Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer
Abstract
:1. Introduction
2. Tryptophan-Microbiota Interactions in the Healthy State
2.1. Bacterial Species Associated with Tryptophan Metabolism and Metabolite Production
2.2. Influence of Diet on the Microbiota-Ryptophan Axis
2.3. Effects of Probiotics and Prebiotics on Tryptophan Metabolism
3. Dysregulation of Gut Microbiota in Disease States
3.1. Evidence for Dysbiosis Disrupting the Axis in Cancer, IBD, Mood Disorders, and ASD
3.2. Potential Mechanisms of Tryptophan Modulation by Specific Microbes
3.3. Contribution to Pathogenesis of Immune, Metabolic, and Disease Progression
4. Therapeutic Opportunities
4.1. Targeting the Microbiota-Tryptophan Axis for Disease Treatment and Prevention
4.2. Combination with Immunotherapies and IDO Inhibitors
5. Challenges and Limitations of Microbiota-Based Therapies
6. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Trp | tryptophan |
| Kyn | kynurenine |
| Kna | kynurenic acid |
| IDO | indoleamine 2,3-dioxygenase |
| TDO | tryptophan 2,3-dioxygenase |
| TPH | tryptophan hydroxylase |
| NAD | nicotinamide adenine dinucleotide |
| TMO | tryptophan 2-monooxygenase |
| TrD | tryptophan decarboxylase |
| ArAT | aromatic amino acid aminotransferase |
| TNA | tryptophanase |
| IAA | indole-3-acetic acid |
| IPA | indole-3-propionic acid |
| ILA | indole-3-lactic acid |
| IAAId | indole-3-acetaldehyde |
| IPA | indole-3-propionic acid |
| AhR | aryl hydrocarbon receptor |
| I3C | indole-3-carbinole |
| ISA | indole sulfuric acid |
| ROS | reactive oxygen species |
| IPYA | indole-3-pyruvate |
| IA | indole-acrylic acid |
| IAld | indole-3-aldehyde |
| I3A | indole-3-acetate |
References
- Le, N.; Otten, W. Tryptophan Metabolism, from Nutrition to Potential Therapeutic Applications. Amino Acids 2011, 41, 1195–1205. [Google Scholar] [CrossRef]
- Keszthelyi, D.; Troost, F.J.; Masclee, A.A.M. Understanding the Role of Tryptophan and Serotonin Metabolism in Gastrointestinal Function. Neurogastroenterol. Motil. 2009, 21, 1239–1249. [Google Scholar] [CrossRef]
- Badawy, A.A. Neuropharmacology Tryptophan Availability for Kynurenine Pathway Metabolism across the Life Span: Control Mechanisms and Focus on Aging, Exercise, Diet and Nutritional Supplements. Neuropharmacology 2017, 112, 248–263. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W.; Darlington, L.G. The Kynurenine Pathway as a Therapeutic Target in Cognitive and Neurodegenerative Disorders. Br. J. Pharmacol. 2013, 169, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Wick, W.; Van Den Eynde, B.J. Tryptophan Catabolism in Cancer: Beyond IDO and Tryptophan Depletion. Cancer Res. 2012, 72, 5435–5440. [Google Scholar] [CrossRef] [PubMed]
- Mellor, A.L.; Munn, D.H. IDO Expression by Dendritic Cells: Tolerance and Tryptophan Catabolism. Nat. Rev. Immunol. 2004, 4, 762–774. [Google Scholar] [CrossRef]
- Badawy, A.A.B. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int. J. Tryptophan Res. 2017, 10, 1178646917691938. [Google Scholar]
- Taleb, S. Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. Front. Immunol. 2019, 10, 2113. [Google Scholar] [CrossRef]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, Tryptophan Metabolism and the Brain-Gut-Microbiome Axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial Tryptophan Catabolites in Health and Disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Elaine, Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and Characterization of Gut Microbiota Decarboxylases That Can Produce the Neurotransmitter Tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Di Luccia, B.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus Reuteri Induces Gut Intraepithelial CD4(+)CD8alphaalpha(+) T Cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Liu, J.X.; Wang, Y.Z.; Wu, Y.M.; Wang, J.K.; Zhou, Y.Y. Influence of Differing Carbohydrate Sources on L-Tryptophan Metabolism by Porcine Fecal Microbiota Studied in Vitro. Livest. Sci. 2009, 120, 43–50. [Google Scholar] [CrossRef]
- Liu, M.; Nieuwdorp, M.; de Vos, W.M.; Rampanelli, E. Microbial Tryptophan Metabolism Tunes Host Immunity, Metabolism, and Extraintestinal Disorders. Metabolites 2022, 12, 834. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar]
- Liu, J.R.; Miao, H.; Deng, D.Q.; Vaziri, N.D.; Li, P.; Zhao, Y.Y. Gut Microbiota-Derived Tryptophan Metabolism Mediates Renal Fibrosis by Aryl Hydrocarbon Receptor Signaling Activation. Cell. Mol. Life Sci. 2021, 78, 909–922. [Google Scholar] [CrossRef]
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; van Raalte, D.H.; Herrema, H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731. [Google Scholar] [CrossRef]
- Sjögren, K.; Engdahl, C.; Henning, P.; Lerner, U.H.; Tremaroli, V.; Lagerquist, M.K.; Bäckhed, F.; Ohlsson, C. The Gut Microbiota Regulates Bone Mass in Mice. J. Bone Miner. Res. 2012, 27, 1357–1367. [Google Scholar] [CrossRef]
- Marcobal, A.; Kashyap, P.C.; Nelson, T.A.; Aronov, P.A.; Donia, M.S.; Spormann, A.; Fischbach, M.A.; Sonnenburg, J.L. A Metabolomic View of How the Human Gut Microbiota Impacts the Host Metabolome Using Humanized and Gnotobiotic Mice. ISME J. 2013, 7, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Mu, C.L.; Farzi, A.; Zhu, W.Y. Tryptophan Metabolism: A Link between the Gut Microbiota and Brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Pi, Y.; Mu, C.L.; Farzi, A.; Liu, Z.; Zhu, W.Y. Increasing Carbohydrate Availability in the Hindgut Promotes Hypothalamic Neurotransmitter Synthesis: Aromatic Amino Acids Linking the Microbiota–Brain Axis. J. Neurochem. 2019, 149, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Evenepoel, P.; Claus, D.; Geypens, B.; Hiele, M.; Geboes, K.; Rutgeerts, P.; Ghoos, Y. Amount and Fate of Egg Protein Escaping Assimilation in the Small Intestine of Humans. Am. J. Physiol. Gastrointest. Liver Physiol. 1999, 277, 935–943. [Google Scholar] [CrossRef]
