid Res. 2013, 54, 1044–1057. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2008, 37, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB, the Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Kölker, S.; Okun, J.G.; Hörster, F.; Assmann, B.; Ahlemeyer, B.; Kohlmüller, D.; Exner-Camps, S.; Mayatepek, E.; Krieglstein, J.; Hoffmann, G.F. 3-Ureidopropionate contributes to the neuropathology of 3-ureidopropionase deficiency and severe propionic aciduria: A hypothesis. J. Neurosci. Res. 2001, 66, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Carreras, M.; Del Hoyo, P.; Martín, M.A.; Rubio, J.C.; Martín, A.; Castellano, G.; Colina, F.; Arenas, J.; Solis-Herruzo, J.A. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 2003, 38, 999–1007. [Google Scholar] [CrossRef]
- Santamaria, E.; Avila, M.A.; Latasa, M.U.; Rubio, A.; Martin-Duce, A.; Lu, S.C.; Mato, J.M.; Corrales, F.J. Functional pro-teomics of nonalcoholic steatohepatitis, mitochondrial proteins as targets of S-adenosylmethionine. Proc. Natl. Acad. Sci. USA 2003, 100, 3065–3070. [Google Scholar] [CrossRef]
- Dasarathy, S.; Yang, Y.; McCullough, A.J.; Marczewski, S.; Bennett, C.; Kalhan, S.C. Elevated hepatic fatty acid oxidation, high plasma fibroblast growth factor 21, and fasting bile acids in nonalcoholic steatohepatitis. Eur. J. Gastroenterol. Hepatol. 2011, 23, 382–388. [Google Scholar] [CrossRef]
- Masoodi, M.; Gastaldelli, A.; Hyötyläinen, T.; Arretxe, E.; Alonso, C.; Gaggini, M.; Brosnan, J.; Anstee, Q.M.; Millet, O.; Ortiz, P.; et al. Metabolomics and lipidomics in NAFLD, biomarkers and non-invasive diagnostic tests. Nat. Rev. Gastroenterol. Hepatol. 2021, 23, 382–388. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Zhou, Y.; Nidhina Haridas, P.A.; Dwivedi, O.P.; Hyötyläinen, T.; Ali, A.; Juuti, A.; Leivonen, M.; Tukiainen, T.; Ahonen, L.; et al. Impaired hepatic lipid synthesis from polyunsaturated fatty acids in TM6SF2 E167K variant carriers with NAFLD. J. Hepatol. 2017, 67, 128–136. [Google Scholar] [CrossRef]
- Israelsen, M.; Juel, H.B.; Detlefsen, S.; Madsen, B.S.; Rasmussen, D.N.; Larssn, T.R.; Kjærgaard, M.; Fernandes Jensen, M.J.; Stender, S.; Hansen, T.; et al. Metabolic and genetic risk factors are the strongest predictors of Severity of alcohol-related liver fibrosis. Clin. Gastroenterol. Hepatol. 2022, 20, 1784-1794.e9. [Google Scholar] [CrossRef]
- Shindou, H.; Hishikawa, D.; Harayama, T.; Yuki, K.; Shimizu, T. Recent progress on acyl CoA, lysophospholipid acyltrans-ferase research. J. Lipid. Res. 2009, 50, S46–S51. [Google Scholar] [CrossRef]
- Loomba, R.; Quehenberger, O.; Armando, A.; Dennis, E.A. Polyunsaturated fatty acid metabolites as novel lipidomic bi-omarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J. Lipid. Res. 2015, 56, 185–192. [Google Scholar] [CrossRef]
- Helsley, R.N.; Varadharajan, V.; Brown, A.L.; Gromovsky, A.D.; Schugar, R.C.; Ramachandiran, I.; Fung, K.; Kabbany, M.N.; Banerjee, R.; Neumann, C.K.; et al. Obesity-linked suppression of membrane-bound O-acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. Elife 2019, 8, e49882. [Google Scholar] [CrossRef] [PubMed]
- Fondevila, M.F.; Fernandez, U.; Gonzalez-Rellan, M.J.; Da Silva Lima, N.; Buque, X.; Gonzalez-Rodriguez, A.; Alonso, C.; Iruarrizaga-Lejarreta, M.; Delgado, T.C.; Varela-Rey, M.; et al. The L-α-Lysophosphatidylinositol/G Protein-Coupled Receptor 55 System induces the development of nonalcoholic steatosis and steatohepatitis. Hepatology 2021, 73, 606–624. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.A.; Freeze, H.H. Studies of mannose metabolism and effects of long-term mannose ingestion in the mouse. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2001, 1528, 116–126. [Google Scholar] [CrossRef]