- Cranwell, P.D.; Noakes, D.E.; Hill, K.J. Gastric Secretion and Fermentation in the Suckling Pig. Br. J. Nutr. 1976, 36, 71–86. [Google Scholar] [CrossRef]
- Smith, E.A.; Macfarlane, G.T. Enumeration of Human Colonie Bacteria Producing Phenolic and Indolic Compounds: Effects of PH, Carbohydrate Availability and Retention Time on Dissimilatory Aromatic Amino Acid Metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Cummings, J.H.; Macfarlane, S.; Gibson, G.R. Influence of Retention Time on Degradation of Pancreatic Enzymes by Human Colonic Bacteria Grown in a 3-stage Continuous Culture System. J. Appl. Bacteriol. 1989, 67, 521–527. [Google Scholar] [CrossRef]
- Roager, H.M.; Hansen, L.B.S.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gøbel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic Transit Time Is Related to Bacterial Metabolism and Mucosal Turnover in the Gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Falony, G.; Darzi, Y.; Lima-Mendez, G.; Garcia Yunta, R.; Okuda, S.; Vandeputte, D.; Valles-Colomer, M.; Hildebrand, F.; Chaffron, S.; et al. Species-Function Relationships Shape Ecological Properties of the Human Gut Microbiome. Nat. Microbiol. 2016, 1, 16088. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; DeLuca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective Increases of Bifidobacteria in Gut Microflora Improve High-Fat-Diet-Induced Diabetes in Mice through a Mechanism Associated with Endotoxaemia. Diabetologia 2007, 50, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Dong, Y.; Zhang, K.; Ma, J.; He, L.; Wei, J.; Jiang, J.; Zhao, Q.; Zhao, Q.; Cao, H. DOP35 High-Fat Diet Reduces Gut Microbiota-Derived Metabolite Indole-Acetic Acid and Aggravates Colitis. J. Crohn’s Colitis 2023, 17, i97–i99. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J. Indole as an Intercellular Signal in Microbial Communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Martínez, I.; Kim, J.; Duffy, P.R.; Schlegel, V.L.; Walter, J. Resistant Starches Types 2 and 4 Have Differential Effects on the Composition of the Fecal Microbiota in Human Subjects. PLoS ONE 2010, 5, e15046. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and Diet-Responsive Groups of Bacteria within the Human Colonic Microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, Q.; Wang, Q.; Chen, Y.; Lv, L.; Zheng, B.; Yan, R.; Jiang, H.; Shen, J.; Wang, S.; et al. The Impact of Dietary Fibers on Clostridioides Difficile Infection in a Mouse Model. Front. Cell. Infect. Microbiol. 2022, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, B.; Ren, L.; Du, H.; Fei, C.; Qian, C.; Li, B.; Zhang, R.; Liu, H.; Li, Z.; et al. High-Fiber Diet Ameliorates Gut Microbiota, Serum Metabolism and Emotional Mood in Type 2 Diabetes Patients. Front. Cell. Infect. Microbiol. 2023, 13, 66. [Google Scholar] [CrossRef]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Laird, T.S.; Flores, N.; Leveau, J.H.J. Bacterial Catabolism of Indole-3-Acetic Acid. Appl. Microbiol. Biotechnol. 2020, 104, 9535–9550. [Google Scholar] [CrossRef]
- Hendrikx, T.; Schnabl, B. Indoles: Metabolites Produced by Intestinal Bacteria Capable of Controlling Liver Disease Manifestation. J. Intern. Med. 2019, 286, 32–40. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, S.; Li, S.; Zhang, Q.; Cai, Y.; Li, P.; Li, H.; Shen, B.; Liao, Q.; Hong, Y.; et al. Indoleacrylic Acid Produced by Parabacteroides Distasonis Alleviates Type 2 Diabetes via Activation of AhR to Repair Intestinal Barrier. BMC Biol. 2023, 21, 90. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Bian, X.; Liu, J.; Zhu, M.; Li, L.; Yao, T.; Tang, C.; Ravichandran, V.; Liao, P.; Papadimitriou, K. Dietary Components, Microbial Metabolites and Human Health: Reading between the Lines. Foods 2020, 9, 1045. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.H.; Xin, F.Z.; Xue, Y.; Hu, Z.; Han, Y.; Ma, F.; Zhou, D.; Liu, X.L.; Cui, A.; Liu, Z.; et al. Indole-3-Propionic Acid Inhibits Gut Dysbiosis and Endotoxin Leakage to Attenuate Steatohepatitis in Rats. Exp. Mol. Med. 2019, 50, 1–14. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, C.; Gao, J. Extensive Summary of the Important Roles of Indole Propionic Acid, a Gut Microbial Metabolite in Host Health and Disease. Nutrients 2022, 15, 151. [Google Scholar] [CrossRef]
- Kapsetaki, S.E.; Alcaraz, G.M.; Maley, C.C.; Whisner, C.M.; Aktipis, A. Diet, Microbes, and Cancer Across the Tree of Life: A Systematic Review. Curr. Nutr. Rep. 2022, 11, 508–525. [Google Scholar] [CrossRef]
- Maukonen, J.; Saarela, M. Human Gut Microbiota: Does Diet Matter? Proc. Nutr. Soc. 2015, 74, 23–36. [Google Scholar] [CrossRef]
- Zhang, X.; Coker, O.O.; Chu, E.S.H.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary Cholesterol Drives Fatty Liver-Associated Liver Cancer by Modulating Gut Microbiota and Metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhang, J.; Jiang, T.; Zhang, Z.; Wang, Z.; Gong, M.; Zhao, L.; Zhang, C. Ketogenic Diets Induced Glucose Intolerance and Lipid Accumulation in Mice with Alterations in Gut Microbiota And. MBio 2021, 12, e03601-20. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Biosynthesis in Colon Cells to Induce Proliferation and Causes. Gastroenterology 2017, 152, 1419–1433.e5. [Google Scholar] [CrossRef]
- Serger, E.; Luengo-gutierrez, L.; Chadwick, J.S.; Kong, G.; Zhou, L.; Crawford, G.; Danzi, M.C.; Myridakis, A.; Brandis, A.; Bello, A.T.; et al. The Gut Metabolite Indole-3 Propionate Promotes Nerve Regeneration and Repair. Nature 2022, 607, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xiao, Y.; Li, D.; Zhang, S.; Wu, Y.; Zhang, Q.; Bai, W. New Insights into the Mechanisms of High-Fat Diet Mediated Gut Microbiota in Chronic Diseases. iMeta 2023, 2, e69. [Google Scholar] [CrossRef]
- Su, X.; Gao, Y.; Yang, R. Gut Microbiota-Derived Tryptophan Metabolites Maintain Gut and Systemic Homeostasis. Cells 2022, 11, 2296. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Boekhorst, J.; Fogliano, V.; Capuano, E.; Wells, J.M. Impact of High-Fiber or High-Protein Diet on the Capacity of Human Gut Microbiota To Produce Tryptophan Catabolites. J. Agric. Food Chem. 2023, 71, 6956–6966. [Google Scholar] [CrossRef]
- Koga, J.; Adachi, T.; Hidaka, H. Molecular Cloning of the Gene for Indolepyruvate Decarboxylase from Enterobacter Cloacae. MGG Mol. Gen. Genet. 1991, 226, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.T.; Cox, R.P.; Jensen, B.B. 3-Methylindole (Skatole) and Indole Production by Mixed Populations of Pig Fecal Bacteria. Appl. Environ. Microbiol. 1995, 61, 3180–3184. [Google Scholar] [CrossRef]
- Li, S. Modulation of Immunity by Tryptophan Microbial Metabolites. Front. Nutr. 2023, 10, 1–20. [Google Scholar] [CrossRef]
- Dai, X.; Zhu, B.T. Indoleamine 2,3-Dioxygenase Tissue Distribution and Cellular Localization in Mice: Implications for Its Biological Functions. J. Histochem. Cytochem. 2010, 58, 17–28. [Google Scholar] [CrossRef]
- Theate, I.; Van Baren, N.; Pilotte, L.; Moulin, P.; Larrieu, P.; Renauld, J.C.; Herve, C.; Gutierrez-Roelens, I.; Marbaix, E.; Sempoux, C.; et al. Extensive Profiling of the Expression of the Indoleamine 2,3-Dioxygenase 1 Protein in Normal and Tumoral Human Tissues. Cancer Immunol. Res. 2015, 3, 161–172. [Google Scholar] [CrossRef]
- Platten, M.; von Knebel Doeberitz, N.; Oezen, I.; Wick, W.; Ochs, K. Cancer Immunotherapy by Targeting IDO1/TDO and Their Downstream Effectors. Front. Immunol. 2015, 6, 673. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Smith, C.; Thomas, S.; Mandik-Nayak, L.; Laury-Kleintop, L.; Metz, R.; Muller, A.J. Indoleamine 2,3-Dioxygenase Pathways of Pathogenic Inflammation and Immune Escape in Cancer. Cancer Immunol. Immunother. 2014, 63, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Malachowski, W.P.; DuHadaway, J.B.; Muller, A.J. Discovery of IDO1 Inhibitors: From Bench to Bedside. Cancer Res. 2017, 77, 6795–6811. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3-Dioxygenase and Tumor-Induced Tolerance. J. Clin. Investig. 2007, 117, 1147–1154. [Google Scholar] [CrossRef]
- Ricciuti, B.; Leonardi, G.C.; Puccetti, P.; Fallarino, F.; Bianconi, V.; Sahebkar, A.; Baglivo, S.; Chiari, R.; Pirro, M. Targeting Indoleamine-2,3-Dioxygenase in Cancer: Scientific Rationale and Clinical Evidence. Pharmacol. Ther. 2019, 196, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.W.; Hajjar, J.; Hwu, P.; Naing, A. Targeting the Indoleamine 2,3-Dioxygenase Pathway in Cancer. J. Immunother. Cancer 2015, 3, 51. [Google Scholar] [CrossRef]
- Harden, J.L.; Egilmez, N.K. Indoleamine 2,3-Dioxygenase and Dendritic Cell Tolerogenicity. Immunol. Investig. 2012, 41, 738–764. [Google Scholar] [CrossRef]
- Liu, M.; Wang, X.; Wang, L.; Ma, X.; Gong, Z.; Zhang, S.; Li, Y. Targeting the IDO1 Pathway in Cancer: From Bench to Bedside. J. Hematol. Oncol. 2018, 11, 100. [Google Scholar] [CrossRef]
- Liu, X.; Shin, N.; Koblish, H.K.; Yang, G.; Wang, Q.; Wang, K.; Leffet, L.; Hansbury, M.J.; Thomas, B.; Rupar, M.; et al. Selective Inhibition of IDO1 Effectively Regulates Mediators of Antitumor Immunity. Blood 2010, 115, 3520–3530. [Google Scholar] [CrossRef]
- Walczak, K.; Da̧browski, W.; Langner, E.; Zgrajka, W.; Piłat, J.; Kocki, T.; Rzeski, W.; Turski, W.A. Kynurenic Acid Synthesis and Kynurenine Aminotransferases Expression in Colon Derived Normal and Cancer Cells. Scand. J. Gastroenterol. 2011, 46, 903–912. [Google Scholar] [CrossRef]
- Engin, A.B.; Karahalil, B.; Karakaya, A.E.; Engin, A. Helicobacter Pylori and Serum Kynurenine-Tryptophan Ratio in Patients with Colorectal Cancer. World J. Gastroenterol. 2015, 21, 3636–3643. [Google Scholar] [CrossRef]
- Teunis, C.; Nieuwdorp, M.; Hanssen, N. Interactions between Tryptophan Metabolism, the Gut Microbiome and the Immune System as Potential Drivers of Non-Alcohol Liver Disease (NAFLD) and Metabolic Diseases. Metabolites 2022, 12, 514. [Google Scholar] [CrossRef]
- Ju, T.; Kong, J.Y.; Stothard, P.; Willing, B.P. Defining the Role of Parasutterella, a Previously Uncharacterized Member of the Core Gut Microbiota. ISME J. 2019, 13, 1520–1534. [Google Scholar] [CrossRef]
- Yang, C.; Du, Y.; Ren, D.; Yang, X.; Zhao, Y. Gut Microbiota-Dependent Catabolites of Tryptophan Play a Predominant Role in the Protective Effects of Turmeric Polysaccharides against DSS-Induced Ulcerative Colitis. Food Funct. 2021, 12, 9793–9807. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A Gut Bacterial Pathway Metabolizes Aromatic Amino Acids into Nine Circulating Metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Hao, Z.; Du, J.; Gao, Y.; Yang, S.; Zhou, Y. Bacteroides Thetaiotaomicron Relieves Colon Inflammation by Activating Aryl Hydrocarbon Receptor and Modulating CD4+T Cell Homeostasis. Int. Immunopharmacol. 2021, 90, 107183. [Google Scholar] [CrossRef]