- Taguchi, T.; Yamashita, E.; Mizutani, T.; Nakajima, H.; Yabuuchi, M.; Asano, N.; Miwa, I. Hepatic glycogen breakdown is implicated in the maintenance of plasma mannose concentration. Am. J. Physiol. Metab. 2005, 288, E534–E540. [Google Scholar] [CrossRef] [PubMed]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metab. Clin. Exp. 2011, 60, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.F.; Vangipurapu, J.; Kuulasmaa, T.; Laakso, M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Goodman, R.P.; Markhard, A.L.; Shah, H.; Sharma, R.; Skinner, O.S.; Clish, C.B.; Deik, A.; Patgiri, A.; Hsu, Y.-H.H.; Masia, R.; et al. Hepatic NADH reductive stress underlies common variation in metabolic traits. Nature 2020, 583, 122–126. [Google Scholar] [CrossRef]
- Hwang, E.S. Pharmacological Nicotinamide, Mechanisms centered around SIRT1 activity. In Translational Epigenetics, Pharmacoepigenetics; Academic Press: Cambridge, MA, USA, 2019; Volume 10, pp. 781–799. [Google Scholar]
- Lake, A.D.; Novak, P.; Shipkova, P.; Aranibar, N.; Robertson, D.G.; Reily, M.D.; Lehman-McKeeman, L.D.; Vaillancourt, R.R.; Cherrington, N.J. Branched chain amino acid metabolism profiles in progressive human nonalcoholic fatty liver disease. Amino Acids 2015, 47, 603–615. [Google Scholar] [CrossRef]
- Orlowski, M.; Meister, A. The gamma-glutamyl cycle, a possible transport system for amino acids. Proc. Natl. Acad. Sci. USA 1970, 67, 1248–1255. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2017, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Shigiyama, F.; Kumashiro, N.; Furukawa, Y.; Funayama, T.; Takeno, K.; Wakui, N.; Ikehara, T.; Nagai, H.; Taka, H.; Fujimura, T.; et al. Characteristics of hepatic insulin-sensitive nonalcoholic fatty liver disease. Hepatol. Commun. 2017, 1, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Teslovich, T.M.; Kim, D.S.; Yin, X.; Stancáková, A.; Jackson, A.U.; Wielscher, M.; Naj, A.; Perry, J.R.B.; Huyghe, J.R.; Stringham, H.M.; et al. Identification of seven novel loci associated with amino acid levels using single-variant and gene-based tests in 8545 Finnish men from the METSIM study. Hum. Mol. Genet. 2018, 27, 1664–1674. [Google Scholar] [CrossRef]
- Zuo, H.; Ueland, P.M.; Ulvik, A.; Eussen, S.J.; Vollset, S.E.; Nygård, O.; Midttun, Ø.; Theofylaktopoulou, D.; Meyer, K.; Tell, G.S. Plasma biomarkers of inflammation, the kynurenine pathway, and risks of all-cause, cancer, and cardiovascular disease mortality, The Hordaland Health Study. Am. J. Epidemiol. 2016, 183, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Luukkonen, P.K.; Tukiainen, T.; Juuti, A.; Sammalkorpi, H.; Haridas, P.A.N.; Niemelä, O.; Arola, J.; Orho-Melander, M.; Hakkarainen, A.; Kovanen, P.T.; et al. Hydroxysteroid 17-β dehydrogenase 13 variant increases phospholipids and protects against fibrosis in nonalcoholic fatty liver disease. J. Clin. Investig. 2020, 5, e132158. [Google Scholar] [CrossRef]
| Men without NAFLD (n = 2339) | Men with NAFLD (n = 2535) | ||||
|---|---|---|---|---|---|
| Mean or % | SD | Mean or % | SD | p Value | |
| Age, years | 57.47 | 7.29 | 57.24 | 6.9 | 0.267 |
| Body mass index, kg/m2 | 23.24 | 1.81 | 32.18 | 4.20 | <0.001 |
| Waist circumference, cm | 86.79 | 5.38 | 112.28 | 10.48 | <0.001 |
| ALT, U/L | 22.90 | 10.26 | 45.09 | 27.20 | <0.001 |
| LDL cholesterol, mmol/L | 3.12 | 0.79 | 3.36 | 0.98 | <0.001 |
| HDL cholesterol, mmol/L | 1.65 | 0.41 | 1.25 | 0.33 | <0.001 |
| Total triglycerides, mmol/L | 0.89 | 0.30 | 2.24 | 1.54 | <0.001 |