- Xiao, L.; Yan, J.; Yang, T.; Zhu, J.; Li, T.; Wei, H.; Chen, J. Fecal Microbiome Transplantation from Children with Autism Spectrum Disorder Modulates Tryptophan and Serotonergic Synapse Metabolism and Induces Altered Behaviors in Germ-Free Mice. mSystems 2021, 6, e01343-20. [Google Scholar] [CrossRef]
- Liu, Z.; Ling, Y.; Peng, Y.; Han, S.; Ren, Y.; Jing, Y.; Fan, W.; Su, Y.; Mu, C.; Zhu, W. Regulation of Serotonin Production by Specific Microbes from Piglet Gut. J. Anim. Sci. Biotechnol. 2023, 14, 1–15. [Google Scholar] [CrossRef]
- Chen, C.M.; Wu, C.C.; Huang, C.L.; Chang, M.Y.; Cheng, S.H.; Lin, C.T.; Tsai, Y.C. Lactobacillus Plantarum PS128 Promotes Intestinal Motility, Mucin Production, and Serotonin Signaling in Mice. Probiotics Antimicrob. Proteins 2022, 14, 535–545. [Google Scholar] [CrossRef]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The Bacterial Signal Indole Increases Epithelial-Cell Tight-Junction Resistance and Attenuates Indicators of Inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef]
- Whisner, C.M.; Athena Aktipis, C. The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One Another’s Growth. Curr. Nutr. Rep. 2019, 8, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Amiot, A.; Baleur, Y.; Levy, M.; Auriault, M.L.; Van Nhieu, J.T.; Delchier, J.C. Microbial Dysbiosis and Colon Carcinogenesis: Could Colon Cancer Be Considered a Bacteria-Related Disease? Therap. Adv. Gastroenterol. 2013, 6, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The Microbiome and Human Cancer. Science 2021, 371. [Google Scholar] [CrossRef] [PubMed]
- Dzutsev, A.; Goldszmid, R.S.; Viaud, S.; Zitvogel, L.; Trinchieri, G. The Role of the Microbiota in Inflammation, Carcinogenesis, and Cancer Therapy. Eur. J. Immunol. 2015, 45, 17–31. [Google Scholar] [CrossRef]
- Makarem, N.; Lin, Y.; Bandera, E.V.; Jacques, P.F.; Parekh, N. Concordance with World Cancer Research Fund/American Institute for Cancer Research (WCRF/AICR) Guidelines for Cancer Prevention and Obesity-Related Cancer Risk in the Framingham Offspring Cohort (1991–2008). Cancer Causes Control 2015, 26, 277–286. [Google Scholar] [CrossRef]
- Romagnolo, D.F.; Selmin, O.I. Flavonoids and Cancer Prevention: A Review of the Evidence. J. Nutr. Gerontol. Geriatr. 2012, 31, 206–238. [Google Scholar] [CrossRef]
- Abdull Razis, A.F.; Mohd Noor, N. Cruciferous Vegetables: Dietary Phytochemicals for Cancer Prevention. Asian Pacific J. Cancer Prev. 2013, 14, 1565–1570. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Sánchez-Quesada, C.; Algarra, I.; Gaforio, J.J. The High-Fat Diet Based on Extra-Virgin Olive Oil Causes Dysbiosis Linked to Colorectal Cancer Prevention. Nutrients 2020, 12, 1705. [Google Scholar] [CrossRef]
- Oostindjer, M.; Alexander, J.; Amdam, G.V.; Andersen, G.; Bryan, N.S.; Chen, D.; Corpet, D.E.; De Smet, S.; Dragsted, L.O.; Haug, A.; et al. The Role of Red and Processed Meat in Colorectal Cancer Development: A Perspective. Meat Sci. 2014, 97, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.J.; Ferrucci, L.M.; Risch, A.; Graubard, B.I.; Ward, M.H.; Park, Y.; Hollenbeck, A.R.; Schatzkin, A.; Sinha, R. A Large Prospective Study of Meat Consumption and Colorectal Cancer Risk: An Investigation of Potential Mechanisms Underlying This Association. Cancer Res. 2010, 70, 2406–2414. [Google Scholar] [CrossRef] [PubMed]
- Foegeding, N.J.; Jones, Z.S.; Byndloss, M.X. Western Lifestyle as a Driver of Dysbiosis in Colorectal Cancer. DMM Dis. Model. Mech. 2021, 14, 1–6. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a Bacterial World, a New Imperative for the Life Sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1–23. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The Microbiome and Innate Immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Laurans, L.; Venteclef, N.; Haddad, Y.; Chajadine, M.; Alzaid, F.; Metghalchi, S.; Sovran, B.; Denis, R.G.P.; Dairou, J.; Cardellini, M.; et al. Genetic Deficiency of Indoleamine 2,3-Dioxygenase Promotes Gut Microbiota-Mediated Metabolic Health. Nat. Med. 2018, 24, 1113–1120. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In The Impact of Food Bioactives on Health. In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Mackie, T.L.A., Eds.; Springer: Cham, Switzerland, 2015; Chapter 27. [Google Scholar] [CrossRef]
- Choi, Y.; Yanagawa, Y.; Kim, S.; Park, T. Involvement of SIRT1-AMPK Signaling in the Protective Action of Indole-3-Carbinol against Hepatic Steatosis in Mice Fed a High-Fat Diet. J. Nutr. Biochem. 2013, 24, 1393–1400. [Google Scholar] [CrossRef]
- Desbonnet, L.; Garrett, L.; Clarke, G.; Kiely, B.; Cryan, J.F.; Dinan, T.G. Effects of the Probiotic Bifidobacterium Infantis in the Maternal Separation Model of Depression. Neuroscience 2010, 170, 1179–1188. [Google Scholar] [CrossRef]
- Desbonnet, L.; Garrett, L.; Clarke, G.; Bienenstock, J.; Dinan, T.G. The Probiotic Bifidobacteria Infantis: An Assessment of Potential Antidepressant Properties in the Rat. J. Psychiatr. Res. 2008, 43, 164–174. [Google Scholar] [CrossRef]
- Tandon, D.; Haque, M.M.; Gote, M.; Jain, M.; Bhaduri, A.; Dubey, A.K.; Mande, S.S. A Prospective Randomized, Double-Blind, Placebo-Controlled, Dose-Response Relationship Study to Investigate Efficacy of Fructo-Oligosaccharides (FOS) on Human Gut Microflora. Sci. Rep. 2019, 9, 5473. [Google Scholar] [CrossRef] [PubMed]
- Radford-Smith, D.E.; Anthony, D.C. Prebiotic and Probiotic Modulation of the Microbiota–Gut–Brain Axis in Depression. Nutrients 2023, 15, 1880. [Google Scholar] [CrossRef]
- Kouchaki, E.; Tamtaji, O.R.; Salami, M.; Bahmani, F.; Daneshvar Kakhaki, R.; Akbari, E.; Tajabadi-Ebrahimi, M.; Jafari, P.; Asemi, Z. Clinical and Metabolic Response to Probiotic Supplementation in Patients with Multiple Sclerosis: A Randomized, Double-Blind, Placebo-Controlled Trial. Clin. Nutr. 2017, 36, 1245–1249. [Google Scholar] [CrossRef]
- Zhai, L.; Spranger, S.; Binder, D.C.; Gritsina, G.; Lauing, K.L.; Giles, F.J.; Wainwright, D.A. Molecular Pathways: Targeting IDO1 and Other Tryptophan Dioxygenases for Cancer Immunotherapy. Clin. Cancer Res. 2015, 21, 5427–5433. [Google Scholar] [CrossRef] [PubMed]
- Essa, M.M.; Subash, S.; Braidy, N.; Al-Adawi, S.; Lim, C.K.; Manivasagam, T.; Guillemin, G.J. Role of NAD+, Oxidative Stress, and Tryptophan Metabolism in Autism Spectrum Disorders. Int. J. Tryptophan Res. 2013, 6, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Palego, L.; Betti, L.; Rossi, A.; Giannaccini, G. Tryptophan Biochemistry: Structural, Nutritional, Metabolic, and Medical Aspects in Humans. J. Amino Acids 2016, 2016, 8952520. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Raynal, S.; Bailbé, D.; Gausseres, B.; Carbonne, C.; Autier, V.; Movassat, J.; Kergoat, M.; Portha, B. Expression of the Kynurenine Pathway Enzymes in the Pancreatic Islet Cells. Activation by Cytokines and Glucolipotoxicity. Biochim. Biophys. Acta-Mol. Basis Dis. 2015, 1852, 980–991. [Google Scholar] [CrossRef]
- Heng, B.; Lim, C.K.; Lovejoy, D.B.; Bessede, A.; Gluch, L.; Guillemin, G.J. Understanding the Role of the Kynurenine Pathway in Human Breast Cancer Immunobiology. Oncotarget 2016, 7, 6506–6520. [Google Scholar] [CrossRef]
- Adams, S.; Braidy, N.; Bessesde, A.; Brew, B.J.; Grant, R.; Teo, C.; Guillemin, G.J. The Kynurenine Pathway in Brain Tumor Pathogenesis. Cancer Res. 2012, 72, 5649–5657. [Google Scholar] [CrossRef]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An Endogenous Tumour-Promoting Ligand of the Human Aryl Hydrocarbon Receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, X.; Dong, W.; Fang, Y.; Lv, J.; Zhang, T.; Fiskesund, R.; Xie, J.; Liu, J.; Yin, X.; et al. Tumor-Repopulating Cells Induce PD-1 Expression in CD8+ T Cells by Transferring Kynurenine and AhR Activation. Cancer Cell 2018, 33, 480–494.e7. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.; Briercheck, E.L.; Freud, A.G.; Trotta, R.; McClory, S.; Scoville, S.D.; Keller, K.; Deng, Y.; Cole, J.; Harrison, N.; et al. AHR Prevents Human IL-1R1hi ILC3 Differentiation to Natural Killer Cells. Cell Rep. 2014, 8, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Murray, I.A.; Patterson, A.D.; Perdew, G.H. Aryl Hydrocarbon Receptor Ligands in Cancer: Friend and Foe. Nat. Rev. Cancer 2014, 14, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Stanford, E.A.; Ramirez-Cardenas, A.; Wang, Z.; Novikov, O.; Alamoud, K.; Koutrakis, P.; Mizgerd, J.P.; Genco, C.A.; Kukuruzinska, M.; Monti, S.; et al. Role for the Aryl Hydrocarbon Receptor and Diverse Ligands in Oral Squamous Cell Carcinoma Migration and Tumorigenesis. Mol. Cancer Res. 2016, 14, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Andersson, P.; McGuire, J.; Rubio, C.; Gradin, K.; Whitelaw, M.L.; Pettersson, S.; Hanberg, A.; Poellinger, L. A Constitutively Active Dioxin/Aryl Hydrocarbon Receptor Induces Stomach Tumors. Proc. Natl. Acad. Sci. USA 2002, 99, 9990–9995. [Google Scholar] [CrossRef] [PubMed]
- Moennikes, O.; Loeppen, S.; Buchmann, A.; Andersson, P.; Ittrich, C.; Poellinger, L.; Schwarz, M. A Constitutively Active Dioxin/Aryl Hydrocarbon Receptor Promotes Hepatocarcinogenesis in Mice. Cancer Res. 2004, 64, 4707–4710. [Google Scholar] [CrossRef]
- Richmond, O.; Ghotbaddini, M.; Allen, C.; Walker, A.; Zahir, S.; Powell, J.B. The Aryl Hydrocarbon Receptor Is Constitutively Active in Advanced Prostate Cancer Cells. PLoS ONE 2014, 9, e95058. [Google Scholar] [CrossRef]
- Vacher, S.; Castagnet, P.; Chemlali, W.; Lallemand, F.; Meseure, D.; Pocard, M.; Bieche, I.; Perrot-Applanat, M. High AHR Expression in Breast Tumors Correlates with Expression of Genes from Several Signaling Pathways Namely Inflammation and Endogenous Tryptophan Metabolism. PLoS ONE 2018, 13, e0190619. [Google Scholar] [CrossRef]
- Corre, S.; Tardif, N.; Mouchet, N.; Leclair, H.M.; Boussemart, L.; Gautron, A.; Bachelot, L.; Perrot, A.; Soshilov, A.; Rogiers, A.; et al. Sustained Activation of the Aryl Hydrocarbon Receptor Transcription Factor Promotes Resistance to BRAF-Inhibitors in Melanoma. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural Segregation of Gut Microbiota between Colorectal Cancer Patients and Healthy Volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-Cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria across Populations and Universal Bacterial Markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, N.; Li, Q.; Chu, E.S.H.; Lau, H.C.H.; Fong, W.; Liu, W.; Liang, C.; Nakatsu, G.; Su, A.C.Y.; Coker, O.O.; et al. Lactobacillus Gallinarum Modulates the Gut Microbiota and Produces Anti-Cancer Metabolites to Protect against Colorectal Tumourigenesis. Gut 2022, 71, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Fong, W.; Li, Q.; Ji, F.; Liang, W.; Lau, H.C.H.; Kang, X.; Liu, W.; To, K.K.-W.; Zuo, Z.; Li, X.; et al. Lactobacillus Gallinarum-Derived Metabolites Boost Anti-PD1 Efficacy in Colorectal Cancer by Inhibiting Regulatory T Cells through Modulating IDO1/Kyn/AHR Axis. Gut 2023. [Google Scholar] [CrossRef]