| Matsuda ISI | 10.33 | 4.59 | 3.40 | 2.03 | <0.001 |
| Plasma adiponectin, ug/mL | 9.22 | 5.07 | 6.82 | 3.61 | <0.001 |
| hs-CRP, mg/l | 1.34 | 3.24 | 3.44 | 6.46 | <0.001 |
| Fasting plasma glucose, mmol/L | 5.60 | 0.59 | 6.48 | 1.57 | <0.001 |
| 2-h plasma glucose, mmol/L | 5.53 | 1.64 | 7.80 | 3.04 | <0.001 |
| Fasting plasma insulin, mU/L | 4.83 | 2.46 | 17.72 | 20.85 | <0.001 |
| 2-h plasma insulin, mU/L | 27.93 | 24.68 | 96.21 | 81.56 | <0.001 |
| Fasting plasma FFA, mmol/L | 0.35 | 0.15 | 0.43 | 0.17 | <0.001 |
| Type 2 diabetes, % | 4.20 | - | 29.8 | - | <0.001 |
| PNPLA3 rs738409-G | Direct Parent | Beta | SE | p Value | Novel |
|---|---|---|---|---|---|
| Organooxygen compound | |||||
| Nicotinate ribonucleoside | Glycosylamine | −0.093 | 0.037 | 3.2 × 10−5 | Yes |
| TM6SF2rs58542926-T | |||||
| Glycerolipid | |||||
| Oleoyl-arachidonoyl-glycerol (18:1/20:4 [2]) | Diacylglycerol | −0.107 | 0.062 | 3.7 × 10−6 | Yes |
| MBOAT7rs641738-T | |||||
| Glycerophospholipid | |||||
| 1-stearoyl-2-arachidonoyl-GPI (18:0/20:4) | PI | −0.207 | 0.029 | <1 × 10−50 | Yes |
| 1-palmitoyl-2-linoleoyl-GPI (16:0/18:2) | PI | 0.177 | 0.029 | 3.1 × 10−16 | Yes |
| 1-palmitoyl-2-oleoyl-GPI (16:0/18:1) | PI | 0.173 | 0.028 | 1.2 × 10−15 | Yes |
| 1-stearoyl-2-linoleoyl-GPI (18:0/18:2) | PI | 0.170 | 0.030 | 1.6 × 10−15 | Yes |
| 1-palmitoyl-2-arachidonoyl-GPI (16:0/20:4) | PI | −0.133 | 0.028 | 1.8x10−9 | Yes |
| 1-stearoyl-2-oleoyl-GPI (18:0/18:1) | PI | 0.101 | 0.037 | 7.9 × 10−05 | Yes |
| 1-linoleoyl-GPI (18:2) | Lyso-PI | 0.183 | 0.030 | 1.0 × 10−17 | Yes |
| 1-palmitoleoyl-GPI (16:1) | Lyso-PI | 0.170 | 0.035 | 8.6 × 10−11 | Yes |
| 1-arachidonoyl-GPI (20:4) | Lyso-PI | −0.120 | 0.030 | 2.4 × 10−8 | Yes |
| 1-oleoyl-GPI (18:1) | Lyso-PI | 0.114 | 0.030 | 1.0 × 10−7 | Yes |
| GCKRrs780094-T | |||||
| Carbohydrate | |||||
| Mannose | - | −0.277 | 0.027 | <1 × 10−50 | No |
| Other metabolites | |||||
| Mannonate | - | −0.181 | 0.027 | 1.0 × 10−17 | Yes |
| PPP1R3Brs4841132-A | |||||
| Amino acids | |||||
| Glycine | - | 0.138 | 0.039 | 9.7 × 10−11 | No |
| Hexanoylglutamine | - | −0.114 | 0.043 | 1.6 × 10−7 | Yes |
| Hydroxy acid | |||||
| 3-hydroxybutyrate (BHBA) | β-hydroxy acid | −0.115 | 0.043 | 6.1 × 10−8 | Yes |
| Other metabolites | |||||
| Retinol (Vitamin A) | Vitamin | −0.106 | 0.036 | 6.9 × 10−7 | Yes |
| Hydroxypalmitoyl sphingomyelin (d18:1/16:0 (OH)) | - | −0.099 | 0.036 | 3.9 × 10−6 | Yes |
| Glutamine conjugate of C6H10O22 | - | −0.089 | 0.044 | 3.7 × 10−5 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes Silva, L.; Vangipurapu, J.; Oravilahti, A.; Männistö, V.; Laakso, M. Plasma Metabolite Signatures in Male Carriers of Genetic Variants Associated with Non-Alcoholic Fatty Liver Disease. Metabolites 2023, 13, 267. https://doi.org/10.3390/metabo13020267
Fernandes Silva L, Vangipurapu J, Oravilahti A, Männistö V, Laakso M. Plasma Metabolite Signatures in Male Carriers of Genetic Variants Associated with Non-Alcoholic Fatty Liver Disease. Metabolites. 2023; 13(2):267. https://doi.org/10.3390/metabo13020267
Chicago/Turabian StyleFernandes Silva, Lilian, Jagadish Vangipurapu, Anniina Oravilahti, Ville Männistö, and Markku Laakso. 2023. "Plasma Metabolite Signatures in Male Carriers of Genetic Variants Associated with Non-Alcoholic Fatty Liver Disease" Metabolites 13, no. 2: 267. https://doi.org/10.3390/metabo13020267
APA StyleFernandes Silva, L., Vangipurapu, J., Oravilahti, A., Männistö, V., & Laakso, M. (2023). Plasma Metabolite Signatures in Male Carriers of Genetic Variants Associated with Non-Alcoholic Fatty Liver Disease. Metabolites, 13(2), 267. https://doi.org/10.3390/metabo13020267