- Shiomi, Y.; Nishiumi, S.; Ooi, M.; Hatano, N.; Shinohara, M.; Yoshie, T.; Kondo, Y.; Furumatsu, K.; Shiomi, H.; Kutsumi, H.; et al. GCMS-Based Metabolomic Study in Mice with Colitis Induced by Dextran Sulfate Sodium. Inflamm. Bowel Dis. 2011, 17, 2261–2274. [Google Scholar] [CrossRef]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 Impacts Colitis by Altering Gut Microbiota Metabolism of Tryptophan into Aryl Hydrocarbon Receptor Ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Sofia, M.A.; Ciorba, M.A.; Meckel, K.; Lim, C.K.; Guillemin, G.J.; Weber, C.R.; Bissonnette, M.; Pekow, J.R. Tryptophan Metabolism through the Kynurenine Pathway Is Associated with Endoscopic Inflammation in Ulcerative Colitis. Inflamm. Bowel Dis. 2018, 24, 1471–1480. [Google Scholar] [CrossRef]
- Salimi Elizei, S.; Poormasjedi-Meibod, M.S.; Wang, X.; Kheirandish, M.; Ghahary, A. Kynurenic Acid Downregulates IL-17/1L-23 Axis in Vitro. Mol. Cell. Biochem. 2017, 431, 55–65. [Google Scholar] [CrossRef]
- Leclerc, D.; Staats Pires, A.C.; Guillemin, G.J.; Gilot, D. Detrimental Activation of AhR Pathway in Cancer: An Overview of Therapeutic Strategies. Curr. Opin. Immunol. 2021, 70, 15–26. [Google Scholar] [CrossRef]
- Gabriely, G.; Wheeler, M.A.; Takenaka, M.C.; Quintana, F.J. Role of AHR and HIF-1α in Glioblastoma Metabolism. Trends Endocrinol. Metab. 2017, 28, 428–436. [Google Scholar] [CrossRef]
- Pernomian, L.; Duarte-Silva, M.; de Barros Cardoso, C.R. The Aryl Hydrocarbon Receptor (AHR) as a Potential Target for the Control of Intestinal Inflammation: Insights from an Immune and Bacteria Sensor Receptor. Clin. Rev. Allergy Immunol. 2020, 59, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.L.; Ley, R.E. The Human Gut Bacteria Christensenellaceae Are Widespread, Heritable, and Associated with Health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhang, M.; Qi, H.; Gao, Y.; Yang, Y.; Yun, H.; Zhang, Q.; Yang, X.; Zhang, Y.; He, J.; et al. Gut Microbiota–Derived Metabolite 3-Idoleacetic Acid Together with LPS Induces IL-35+ B Cell Generation. Microbiome 2022, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New Evidences on the Altered Gut Microbiota in Autism Spectrum Disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.J.; Bassis, C.M.; Hein, R.; Assari, S.; Flowers, S.A.; Kelly, M.B.; Young, V.B.; Ellingrod, V.E.; McInnis, M.G. The Gut Microbiome Composition Associates with Bipolar Disorder and Illness Severity. J. Psychiatr. Res. 2017, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A.B.; Guillemin, G.J. Species Differences in Tryptophan Metabolism and Disposition. Int. J. Tryptophan Res. 2022, 15. [Google Scholar] [CrossRef]
- Ramos-Chávez, L.A.; Lugo Huitrón, R.; González Esquivel, D.; Pineda, B.; Ríos, C.; Silva-Adaya, D.; Sánchez-Chapul, L.; Roldán-Roldán, G.; Pérez de la Cruz, V. Relevance of Alternative Routes of Kynurenic Acid Production in the Brain. Oxid. Med. Cell. Longev. 2018, 2018, 5272741. [Google Scholar] [CrossRef]
- Lukić, I.; Ivković, S.; Mitić, M.; Adžić, M. Tryptophan Metabolites in Depression: Modulation by Gut Microbiota. Front. Behav. Neurosci. 2022, 16, 1–17. [Google Scholar] [CrossRef]
- Anesi, A.; Rubert, J.; Oluwagbemigun, K.; Orozco-Ruiz, X.; Nöthlings, U.; Breteler, M.M.B.; Mattivi, F. Metabolic Profiling of Human Plasma and Urine, Targeting Tryptophan, Tyrosine and Branched Chain Amino Acid Pathways. Metabolites 2019, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Chappell, C.; Gonzales, C.; Okhuysen, P. A Rapid and Specific Method for the Detection of Indole in Complex Biological Samples. Appl. Environ. Microbiol. 2015, 81, 8093–8097. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, T.; Vidova, V.; Bienertova-Vasku, J.; Janku, P.; Almasi, M.; Klanova, J.; Spacil, Z. Urinary Intermediates of Tryptophan as Indicators of the Gut Microbial Metabolism. Anal. Chim. Acta 2017, 987, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Mosterd, C.M.; Kanbay, M.; van den Born, B.J.H.; van Raalte, D.H.; Rampanelli, E. Intestinal Microbiota and Diabetic Kidney Diseases: The Role of Microbiota and Derived Metabolites Inmodulation of Renal Inflammation and Disease Progression. Best Pract. Res. Clin. Endocrinol. Metab. 2021, 35, 101484. [Google Scholar] [CrossRef] [PubMed]
- Jennis, M.; Cavanaugh, C.R.; Leo, G.C.; Mabus, J.R.; Lenhard, J.; Hornby, P.J. Microbiota-Derived Tryptophan Indoles Increase after Gastric Bypass Surgery and Reduce Intestinal Permeability in Vitro and in Vivo. Neurogastroenterol. Motil. 2018, 30, e13178. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal Bacteria-Dependent Indole Production Enhances Epithelial Barrier Function in the Colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef]
- Hubbard, T.D.; Murray, I.A.; Bisson, W.H.; Lahoti, T.S.; Gowda, K.; Amin, S.G.; Patterson, A.D.; Perdew, G.H. Adaptation of the Human Aryl Hydrocarbon Receptor to Sense Microbiota-Derived Indoles. Sci. Rep. 2015, 5, 12689. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut Microbiota-Derived Metabolites as Key Actors in Inflammatory Bowel Disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Kurata, K.; Kawahara, H.; Nishimura, K.; Jisaka, M.; Yokota, K.; Shimizu, H. Skatole Regulates Intestinal Epithelial Cellular Functions through Activating Aryl Hydrocarbon Receptors and P38. Biochem. Biophys. Res. Commun. 2019, 510, 649–655. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, L.; Yang, T.; Feng, Y.L.; Vaziri, N.D.; Liu, B.L.; Liu, Q.Q.; Guo, Y.; Zhao, Y.Y. Aryl Hydrocarbon Receptor Activation Mediates Kidney Disease and Renal Cell Carcinoma. J. Transl. Med. 2019, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.B.; Tanaka, A.; Kuhara, T.; Xiao, J.Z. Potential Effects of Indole-3-Lactic Acid, a Metabolite of Human Bifidobacteria, on NGF-Induced Neurite Outgrowth in PC12 Cells. Microorganisms 2020, 8, 398. [Google Scholar] [CrossRef] [PubMed]
- Aoki, R.; Aoki-Yoshida, A.; Suzuki, C.; Takayama, Y. Indole-3-Pyruvic Acid, an Aryl Hydrocarbon Receptor Activator, Suppresses Experimental Colitis in Mice. J. Immunol. 2018, 201, 3683–3693. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota Metabolite Short Chain Fatty Acids, GPCR, and Inflammatory Bowel Diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic Short Chain Fatty Acids Limit Antitumor Effect of CTLA-4 Blockade in Hosts with Cancer. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kles, K.A.; Chang, E.B. Short-Chain Fatty Acids Impact on Intestinal Adaptation, Inflammation, Carcinoma, and Failure. Gastroenterology 2006, 130, 100–105. [Google Scholar] [CrossRef]
- Martin-Gallausiaux, C.; Larraufie, P.; Jarry, A.; Béguet-Crespel, F.; Marinelli, L.; Ledue, F.; Reimann, F.; Blottière, H.M.; Lapaque, N. Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells. Front. Immunol. 2018, 9, 2838. [Google Scholar] [CrossRef]
- Gouasmi, R.; Ferraro-Peyret, C.; Nancey, S.; Coste, I.; Renno, T.; Chaveroux, C.; Aznar, N.; Ansieau, S. The Kynurenine Pathway and Cancer: Why Keep It Simple When You Can Make It Complicated. Cancers 2022, 14, 2793. [Google Scholar] [CrossRef]
- Kudo, T.; Prentzell, M.T.; Mohapatra, S.R.; Sahm, F.; Zhao, Z.; Grummt, I.; Wick, W.; Opitz, C.A.; Platten, M.; Green, E.W. Constitutive Expression of the Immunosuppressive Tryptophan Dioxygenase TDO2 in Glioblastoma Is Driven by the Transcription Factor C/EBPβ. Front. Immunol. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Novikov, O.; Wang, Z.; Stanford, E.A.; Parks, A.J.; Ramirez-Cardenas, A.; Landesman, E.; Laklouk, I.; Sarita-Reyes, C.; Gusenleitner, D.; Li, A.; et al. An Aryl Hydrocarbon Receptor-Mediated Amplification Loop That Enforces Cell Migration in ER-/PR-/Her2- Human Breast Cancer Cells. Mol. Pharmacol. 2016, 90, 674–688. [Google Scholar] [CrossRef]
- Yu, C.P.; Fu, S.F.; Chen, X.; Ye, J.; Ye, Y.; Kong, L.D.; Zhu, Z. The Clinicopathological and Prognostic Significance of IDO1 Expression in Human Solid Tumors: Evidence from a Systematic Review and Meta-Analysis. Cell. Physiol. Biochem. 2018, 49, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, J.; Shen, H.; Wang, J. The Prognostic Value of IDO Expression in Solid Tumors: A Systematic Review and Meta-Analysis. BMC Cancer 2020, 20, 471. [Google Scholar] [CrossRef] [PubMed]
- Currier, A.R.; Ziegler, M.H.; Riley, M.M.; Babcock, T.A.; Telbis, V.P.; Carlin, J.M. Tumor necrosis factor-and lipopolysaccharide Enhance Interferon-Induced antichlamydial indoleamine Dioxygenase Activity Independently. J. Interf. Cytokine Res. 2000, 20, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.M.; Hale, P.T.; Carlin, J.M. The Role of IFN-γ and TNF-α-Responsive Regulatory Elements in the Synergistic Induction of Indoleamine Dioxygenase. J. Interf. Cytokine Res. 2005, 25, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Litzenburger, U.M.; Opitz, C.A.; Sahm, F.; Rauschenbach, K.J.; Trump, S.; Winter, M.; Ott, M.; Ochs, K.; Lutz, C.; Liu, X.; et al. Constitutive IDO Expression in Human Cancer Is Sustained by an Autocrine Signaling Loop Involving IL-6, STAT3 and the AHR. Oncotarget 2014, 5, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, N.; Ge, Y.; Yang, Y.; Ren, F.; Wu, Z. Tryptophan and the Innate Intestinal Immunity: Crosstalk between Metabolites, Host Innate Immune Cells, and Microbiota. Eur. J. Immunol. 2022, 52, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl Hydrocarbon Receptor-Induced Signals up-Regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterology 2011, 141, 237–248.e1. [Google Scholar] [CrossRef]
- Ding, Y.; Yanagi, K.; Yang, F.; Callaway, E.; Cheng, C.; Hensel, M.E.; Menon, R.; Alaniz, R.C.; Lee, K.; Jayaraman, A.; et al. Oral Supplementation of Gut Microbial Metabolite Indole-3-Acetate Alleviates Diet-Induced Steatosis and Inflammation in Mice. Elife 2023, 12. [Google Scholar] [CrossRef]
- Özoğul, F.; Kuley, E.; Özoğul, Y.; Özoğul, İ. The Function of Lactic Acid Bacteria on Biogenic Amines Production by Food-Borne Pathogens in Arginine Decarboxylase Broth. Food Sci. Technol. Res 2012, 18, 795–804. [Google Scholar] [CrossRef]
- Lin, R.; Sun, Y.; Mu, P.; Zheng, T.; Mu, H.; Deng, F.; Deng, Y.; Wen, J. Lactobacillus Rhamnosus GG Supplementation Modulates the Gut Microbiota to Promote Butyrate Production, Protecting against Deoxynivalenol Exposure in Nude Mice. Biochem. Pharmacol. 2020, 175, 113868. [Google Scholar] [CrossRef]
- Scholtens, P.A.; Goossens, D.A.M.; Staiano, A. Stool Characteristics of Infants Receiving Short-Chain Galacto-Oligosaccharides and Long-Chain Fructo-Oligosaccharides: A Review. World J. Gastroenterol. 2014, 20, 13446–13452. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Guo, Y.; Zhang, S.; Chen, Z.; Wu, K.; Liu, Q.; Liu, K.; Wen, L.; Wei, Y.; Wang, B.; et al. Fecal Microbiota Transplantation Can Alleviate Gastrointestinal Transit in Rats with High-Fat Diet-Induced Obesity via Regulation of Serotonin Biosynthesis. Biomed Res. Int. 2018, 2018, 8308671. [Google Scholar] [CrossRef] [PubMed]
- Vujkovic-Cvijin, I.; Swainson, L.A.; Chu, S.N.; Ortiz, A.M.; Santee, C.A.; Petriello, A.; Dunham, R.M.; Fadrosh, D.W.; Lin, D.L.; Faruqi, A.A.; et al. Gut-Resident Lactobacillus Abundance Associates with IDO1 Inhibition and Th17 Dynamics in SIV-Infected Macaques. Cell Rep. 2015, 13, 1589–1597. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Santoni, M.; Piva, F.; Conti, A.; Santoni, A.; Cimadamore, A.; Scarpelli, M.; Battelli, N.; Montironi, R. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Eur. Urol. 2018, 74, 521–522. [Google Scholar] [CrossRef]
- Opitz, C.A.; Somarribas Patterson, L.F.; Mohapatra, S.R.; Dewi, D.L.; Sadik, A.; Platten, M.; Trump, S. The Therapeutic Potential of Targeting Tryptophan Catabolism in Cancer. Br. J. Cancer 2020, 122, 30–44. [Google Scholar] [CrossRef]
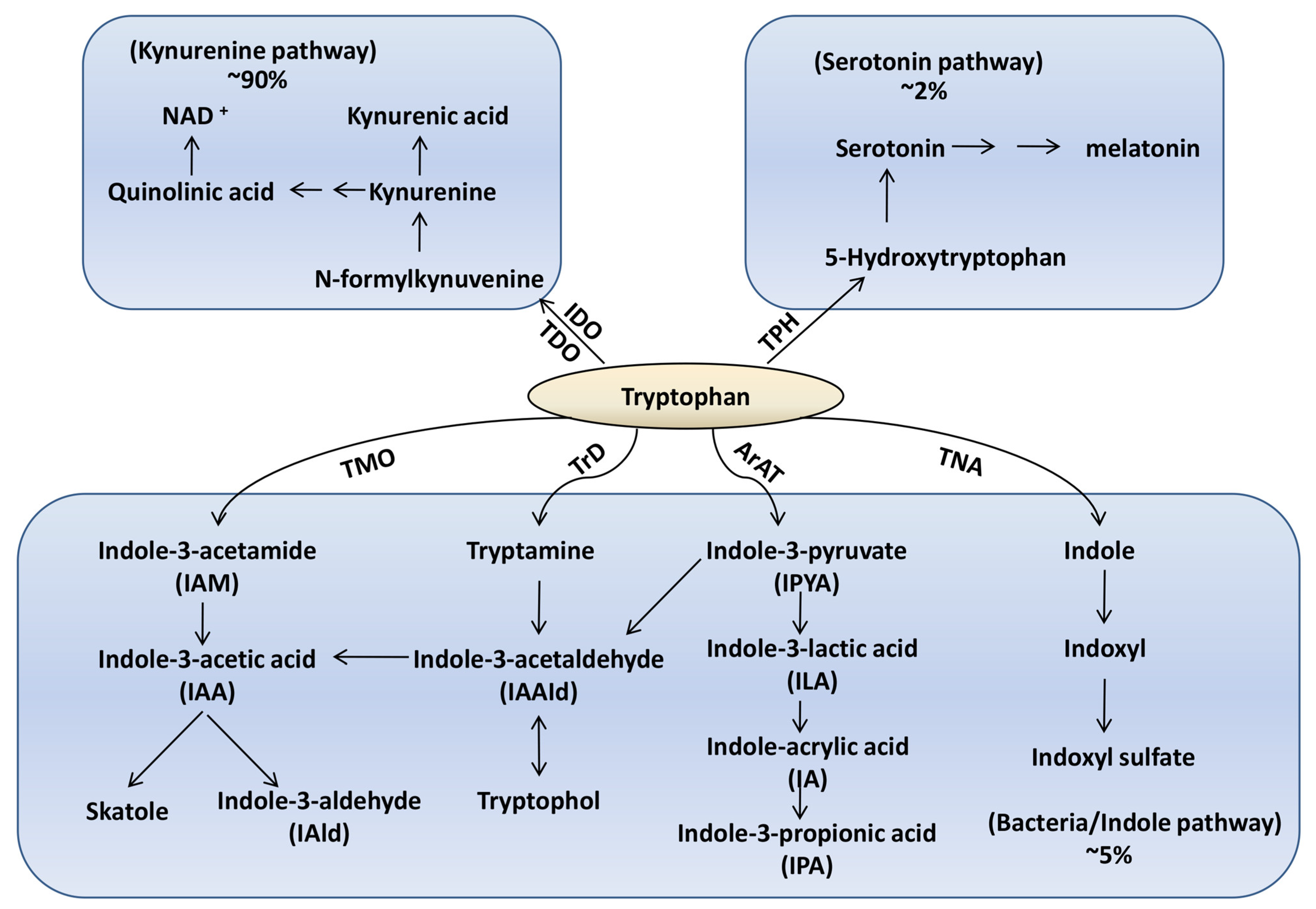
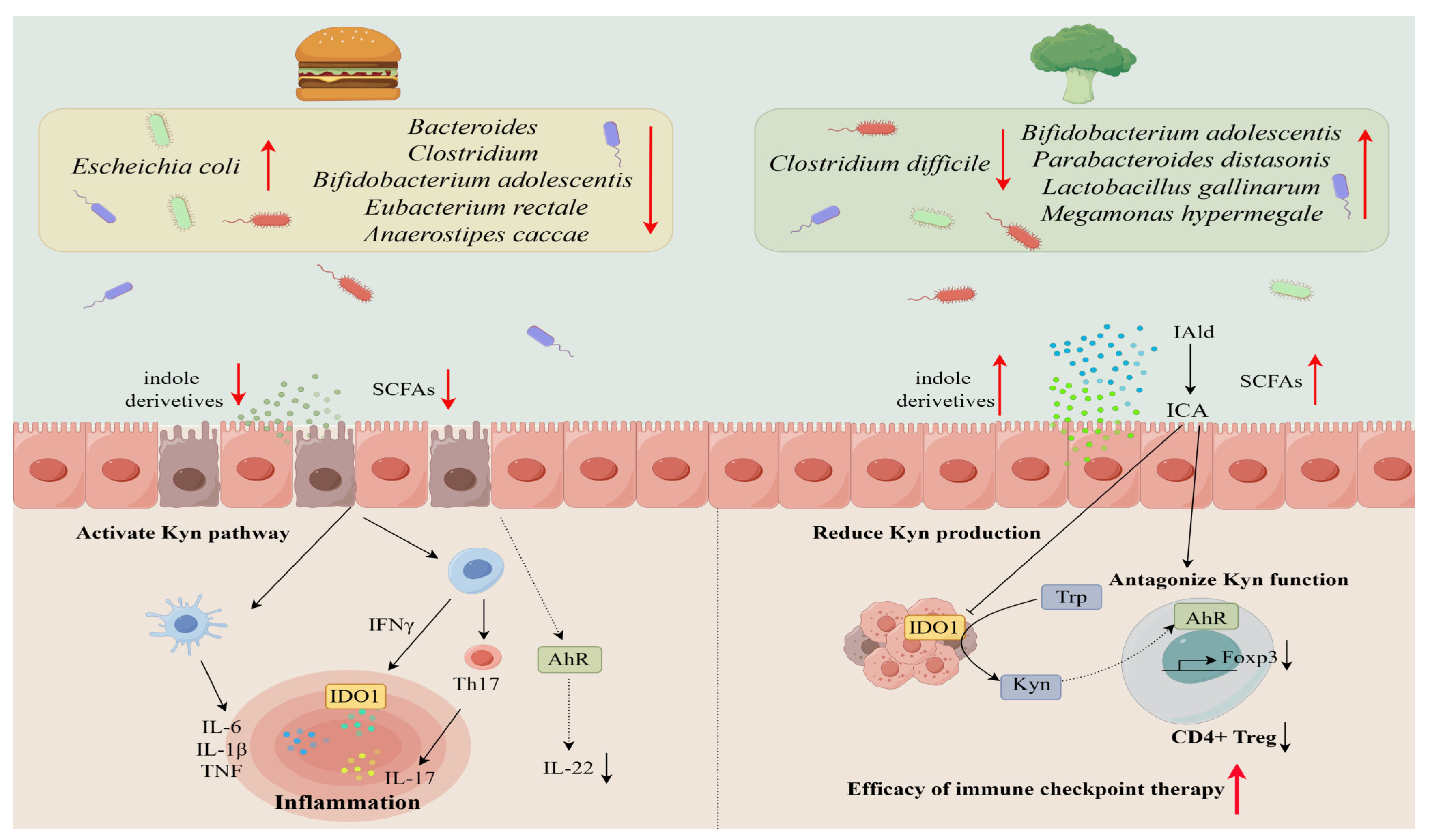
| Tryptophan Metabolism | Producers | Diet Influence | Impact on Metabolism | Impact on Microbes | Reference |
|---|---|---|---|---|---|
| Indole | Bacteroides thetaiotaomicron | High-fat diet | Indole Production↓ | Bacteroides spp.↓ Escheichia coli↑ Clostridium↓ | [11,31,32] |
| Bacteroides ovatus | |||||
| Clostridium limosum | |||||
| Clostridium bifermentans | |||||
| Clostridium malenomenatum | |||||
| Clostridium lentoputrescens | |||||
| Clostridium tetani | |||||
| Clostridium tetanomorphum | |||||
| Enterococcus faecalis | |||||
| Escheichia coli | |||||
| Fusobacterium nucleatum | |||||
| Haemophilus influenza | |||||
| Proteus vulgaris | |||||
| Paracolobactrumcoliforme | |||||
| Salmonella enterica | |||||
| …for more see [33] | |||||
| Indole-3-acetic acid (IAA) | Bacteroides thetaiotaomicron | High-fat diet | IAA Production↓ | Bifidobacterium spp.↓ Bacteroides↓ Bifidobacterium adolescentis↓ | [31,34,35,36,37,38] |
| Bacteroides ovatus | |||||
| Bacteroides fragilis | |||||
| Bifidobacterium adolescentis | |||||
| Bifidobacterium pseudolongum | |||||
| Clostridium difficile | |||||
| Clostridium lituseburense | High-fiber diet | IAA Production↑ | Bifidobacteriumadolescentis↑ Clostridium difficile↓ | ||
| Clostridium sporogenes | |||||
| Escherichia coli | |||||
| Eubacterium hallii | |||||
| Eubacterium cylindroides | |||||
| …for more see [39] | |||||
| Indole-3-acrylic acid (IA) | Clostridium sporogenes | High-fiber diet | IA production ↑ | Parabacteroides distasonis↑ | [40,41] |
| Peptostreptococcusrussellii | |||||
| Peptostreptococcusanaerobius | |||||
| Peptostreptococcusstomatis | |||||
| Parabacteroides distasonis | |||||
| …for more see [42] | |||||
| Indole-3-propionic acid (IPA) | Clostridium sporogenes | High-fat diet | IPA Production↓ | Clostridiumsporogenes↓ | [43,44,45,46,47,48,49,50] |
| Clostridium caloritolerans | |||||
| Clostridium botulinum | High-fiber diet | IPA Production↑ | Clostridium↑ Bifidobacterium↑ Lactobacillus↑ Peptostreptococcus↑ | ||
| Peptostreptococcusasaccharolyticus | |||||
| Peptostreptococcusrussellii | |||||
| PeptostreptococcusanaerobiusCC14N | |||||
| Peptostreptococcusstomatis | Ketogenic diet | IPA Production↓ | Lactobacillus murinus ↓ | ||
| …for more see [51] | |||||
| Indole-3-lactic acid (ILA) | Anaerostipeshadrus | High-fat diet | ILA Production↓ | Eubacterium↓ Eubacterium rectale↓ Anaerostipescaccae↓ Bifidobacterium adolescentis↓ | [37,52] |
| Anaerostipescaccae | |||||
| Bacteroides thetaiotaomicron | |||||
| Bacteroides eggerthii | |||||
| Bacteroides ovatus | |||||
| Bifidobacterium adolescentis | |||||
| Bifidobacterium bifidum | |||||
| Bifidobacterium pseudolongum | |||||
| Clostridium bartlettii | |||||
| Clostridium sporogenes | |||||
| Escherichia coli | High-fiber diet | ILA Production↑ | Lactobacillus↑ Megamonas↑ | ||
| Eubacterium rectale | |||||
| Eubacterium cylindroides | |||||
| Faecalibacteriumprausnitzii | |||||
| Lactobacillus murinus | |||||
| Lactobacillus paracasei | |||||
| Lactobacillus reuteri | |||||
| Megamonas hypermegale | |||||
| … for more see [53] | |||||
| Indole-3-aldehyde(IAld) | Lactobacillus johnsonii | High-fiber diet | IAId Production↑ | Lactobacillus↑ | [54] |
| Lactobacillusreuteri | |||||
| Lactobacillusacidophilus | |||||
| Lactobacillusgallinarum | |||||
| … for more see [30] | |||||
| Indole-3-acetaldehyde (IAAld) | Escherichia coli | / | / | / | [55] |
| Tryptamine | Firmicutes C. sporogenes | High-fat diet | Tryptamine↓ | Bacteroides↓ Escheichia coli↑ | [52,56] |
| Clostridium sporogenes | |||||
| Escherichia. coli | |||||
| Ruminococcusgnavus | |||||
| Bacteroides | |||||
| 3-methylindole (skatole) | Bacteroides thetaiotaomicron | High-fat diet | complex manner | / | [57,58] |
| Butyrivibriofibrisolvens | |||||
| Clostridium bartlettii | |||||
| Clostridium drakei | |||||
| Eubacterium rectale | |||||
| Megamonas hypermegale | |||||
| Parabacteroides distasonis | |||||
| … for more see [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Li, J.; Ying, S. Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer. Metabolites 2023, 13, 1166. https://doi.org/10.3390/metabo13111166
Hou Y, Li J, Ying S. Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer. Metabolites. 2023; 13(11):1166. https://doi.org/10.3390/metabo13111166
Chicago/Turabian StyleHou, Yingjian, Jing Li, and Shuhuan Ying. 2023. "Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer" Metabolites 13, no. 11: 1166. https://doi.org/10.3390/metabo13111166
APA StyleHou, Y., Li, J., & Ying, S. (2023). Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer. Metabolites, 13(11), 1166. https://doi.org/10.3390/metabo13111166





